Abstract
Aquatic bacteria belonging to the deep-branching phylum Planctomycetes play a major role in global carbon and nitrogen cycles. However, their uncommon morphology and physiology, and their roles and survival on biotic surfaces in marine environments, are only partially understood. Access to axenic cultures of different planctomycetal genera is key to study their complex lifestyles, uncommon cell biology and primary and secondary metabolism in more detail. Here, we describe the characterisation of strain Enr8T isolated from a marine biotic surface in the seawater close to the shallow-sea hydrothermal vent system off Panarea Island, an area with high temperature and pH gradients, and high availability of different sulphur and nitrogen sources resulting in a great microbial diversity. Strain Enr8T showed typical planctomycetal traits such as division by polar budding, aggregate formation and presence of fimbriae and crateriform structures. Growth was observed at ranges of 15–33 °C (optimum 30 °C), pH 6.0–8.0 (optimum 7.0) and at NaCl concentrations from 100 to 1200 mM (optimum 350–700 mM). Strain Enr8T forms white colonies on solid medium and white flakes in liquid culture. Its genome has a size of 6.20 Mb and a G + C content of 59.2%. Phylogenetically, the strain belongs to the genus Blastopirellula. We propose the name Blastopirellula retiformator sp. nov. for the novel species, represented by the type strain Enr8T (DSM 100415T = LMG 29081T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Members of the family Planctomycetaceae, which belong to the environmentally, medically and biotechnologically relevant PVC superphylum (Planctomycetes-Verrucomicrobia-Chlamydiae, and others) (Spring et al. 2016; Wagner and Horn 2006), are ubiquitous microorganisms dwelling mostly in aquatic environments, in which they play key roles in cycling of carbon and nitrogen. However, the mechanisms as to how these microorganisms gain nutrients from biotic material in the oceans (in particular carbon and nitrogen for biomass and energy formation) are only partially understood. Species belonging to the family Planctomycetaceae are typically found to be attached to marine biotic surfaces, such as algae or kelp, and probably form biomass by using algal compounds as carbon and energy sources (Bondoso et al. 2015; Wiegand et al. 2019). Such nutrient-rich environments—in contrast to the oligotrophic seawater—attract also faster-growing competing microorganisms, e.g. Roseobacter sp. (Frank et al. 2014; Wiegand et al. 2018), but the latter fail to outcompete the slower-growing species of the family Planctomycetaceae. Two theories provide plausible explanations for this counter-intuitive observation: (I) Planctomycetaceae harbour a specialised machinery for uptake and degradation of complex polysaccharides released by algae or (II) they produce small molecules as a metabolic defence strategy. Both theories are supported by large planctomycetal genomes and the high number of predicted enzymes involved in the catabolism of complex sugars (Naumoff et al. 2014; Reisky et al. 2018; Wegner et al. 2013) (supporting theory I) or for production of secondary metabolites (supporting theory II) (Graça et al. 2016; Jeske et al. 2016; Wiegand et al. 2018, 2019). Uptake and catabolism of polymeric sugars might be facilitated by the unique pili-forming crateriform structures and an enlarged periplasm (Boedeker et al. 2017), while the mostly unexplored secondary metabolism could be a promising source for novel compounds, including those with health-promoting activities in humans (Graça et al. 2016).
Not only from a metabolic perspective, but also structurally, the phylum Planctomycetes is an interesting research topic. Eukaryotic-like morphological traits of Planctomycetes led to the conclusion that they might be beyond the bacterial cell plan (Fuerst and Sagulenko 2011; König et al. 1984; Lonhienne et al. 2010). In recent years, this picture changed with the advent of novel microscopic techniques and detailed physiological analyses of Planctomycetes (Jeske et al. 2015; Jogler et al. 2011; Jogler and Jogler 2013; Rivas-Marin et al. 2016; van Teeseling et al. 2015). The cell envelope of Planctomycetes was ultimately found to resemble that of Gram-negative bacteria (Boedeker et al. 2017; Devos 2014). But still, Planctomycetes remain exceptional. They divide by budding, binary fission or even a combination of both and lack proteins of the canonical divisome (Wiegand et al. 2019). Many strains have been shown to be resistant to several antibiotics (Cayrou et al. 2010; Godinho et al. 2019), either because of their degradation, or intrinsic resistance due to the lack of targets (Jogler et al. 2012; Pilhofer et al. 2008).
In conclusion, Planctomycetes have some characteristic traits (Wiegand et al. 2018, 2019), which encourages more detailed research and motivates us to steadily expand the collection of axenic cultures. In this study, we took samples in the surroundings of Panarea Island in the Tyrrhenian Sea off the southwestern coast of Italy. This region includes a shallow-sea hydrothermal vent system and consists of areas with increased temperatures, steep temperature gradients and high levels of different nitrogen and sulphur sources (Maugeri et al. 2010). Due to the expected microbial diversity (Manini et al. 2008), we considered this location a valuable source of so far unknown species of the family Planctomycetaceae and here describe the characterization of the novel strain Enr8T isolated from a hydrothermal area close to Panarea Island.
Materials and methods
Isolation and cultivation conditions
Strain Enr8T was isolated on the 10th of September 2013 from a marine biotic surface (Fig. 1) in a hydrothermal area in the Tyrrhenian Sea (sampling site 38.6387 N 15.1068 E) 2.5 km east of the port of Panarea Island. The strain was isolated as described previously (Wiegand et al. 2019) and subsequently cultivated in M1 medium with HEPES as buffering agent and supplemented with N-acetyl glucosamine (NAG) and artificial seawater (ASW) (medium designation M1H NAG ASW) (Kallscheuer et al. 2019a).
Physiological analyses
The temperature optimum of strain Enr8T was determined in M1H NAG ASW medium by cultivation at temperatures of 10, 15, 20, 22, 24, 27, 30, 33, 36 and 40 °C in a shaking incubator at 110 rpm with an initial pH of 8.0. For the determination of the pH optimum 100 mM HEPES was used as buffering agent for cultivations at pH 7.0, 7.5 and 8.0. For cultivations at pH 5.0 and 6.0 HEPES was replaced by 100 mM 2-(N-morpholino)ethanesulfonic acid (MES) and at pH 9.0 and 10.0 100 mM N-cyclohexyl-2-aminoethanesulfonic acid (CHES) was used as buffering agent. Cultivations for determination of the pH optimum were performed at 28 °C in a shaking incubator at 110 rpm. For analysis of the salt tolerance, strain Enr8T was cultivated in M1H NAG ASW medium with different concentrations of NaCl. The tested concentrations were 0, 100, 200 (standard NaCl concentration in ASW), 350, 500, 700, 900, 1200 and 1600 mM NaCl. Due to formation of white flakes by aggregation of cells in all experiments, growth was assessed by visual inspection of the culture taking the number and size of white flakes into account. Measurement of the optical density at 600 nm (OD600) was not possible.
Light microscopy and electron microscopy
Microscopic analyses were performed according to a previously published protocol (Kallscheuer et al. 2019a).
Genome information and analysis of genome-encoded features
The genome (accession no. SJPF00000000) and 16S rRNA gene sequence (accession no. MK554541) of strain Enr8T are available from GenBank (Wiegand et al. 2019). The primary metabolism was analysed by examining locally computed InterProScan (Mitchell et al., 2019) results cross-referenced with information from the UniProt database and BLASTp results of ‘typical’ protein sequences.
Phylogenetic analysis
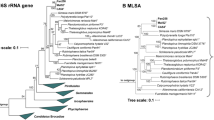
16S rRNA gene phylogeny was computed for strain Enr8T, the type strains of all described planctomycetal species (as available in May 2019) and all isolates recently published (Wiegand et al. 2019), including the strains described recently (Boersma et al. 2019; Kallscheuer et al. 2019a, b, c, d; Kohn et al. 2019). The 16S rRNA gene sequences were aligned with SINA (Pruesse et al. 2012). The phylogenetic analysis was performed with RAxML (Stamatakis 2014) employing a maximum likelihood approach with 1000 bootstraps, the nucleotide substitution model GTR, gamma distributed rate variation and estimation of proportion of invariable sites (GTRGAMMAI option). Three 16S rRNA genes of bacterial strains from the PVC superphylum were used as outgroup. The average nucleotide identity (ANI) was calculated using OrthoANI (Lee et al. 2016) and the average amino acid identity (AAI) was calculated using the aai.rb script of the enveomics collection (Rodriguez-R and Konstantinidis 2016). The percentage of conserved proteins (POCP) was calculated as described (Qin et al. 2014).
Results and discussion
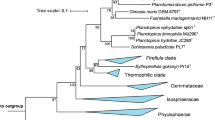
Phylogenetic analysis
According to maximum likelihood-based 16S rRNA gene sequence analysis shown in Fig. 2, the current closest relative of strain Enr8T is Blastopirellula marina, which is also the type species of the genus (Schlesner et al. 2004). The type strain of B. marina, DSM 3645T, was originally isolated from brackish water in the Baltic Sea (Schlesner 1986) and together with Blastopirellula cremea (Lee et al. 2013) forms the current genus Blastopirellula. B. marina DSM 3645T and strain Enr8T share a 16S rRNA similarity of 98.9%. This value is slightly above the proposed species threshold of 98.7% (Stackebrandt and Ebers 2006), which could indicate that strain Enr8T belongs to the species B. marina. However, it was shown before that 16S rRNA genes are not in all cases a reliable marker for Planctomycetes as some strains have high 16S rRNA gene sequence identities, but nevertheless belong to different species (Bondoso et al. 2013; Kohn et al. 2019). Thus, other phylogenetic markers need to be considered in order to achieve a reliable classification of strain Enr8T. For example, the similarity of the analysed partial rpoB sequence is 88.4%, which is clearly below the proposed threshold of 95.5% for strains belonging to the same species (Bondoso et al. 2013). The same is true for ANI values of 78.3% when applying the species threshold of 95–96% (Kim et al. 2014). Taken together, rpoB similarity and ANI values clearly reinforce the conclusion that strain Enr8T belongs to a separate species and does not represent a strain of the species B. marina. Thus, it appears likely that 16S rRNA gene identity comparisons in this clade do not follow the proposed species threshold of 98.7% (Stackebrandt and Ebers 2006). Accordingly, strain Enr8T and B. marina have an AAI of 75.8% and a POCP of 77.6%. Both values are above the respective genus threshold ranges (AAI: 60–80% and POCP: 50%) (Luo et at. 2014; Qin et al. 2014), confirming that both strains belong to the same genus. In order to show that strain Enr8T and B. marina DSM 3645T do not belong to the recently proposed genus ‘Bremerella’ (Rensink et al. 2019), minimal 16S rRNA gene sequence identities, AAI values, rpoB similarities and POCP values of strain Enr8T and ‘Bremerella vulcanica’ Pan97T were compared. Comparison yields a 16S rRNA gene identity of 94.3%, an AAI value of 54.7% and a partial rpoB sequence of 74.7%, which are below the genus thresholds of 94.5%, 60–80% and 75.5–78%, respectively (Kallscheuer et al. 2019d; Luo et al. 2014; Yarza et al. 2014). The POCP value of 60.4% is only slightly above the genus threshold of 50% (Qin et al. 2014).
Maximum likelihood analysis of 16S rRNA gene sequences. Phylogenetic tree showing the position of strain Enr8T. 16S rRNA gene phylogeny was computed as described in the Materials and methods section. Bootstrap values after 1000 re-samplings are given in % at the nodes. The outgroup consists of three 16S rRNA genes from the PVC superphylum. The Gimesia clade includes species of the genera Gimesia, Planctopirus, Fuerstiella, Schlesneria, Rubinisphaera and Planctomicrobium, while the thermophilic clade includes species of the genera Thermostilla, Thermogutta and Thermopirellula
Morphological and physiological analyses
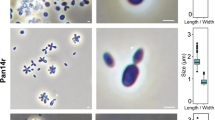
Strain Enr8T was cultivated in M1H NAG ASW medium and exponentially growing cells were used for morphological characterisation using phase contrast microscopy and scanning electron microscopy (Fig. 3). Detailed information on morphology, locomotion and cell division in comparison to the closely related species B. marina is summarised in Table 1. Cells of strain Enr8T appear ovoid to pear-shaped (length: 1.3 ± 0.2 µm, width: 0.7 ± 0.1 µm) (Fig. 3a), form strong aggregates and contain crateriform structures at the cell poles (Fig. 3b–e). Polar budding was observed as mode of cell division with the shape of the bud being similar to that of the mother cell (Fig. 3a). Fibres together with an extracellular matrix facilitate attachment to other cells or surfaces, thereby causing white flake formation visible with the naked eye in broth cultures. Colonies of strain Enr8T are white indicating the lack of carotenoid formation. Size, cell morphology and colour are similar to B. marina (Schlesner et al. 2004) (Table 1).
Phase contrast and scanning electron microscopy images and cell size plot of strain Enr8T. The figure shows the mode of cell division (a) and gives an overview on the cell morphology of strain Enr8T (b, d, e). The scale bar is 1 µm. For determination of the cell size (c) at least 100 representative cells were counted manually or by using a semi-automated object count tool
In cultivation experiments, strain Enr8T was found to grow at a pH range of 6.0–8.0 and at temperatures from 15 to 33 °C (Fig. 4a, b). The optimal conditions were determined to be pH 7.0 and 30 °C. During cultivation, formation of strong aggregates was observed, which became visible as white flakes settling down in the cultivation tubes, for example as shown for the determination of the pH optimum (Fig. 4a). This rendered measurement of cell densities using OD600 impossible. Instead, optimal conditions (pH, temperature and NaCl concentration) were determined by visual inspection taking the observed differences in the number and size of flakes into account. The temperature optimum at 30 °C is the same as for B. marina and both failed to grow at 36–38 °C (Schlesner 1986). Due to aggregate formation, a maximal growth rate could not be calculated for strain Enr8T. In the salt tolerance experiments, the strain showed growth in the presence of 0.6–7.0% (w/v) NaCl (100–1200 mM), but failed to grow without NaCl or with 9.3% (w/v) NaCl (1600 mM) (Fig. 4c). Optimal growth was observed in the range of 2.1–4.1% (w/v) NaCl (350–700 mM), which is in the range of natural seawater in the Mediterranean Sea with a total salt concentration of around 3.8% (w/v). The high salt tolerance of strain Enr8T is a distinctive difference compared to B. marina. While concentrations of 1% NaCl were lethal for B. marina (Schlesner 1986), Enr8T showed optimal growth at 2–4% NaCl and could even grow in the presence of 7% NaCl.
Optimum of pH, temperature and NaCl concentration for strain Enr8T. Cultivations in M1H NAG ASW medium were performed at different pH values (a constant temperature of 28 °C), different temperatures (b at pH 7.5, temperatures given in °C) and different NaCl concentrations (c given in mM) in biological triplicates. Due to strong formation of aggregates during cultivation it was not possible to measure cell density as OD600. Instead, exemplary photographs of the cultures of the pH optimum determination experiment after a cultivation time of 170 h are shown (a). Growth was classified in the range from “−” (no growth) to “+++” (very good growth)
Genomic characteristics
The relevant genome characteristics in comparison to the closely related species B. marina are summarised in Table 1. The genome size of strain Enr8T is 6.20 Mb. The genome is slightly smaller compared to B. marina DSM 3645T (6.66 Mb), but both strains have a similar G + C content (Enr8T: 59.2%, DSM 3645T: 57.4%). Automated annotation yielded 5033 putative protein-encoding genes for strain Enr8T, of which 42% (2107 genes) are annotated as hypothetical proteins. The calculated values, corresponding to 812 protein-coding genes per Mb and a coding density of 86%, are nearly identical to B. marina. Two copies of the 16S rRNA gene and 72 tRNA-encoding genes were detected in strain Enr8T, while B. marina harbours only one 16S rRNA gene and 56 tRNA-encoding genes.
Genome-based analysis of the central carbon metabolism
Based on the genome sequences of Enr8T and B. marina DSM 3645T, we analysed the presence of key metabolic enzymes of the central carbon metabolism. The analysis included glycolytic pathways, gluconeogenesis, the tricarboxylic acid (TCA) cycle and anaplerotic reactions (Table 2). Both strains harbour the genes coding for enzymes involved in glycolytic reactions either using the Embden-Meyerhof-Parnas pathway (referred to as glycolysis) or the alternative Entner-Doudoroff pathway. However, we failed to identify the gene coding for phosphoglycerate mutase (Pgm), which catalyses the reversible isomerisation of 3-phosphoglycerate to 2-phosphoglycerate and is required for the activity of both pathways. As the degradation of glucose by both strains suggests that the glycolysis is active, the two strains either bypass the reaction, e.g. by using 2,3-bisphosphoglycerate as intermediate or the protein sequence of Pgm differs from canonical sequences and escaped our analysis. The pentose phosphate pathway and the TCA cycle appear to be fully functional in both strains since we were able to assign genes to all participating enzymes (Table 2). Both strains also encode phosphoenolpyruvate carboxykinase (ATP) and fructose-1,6-bisphosphatase, the key enzymes required for de novo synthesis of sugar phosphates from the TCA cycle intermediate oxaloacetate. The gluconeogenesis pathway, however, also requires the phosphoglycerate mutase, which we could not identify in the two strains (as discussed above). Bacteria typically perform anaplerosis (replenishing of TCA cycle intermediates) by the glyoxylate shunt or by carboxylation of pyruvate or phosphoenolpyruvate (PEP). The carboxylation reactions rely on additional pathways providing pyruvate or PEP (typically glycolysis), while the glyoxylate shunt converts the TCA cycle intermediate isocitrate and the TCA cycle substrate acetyl-CoA to one molecule of succinate and malate. Both strains apparently lack the genes required for a functional glyoxylate shunt (Table 2), which is required e.g. during growth with acetate or fatty acids as sole carbon and energy source. This would imply that both strains are unable to use acetate or fatty acids as sole carbon and energy source, which may support the hypothesis that species of the family Planctomycetaceae consume complex sugars derived from phototrophs rather than short- or long-chain carboxylic acids.
Putative gene clusters involved in secondary metabolite production
To gain a first insight into the potential of strain Enr8T as a source of secondary metabolites an AntiSMASH analysis based on its genome sequence was conducted (Blin et al. 2019). It appears reasonable to assume that the complex lifestyle and growth in competitive environments such as marine surfaces is related to the production of secondary metabolites allowing strain Enr8T to cope with abiotic and biotic stresses. Of major interest in this case are multi-domain protein complexes of the families of polyketide synthases (PKSs) or non-ribosomal peptide synthetases (NRPSs) as these serve as molecular assembly lines for production of secondary metabolites starting from metabolites of the primary carbon metabolism (e.g. malonyl-CoA or amino acids) (Park et al. 2019). Strain Enr8T harbours three genes or clusters involved in the production of terpenoids. These are probably not involved in the production of carotenoids (a major class of terpenoids) as the strain is white and thus lacks pigmentation. In addition, Enr8T harbours genes coding for a putative type I PKS, a putative NRPS, a putative mixed NRPS-type I PKS and a protein related to bacteriocin biosynthesis. The same set of clusters is also encoded in the genome of B. marina DSM 3645T except for the putative NRPS gene, which was not identified in B. marina. The exact products formed by the identified PKS and NRPS enzymes should be identified in future studies to gain insights into their biological significance.
Taken together, our phylogenetic analysis and the results of the morphological and physiological characterisation support the conclusion that strain Enr8T represents a novel species of the genus Blastopirellula, for which we propose the name Blastopirellula retiformator sp. nov.
Description of Blastopirellula retiformator sp. nov
Blastopirellula retiformator (re.ti.for.ma’tor. L. neut. n. rete a net; L. masc. n. formator a shaper, creator; N.L. masc. n. retiformator corresponding to the characteristic fibre- and extracellular matrix-mediated formation of visible flake-like structures).
Colonies are white. Cells are pear-shaped (length: 1.3 ± 0.2 µm, width: 0.7 ± 0.1 µm) and form large aggregates which become visible as white flakes in liquid culture. Cells divide by polar budding and grow at ranges of 15–33 °C (optimum 30 °C), pH 6.0–8.0 (optimum 7.0) and NaCl concentrations from 100 to 1200 mM (optimum 350–700 mM). The genome (accession no. SJPF00000000) and 16S rRNA gene sequence (accession no. MK554541) of the type strain are available from GenBank. The type strain genome has a size of 6.20 Mb and a G + C content of 59.2%. The type strain is Enr8T (DSM 100415T = LMG 29081T, also designated Enrichment 8), isolated from a hydrothermal area close to the island Panarea, Italy.
References
Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T (2019) antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87
Boedeker C, Schuler M, Reintjes G, Jeske O, van Teeseling MC, Jogler M, Rast P, Borchert D, Devos DP, Kucklick M, Schaffer M, Kolter R, van Niftrik L, Engelmann S, Amann R, Rohde M, Engelhardt H, Jogler C (2017) Determining the bacterial cell biology of Planctomycetes. Nat Commun 8:14853
Boersma A, Kallscheuer N, Wiegand S, Rast R, Peeters S, Mesman R, Heuer A, Boedeker C, Jetten M, Rohde M, Jogler M, Jogler C (2019) Alienimonas californiensis gen. nov. sp. nov., a novel Planctomycete isolated from the kelp forest in Monterey Bay. Antonie van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01367-4
Bondoso J, Harder J, Lage OM (2013) rpoB gene as a novel molecular marker to infer phylogeny in Planctomycetales. Antonie Van Leeuwenhoek 104:477–488
Bondoso J, Albuquerque L, Nobre MF, Lobo-da-Cunha A, da Costa MS, Lage OM (2015) Roseimaritima ulvae gen. nov., sp. nov. and Rubripirellula obstinata gen. nov., sp. nov. two novel planctomycetes isolated from the epiphytic community of macroalgae. Syst Appl Microbiol 38:8–15
Cayrou C, Raoult D, Drancourt M (2010) Broad-spectrum antibiotic resistance of Planctomycetes organisms determined by Etest. J Antimicrob Chemother 65:2119–2122
Devos DP (2014) Re-interpretation of the evidence for the PVC cell plan supports a Gram-negative origin. Antonie Van Leeuwenhoek 105:271–274
Frank O, Michael V, Pauker O, Boedeker C, Jogler C, Rohde M, Petersen J (2014) Plasmid curing and the loss of grip—the 65-kb replicon of Phaeobacter inhibens DSM 17395 is required for biofilm formation, motility and the colonization of marine algae. Syst Appl Microbiol 38:120–127
Fuerst JA, Sagulenko E (2011) Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol 9:403–413
Godinho O, Calisto R, Ovreas L, Quinteira S, Lage OM (2019) Antibiotic susceptibility of marine Planctomycetes. Antonie Van Leeuwenhoek 112:1273–1280
Graça AP, Calisto R, Lage OM (2016) Planctomycetes as novel source of bioactive molecules. Front Microbiol 7:1241
Jeske O, Schüler M, Schumann P, Schneider A, Boedeker C, Jogler M, Bollschweiler D, Rohde M, Mayer C, Engelhardt H (2015) Planctomycetes do possess a peptidoglycan cell wall. Nat Commun 6:7116
Jeske O, Surup F, Ketteniß M, Rast P, Förster B, Jogler M, Wink J, Jogler C (2016) Developing techniques for the utilization of Planctomycetes as producers of bioactive molecules. Front Microbiol 7:1242
Jogler M, Jogler C (2013) Towards the development of genetic tools for Planctomycetes. In: Fuerst JA (ed) Planctomycetes: cell structure, origins and biology. Springer, Berlin, pp 141–164
Jogler C, Glöckner FO, Kolter R (2011) Characterization of Planctomyces limnophilus and development of genetic tools for its manipulation establish it as a model species for the phylum Planctomycetes. Appl Environ Microbiol 77:5826–5829
Jogler C, Waldmann J, Huang X, Jogler M, Glöckner FO, Mascher T, Kolter R (2012) Identification of proteins likely to be involved in morphogenesis, cell division, and signal transduction in Planctomycetes by comparative genomics. J Bacteriol 194:6419–6430
Kallscheuer N, Jogler M, Wiegand S, Peeters S, Heuer A, Boedeker C, Jetten M, Rohde M, Jogler C (2019a) Rubinisphaera italica sp. nov. isolated from a hydrothermal area in the Tyrrhenian Sea close to the volcanic island Panarea. Antonie van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01329-w
Kallscheuer N, Jogler M, Wiegand S, Peeters S, Heuer A, Boedeker C, Jetten M, Rohde M, Jogler C (2019b) Three novel Rubripirellula species isolated from artificial plastic surfaces submerged in the German part of the Baltic Sea and the estuary of the river Warnow. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01368-3
Kallscheuer N, Wiegand S, Jogler M, Boedeker C, Peeters S, Rast P, Heuer A, Jetten M, Rohde MCJ (2019c) Rhodopirellula heiligendammensis sp. nov., Rhodopirellula pilleata sp. nov., and Rhodopirellula solitaria sp. nov. isolated from natural or artificial marine surfaces in Northern Germany and California, USA. Antonie van Leeuwenhoek, USA. https://doi.org/10.1007/s10482-019-01366-5
Kallscheuer N, Wiegand S, Peeters SH, Jogler M, Boedeker C, Heuer A, Rast P, Jetten MSM, Rohde M, Jogler C (2019d) Description of three bacterial strains belonging to the new genus Novipirellula gen. nov., reclassificiation of Rhodopirellula rosea and Rhodopirellula caenicola and readjustment of the genus threshold of the phylogenetic marker rpoB for Planctomycetales. Antonie van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01374-5
Kim M, Oh HS, Park SC, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351
Kohn T, Wiegand S, Boedeker C, Rast P, Heuer A, Schüler M, Rohde C, Müller R-W, Brümmer F, Rohde M, Engelhardt H, Jogler M, Jogler C (2019) Planctopirus ephydatiae, a novel planctomycetal species isolated from the freshwater sponge Ephydatia fluviatilis. Syst Appl Microbiol. https://doi.org/10.1016/j.syapm.2019.126022
König E, Schlesner H, Hirsch P (1984) Cell wall studies on budding bacteria of the Planctomyces/Pasteuria group and on a Prosthecomicrobium sp. Arch Microbiol 138:200–205
Lee H-W, Roh SW, Shin N-R, Lee J, Whon TW, Jung M-J, Yun J-H, Kim M-S, Hyun D-W, Kim D, Bae J-W (2013) Blastopirellula cremea sp. nov., isolated from a dead ark clam. Int J Syst Evol Microbiol 63(Pt 6):2314–2319
Lee I, Ouk Kim Y, Park SC, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103
Lonhienne TG, Sagulenko E, Webb RI, Lee K-C, Franke J, Devos DP, Nouwens A, Carroll BJ, Fuerst JA (2010) Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci USA 107:12883–12888
Luo C, Rodriguez RL, Konstantinidis KT (2014) MyTaxa: an advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res 42:e73
Manini E, Luna G, Corinaldesi C, Zeppilli D, Bortoluzzi G, Caramanna G, Raffa F, Danovaro R (2008) Prokaryote diversity and virus abundance in shallow hydrothermal vents of the Mediterranean Sea (Panarea Island) and the Pacific Ocean (North Sulawesi-Indonesia). Microbial Ecol 55:626–639
Maugeri TL, Bianconi G, Canganella F, Danovaro R, Gugliandolo C, Italiano F, Lentini V, Manini E, Nicolaus B (2010) Shallow hydrothermal vents in the southern Tyrrhenian Sea. Chem Ecol 26:285–298
Mitchell AL, Attwood TK, Babbitt PC et al (2019) InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res 47:D351–D360
Naumoff D, Ivanova A, Dedysh S (2014) Phylogeny of β-xylanases from Planctomycetes. Mol Biol 48:439–447
Park SR, Yoon YJ, Pham JV, Yilma MA, Feliz A, Majid MT, Maffetone N, Walker JR, Kim E, Reynolds JM (2019) A review of the microbial production of bioactive natural products and biologics. Front Microbiol 10:1404
Pilhofer M, Rappl K, Eckl C, Bauer AP, Ludwig W, Schleifer KH, Petroni G (2008) Characterization and evolution of cell division and cell wall synthesis genes in the bacterial phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and phylogenetic comparison with rRNA genes. J Bacteriol 190:3192–3202
Pruesse E, Peplies J, Glöckner FO (2012) SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829
Qin Q-L, Xie B-B, Zhang X-Y, Chen X-L, Zhou B-C, Zhou J, Oren A, Zhang Y-Z (2014) A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol 196:2210–2215
Reisky L, Büchsenschütz HC, Engel J, Song T, Schweder T, Hehemann J-H, Bornscheuer UT (2018) Oxidative demethylation of algal carbohydrates by cytochrome P450 monooxygenases. Nat Chem Biol 14:342
Rensink S, Wiegand S, Kallscheuer N, Rast P, Peeters S, Heuer A, Boedeker C, Jetten MSM, Rohde M, Jogler M, Jogler C (2019) Description of the novel planctomycetal genus Bremerella, containing Bremerella volcania sp. nov., isolated from an active volcanic site, and reclassification of Blastopirellula cremea as Bremerella cremea comb. nov. Antonie van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01378-1
Rivas-Marin E, Canosa I, Santero E, Devos DP (2016) Development of genetic tools for the manipulation of the planctomycetes. Front Microbiol 7:914
Rodriguez-R, LM, Konstantinidis KT (2016) The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints
Schlesner H (1986) Pirella marina sp. nov., a budding, peptidoglycan-less bacterium from brackish water. Syst Appl Microbiol 8:177–180
Schlesner H, Rensmann C, Tindall BJ, Gade D, Rabus R, Pfeiffer S, Hirsch P (2004) Taxonomic heterogeneity within the Planctomycetales as derived by DNA–DNA hybridization, description of Rhodopirellula baltica gen. nov., sp. nov., transfer of Pirellula marina to the genus Blastopirellula gen. nov. as Blastopirellula marina comb. nov. and emended description of the genus Pirellula. Int J Syst Evol Microbiol 54:1567–1580
Spring S, Bunk B, Spröer C, Schumann P, Rohde M, Tindall BJ, Klenk H-P (2016) Characterization of the first cultured representative of Verrucomicrobia subdivision 5 indicates the proposal of a novel phylum. ISME J 10:2801
Stackebrandt E, Ebers J (2006) Taxonomic parameter revisited: tarnished gold standards. Microbiol Today 33:152–155
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Van Teeseling MC, Mesman RJ, Kuru E, Espaillat A, Cava F, Brun YV, VanNieuwenhze MS, Kartal B, Van Niftrik L (2015) Anammox Planctomycetes have a peptidoglycan cell wall. Nat Commun 6:6878
Wagner M, Horn M (2006) The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol 17:241–249
Wegner C-E, Richter-Heitmann T, Klindworth A, Klockow C, Richter M, Achstetter T, Glöckner FO, Harder J (2013) Expression of sulfatases in Rhodopirellula baltica and the diversity of sulfatases in the genus Rhodopirellula. Mar Genomics 9:51–61
Wiegand S, Jogler M, Jogler C (2018) On the maverick Planctomycetes. FEMS Microbiol Rev 42:739–760
Wiegand S, Jogler M, Boedeker C, Pinto D, Vollmers J, Rivas-Marín E, Kohn T, Peeters SH, Heuer A, Rast P, Oberbeckmann S, Bunk B, Jeske O, Meyerdierks A, Storesund JE, Kallscheuer N, Lücker S, Lage OM, Pohl T, Merkel BJ, Hornburger P, Müller R-W, Brümmer F, Labrenz M, Spormann AM, Op den Camp H, Overmann J, Amann R, Jetten MSM, Mascher T, Medema MH, Devos DP, Kaster A-K, Øvreås L, Rohde M, Galperin MY, Jogler C (2019) Cultivation and functional characterization of 79 Planctomycetes uncovers their unique biology. Nat Microbiol. https://doi.org/10.1038/s41564-019-0588-1
Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer K-H, Whitman WB, Euzéby J, Amann R, Rosselló-Móra R (2014) Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12(9):635–645
Acknowledgements
Part of this research was funded by the Deutsche Forschungsgemeinschaft Grants KA 4967/1-1 and JO 893/4-1, Grant ALWOP.308 of the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO), SIAM (Soehngen Institute for Anaerobic Microbiology) Grant No. 024002002 and the Radboud Excellence fellowship. We thank Ina Schleicher for skilful technical assistance. Brian Tindall and Regine Fähnrich from the DSMZ as well as the BCCM/LMG Bacteria collection we thank for support during strain deposition. We thank the Scientific Diving Center of the Bergakademie Freiberg, Germany, Thomas Pohl, Peter Hornburger and all participants of the 2013 Panarea Expedition for sampling support.
Author information
Authors and Affiliations
Contributions
NK wrote the manuscript, analysed the data and prepared the figures, SW performed the genomic and phylogenetic analysis, MJ, AH and PR isolated the strains and performed cultivations and strain deposition, SR and ASB performed the cultivation for determination of the pH optimum, SHP and CB performed the light microscopic analysis, MSMJ contributed to text preparation and revised the manuscript, MR performed the electron microscopic analysis, CJ and MJ took the samples and CJ supervised the study. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kallscheuer, N., Wiegand, S., Heuer, A. et al. Blastopirellula retiformator sp. nov. isolated from the shallow-sea hydrothermal vent system close to Panarea Island. Antonie van Leeuwenhoek 113, 1811–1822 (2020). https://doi.org/10.1007/s10482-019-01377-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-019-01377-2