Abstract
Planctomycetes are a unique and important phylum containing mostly aquatic bacteria, which are often associated with phototrophic surfaces. A complex lifestyle, their potential for the production of bioactive small molecules, their unusual cell biology and a large number of giant and hypothetical genes in their genomes make these microorganisms a fascinating topic for further research. Here, we characterise three novel planctomycetal strains isolated from polystyrene and polyethylene particles that were submerged in the German part of the Baltic Sea and the estuary of the river Warnow. All three strains showed typical planctomycetal traits such as division by polar budding and formation of rosettes. The isolated strains were mesophilic and neutrophilic chemoheterotrophs and reached generation times of 10–25 h during laboratory-scale cultivation. Taxonomically, the three strains belong to the genus Rubripirellula. Based on our analyses all three strains represent novel species, for which we propose the names Rubripirellula amarantea sp. nov., Rubripirellula tenax sp. nov. and Rubripirellula reticaptiva sp. nov. The here characterised strains Pla22T (DSM 102267T = LMG 29691T), Poly51T (DSM 103356T = VKM B-3438T) and Poly59T (DSM 103767T = LMG 29696T) are the respective type strains of these novel species. We also emend the description of the genus Rubripirellula.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Planctomycetes are ubiquitous bacteria often associated with nutrient-rich aquatic surfaces. By occupying these ecological niches, Planctomycetes largely contribute to the vital activity of global carbon and nitrogen cycles (Wiegand et al. 2018). Phylogenetically, Planctomycetes form the PVC superphylum alongside with the phyla of Verrucomicrobia, Lentisphaerae, Kirimatiellaeota, Candidatus Omnitrophica and Chlamydiae (Spring et al. 2016; Wagner and Horn 2006). Based on exceptional traits, such as, presence of compartment-like structures (Lindsay et al. 1997) and lack of peptidoglycan (König et al. 1984), Planctomycetes were thought to be beyond the bacterial cell plan (Devos et al. 2013; Fuerst and Sagulenko 2011). It was even speculated that they might represent the missing link between bacteria and eukaryotes (Devos and Reynaud 2010). In the recent years, however, presence of peptidoglycan was confirmed (Jeske et al. 2015; van Teeseling et al. 2015) and compartment-like structures were re-interpreted as invaginations of the cytoplasmic membrane (Acehan et al. 2013; Boedeker et al. 2017; Lage et al. 2013; Santarella-Mellwig et al. 2013). Planctomycetes were thus classified as Gram-negative bacteria (Devos 2014), but still remain exceptional and enigmatic in comparison to well-characterised bacteria of other phyla. Most Planctomycetes divide unusually by budding, while some also perform binary fission or even a combination of both modes of division (Wiegand et al. 2019). The observed lack of canonical divisome proteins including the otherwise universal FtsZ (Jogler et al. 2012; Pilhofer et al. 2008) might also be part of the explanation why Planctomycetes are resistant to many antibiotics (Cayrou et al. 2010; Godinho et al. 2019).
As a result of the absence of nutrients in oligotrophic seawater, several Planctomycetes attach to nutrient-rich aquatic surfaces (Bengtsson et al. 2012; Bondoso et al. 2014, 2015, 2017; Lage and Bondoso 2014; Vollmers et al. 2017). Attachment typically goes along with a lifestyle switch between planktonic swimmer cells and sessile stalked mother cells (Jogler et al. 2011). Once attached to the surface, they start to degrade polymeric compounds for biomass production, e.g. utilising complex sugars released by algae. This strategy was demonstrated for the model polysaccharide dextran (Jeske et al. 2013; Lachnit et al. 2013). Unique sugar-binding pili attached to crateriform structures and an extremely enlarged periplasm are believed to be involved in the uptake of such high molecular weight sugars (Boedeker et al. 2017), which requires further attention in the next years.
Planctomycetes were found to be predominant members in biofilms on the mentioned nutrient-rich aquatic surfaces (Bengtsson and Øvreås 2010; Kohn et al. 2019), which is counter-intuitive when considering their slow growth compared to other natural competitors, e.g. members of the Roseobacter clade (Frank et al. 2014; Wiegand et al. 2018). It was proposed that this observation is the result of defense strategies involving secondary metabolite production, which, in presence of a competitor, ultimately lead to release of antimicrobial compounds. This notion, that Planctomycetes are ‘talented’ producers of such small bioactive molecules, is substantiated by large genomes (Kohn et al. 2016) as well as several predicted gene clusters involved in small molecule production (Graca et al. 2016; Jeske et al. 2016; Wiegand et al. 2019).
Succinctly, Planctomycetes are amongst the most maverick of all bacteria (Wiegand et al. 2018), which motivated us to expand the collection of Planctomycetes by isolating and characterising 79 novel planctomycetal strains presented in an overview article (Wiegand et al. 2019). Here, we validly describe three novel strains, Pla22T, Poly51T, and Poly59T, isolated from plastic particles submerged in the Baltic Sea and the river Warnow in northern Germany. The genus Rubripirellula, to which the three here proposed species belong, was described earlier with Rubripirellula obstinata as the type species and the only member of the genus so far. The type strain R. obstinata LF1T was isolated from algae (Laminaria sp.) at the coast of Porto, Portugal (Bondoso et al. 2015).
Materials and methods
Preparation of cultivation medium
For strain isolation and subsequent cultivations M1H NAG ASW medium [M1 medium with HEPES as buffering agent (M1H) additionally supplemented with N-acetyl glucosamine (NAG) as carbon and nitrogen source and artificial seawater (ASW)] was used. This medium is originally based on a recipe described earlier (Staley et al. 1992). This recipe was extensively modified to allow cultivation of a broad range of different planctomycetal strains and was prepared as described before (Kallscheuer et al. 2019).
Isolation of the strains
Strain Pla22T was sampled on the 4th of September 2014 from polyethylene (PE) pellets (ExxonMobil HDPE HTA 108, diameter 3 mm), which were incubated in 2 m depth for 14 days in the Warnow river north of Rostock, Germany. The sampling site (54.106 N, 12.096 E) is located close to a wastewater treatment plant discharge. The exact setup of sampling was described earlier (Oberbeckmann et al. 2018). Strain Poly51T and strain Poly59T were sampled on the 8th of October 2015 at Heiligendamm, Germany (54.146 N, 11.843 E) from polystyrene (PS) and PE particles, respectively, stored in separate incubators in the water for 14 days in 2 m depth and accessed via Heiligendamm pier. The strains were isolated from the plastic particles as described before (Wiegand et al. 2019). Plastic was chosen as material for sampling as it is cheap and easily available, provides a high surface area, is non-toxic and inert, i.e. it neither reacts with the seawater nor is degraded. Briefly, for the isolation of Pla22T (September 2014) samples were stored for 8 weeks at 4 °C before isolation of bacterial biofilms by digestion with 2 mg/mL (28 U/mL) β-galactosidase for 30 min at 30 °C and simultaneous vortexing every 5 min followed by 10 min sonication at 30 °C. Separation of plastic from the biofilm was performed by filtration. Biofilms were stored for 11 months at 4 °C prior to plating. For isolation of Poly51T and Poly59T (October 2015) incubated plastic particles were washed three times with sterile natural seawater and stored at 4 °C until cultivation (5 days after sampling). Plastic particles were vortexed and 50 µL of the liquid seawater was used for cultivation on M1H NAG ASW plates containing 8 g/L gellan gum. For selection of Planctomycetes 500 mg/L streptomycin and 200 mg/L ampicillin were added as antibiotic reagents, while 20 mg/L cycloheximide was used to prevent fungal growth. In order to check whether the isolated strains indeed represent Planctomycetes the 16S rRNA gene was amplified by PCR and sequenced as previously described (Rast et al. 2017).
Light microscopy
Phase contrast (Phaco) analyses were performed employing a Nikon Eclipse Ti inverted microscope with a Nikon DS-Ri2 camera (blue LED). Specimens were immobilised in MatTek glass bottom dishes (35 mm, No. 1.5) employing a 1% agarose cushion (Will et al. 2018). Images were analysed using the Nikon NIS-Elements software (version 4.3). To determine the cell size, at least 100 representative cells were counted manually (Annotations and Measurements, NIS-Elements) or by using the NIS-Elements semi-automated Object Count tool (smooth: 4×, clean: 4×, fill holes: on, separate: 4×).
Electron microscopy
For field emission scanning electron microscopy bacteria were fixed in 1% (v/v) formaldehyde in HEPES buffer (3 mM HEPES, 0.3 mM CaCl2, 0.3 mM MgCl2, 2.7 mM sucrose, pH 6.9) for 1 h on ice and washed once employing the same buffer (Rast et al. 2017). Cover slips with a diameter of 12 mm were coated with a poly-l-lysine solution (Sigma-Aldrich) for 10 min, washed in distilled water and air-dried. 50 µL of the fixed bacteria solution was placed on a cover slip and allowed to settle for 10 min. Cover slips were then fixed in 1% glutaraldehyde in TE buffer (20 mM TRIS, 1 mM EDTA, pH 6.9) for 5 min at room temperature and subsequently washed twice with TE buffer before dehydrating in a graded series of acetone (10, 30, 50, 70, 90, 100%) on ice for 10 min at each concentration. Samples from the 100% acetone step were brought to room temperature before placing them in fresh 100% acetone. Samples were then subjected to critical-point drying with liquid CO2 (CPD 300, Leica). Dried samples were covered with a gold/palladium (80/20) film by sputter coating (SCD 500, Bal-Tec) before examination in a field emission scanning electron microscope (Zeiss Merlin) using the Everhart–Thornley HESE2 detector and the inlens SE detector in a 25:75 ratio at an acceleration voltage of 5 kV.
Genome information
Genome information of the three isolated strains is available under accession numbers SJPI00000000 (Pla22T), SJPW00000000 (Poly51T) and SJPX00000000 (Poly59T). The corresponding 16S rRNA gene sequences can be found under accession numbers MK554581 (Pla22T), MK554552 (Poly51T) and MK554553 (Poly59T).
Physiological analysis
For determination of the pH optimum 100 mM HEPES was used for cultivations at pH 7.0, 7.5 and 8.0. For cultivation at pH 5.0 and 6.0 HEPES was replaced by 100 mM 2-(N-morpholino)ethanesulfonic acid (MES), whereas 100 mM N-cyclohexyl-2-aminoethane-sulfonic acid (CHES) served as a buffering agent at pH 9.0 and 10.0. Cultivations for determination of the pH optimum were performed at 28 °C. For determination of the temperature optimum the strains were cultivated at temperatures ranging from 10 to 40 °C in M1H NAG ASW medium at pH 7.5.
Phylogenetic analysis
16S rRNA gene phylogeny was computed for strains Pla22T, Poly51T and Poly59T, the type strains of all described planctomycetal species (available in May 2019) and all isolates recently published (Wiegand et al. 2019). The 16S rRNA gene sequences were aligned with SINA (Pruesse et al. 2012). The phylogenetic analysis was done with RAxML (Stamatakis 2014) employing a maximum likelihood (ML) approach with 1000 bootstraps, the nucleotide substitution model GTR, gamma distributed rate variation and estimation of proportion of invariable sites (GTRGAMMAI option). Three 16S rRNA genes of bacterial strains from the PVC superphylum were used as outgroup. Average nucleotide identities (ANI) were calculated using OrthoANI (Lee et al. 2016) and average amino acid identities (AAI) were calculated using the aai.rb script of the enveomics collection (Rodriguez-R and Konstantinidis 2016). Values of percentage of conserved proteins (POCP) were calculated as described before (Qin et al. 2014). The rpoB nucleotide sequences were taken from the above-mentioned as well as other publicly available genome annotations and the sequence identities were determined as described (Bondoso et al. 2013). Upon extracting only those parts of the sequence that would have been sequenced with the described primer set the alignment and matrix calculation was done with Clustal Omega (Sievers et al. 2011).
Results and discussion
Phylogenetic analysis
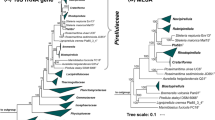
The phylogenetic positions of strains Poly59T, Poly51T and Pla22T were determined by 16S rRNA gene analysis as shown in Fig. 1. The three strains cluster monophyletically with R. obstinata LF1T (Bondoso et al. 2015). An assessment of different phylogenetic markers describing the relationship between the type strain and the novel isolates can be found in Table 1. The 16S rRNA gene similarities of Poly59T, Poly51T and Pla22T to R. obstinata LF1T are all below the species threshold of 98.7% (Stackebrandt and Ebers 2006), but above the proposed genus threshold of 94.5% (Yarza et al. 2014). The comparison of rpoB sequence fragments as described (Bondoso et al. 2013) gave similarity results between 80.9 and 82.9%, which are values below the proposed species threshold, but above the given genus threshold (Bondoso et al. 2013). This implies that none of the novel strains belong to the species Rubripirellula obstinata, but that all are members of the genus Rubripirellula. The ANI values of under 95% support this result of separate species (Kim et al. 2014). Additionally, the two-way AAI in the range of 60–80% (Luo et al. 2014) and a POCP of > 50% (Qin et al. 2014) also indicate that all strains belong to the same genus. The two closely related strains Poly51T and Poly59T have a 16S rRNA gene sequence identity of 98.4%, an rpoB sequence identity of 85.8%, and an ANI of 75.4% implying that they also form two separate species.
16S rRNA gene-based phylogeny. The phylogenetic tree highlights the position of the three here investigated strains in relation to their closest described relatives. 16S rRNA gene phylogeny was computed using the maximum likelihood method. Bootstrap values after 1000 re-samplings are given at the nodes (in %). The outgroup consists of three 16S rRNA genes from the PVC superphylum
Morphological and physiological analysis
Cell morphologies and cell sizes of strains Pla22T, Poly51T and Poly59T were analysed by light microscopy (Fig. 2) and scanning electron microscopy (Fig. 3) during exponential growth. The obtained images were compared to the already described strain R. obstinata LF1T. All four strains form mainly loose aggregates and divide by polar budding. Buds have the same shape as the mother cell. Flagella formation was only observed for LF1T. All four strains contain fimbriae at the budding pole but lack a stalk. A holdfast structure was only observed in strain LF1T. Only Poly51T and LF1T formed a visible capsule. Detailed information on morphology, locomotion and mechanism of cell division is summarised in Table 2.
Light microscopy images and cell size plots of the three isolated strains. The mode of cell division (a, c, e) and a general overview of cell morphology (b, d, f) is shown in the pictures. The scale bar is 1 µm. For determination of the cell size (g, h, i) at least 100 representative cells were counted manually or by using a semi-automated object count tool
Cells of strain Pla22T are round grain rice-shaped with an average size of 1.7 ± 0.3 µm in length and 0.9 ± 0.2 µm in width (Fig. 2a, g). Colonies have an amaranth pink color. The strain is aerobic and grew at pH values ranging from 6.0 to 8.5 with an optimum at pH 7.5 (Fig. 4). Growth was observed at temperatures from 10 to 36 °C with optimal growth at 33 °C. In M1H NAG ASW medium a maximal growth rate of 0.068 h−1 was observed, which corresponds to a doubling time of approximately 10 h (Fig. 4).
Temperature and pH optima of the isolated strains. In the upper panel, the given data points show the average growth rates obtained after cultivation of the three isolated strains in M1H NAG ASW medium in biological triplicates at different temperatures and a constant pH of 7.5. In the bottom panel, the data points show the average growth rates for cultivation at different pH values and a constant temperature of 28 °C
Strain Poly51T has a similar shape and color as strain Pla22T, but the cells are slightly smaller and appear chubbier (Poly51T: 1.4 ± 0.2 × 0.9 ± 0.1 µm) (Figs. 2c, h, 3c). Strain Poly51T grows at a pH range of 6.5–9.0 with an optimum at pH 8.0. The temperature optimum of the strain is between 22 and 24 °C, while cell growth was observed at temperatures ranging from 10 to 28 °C (Fig. 4). The strain did not grow at temperatures of 30 °C or higher. The highest observed growth rate is 0.028 h−1 corresponding to a generation time of 25 h.
Cells of Poly59T appear round grain rice-shaped to round (Figs. 2e, 3e, f) with an average cell size of 1.5 ± 0.3 µm (length) × 1.0 ± 0.2 µm (width), are pink-colored and show the same behavior as Poly51T with regard to pH (growth range 6.5–9.0, optimum 7.5–8.0) (Fig. 4). Comparable to Poly51T, strain Poly59T grows best at 24 °C, but in contrast could also grow at 30 and 33 °C. Its highest grow rate was calculated to 0.043 h−1, corresponding to a doubling time of approx. 16 h. The type species R. obstinata LF1T has a temperature optimum at 25 °C (range 10–30 °C) and grows in a pH range of 7.5–10.5 (Bondoso et al. 2015). Thus, the temperature optimum of LF1T is very similar to Poly51T and Poly59T, but different from Pla22T (33° C). Remarkably, the temperature optimum of Pla22T being 8 °C higher than that of the three other strains is the most striking difference and is also reflected by a higher growth rate of Pla22T at the optimal temperature. While Poly51T, Poly59T and LF1T were isolated from the Baltic Sea or the Atlantic Ocean, Pla22T was isolated from the estuary of the river Warnow close to a wastewater treatment plant. The water temperature profile in the sampling areas might be considerably different and allows at least to speculate that the higher temperature optimum of Pla22T might be related to the sampling location. With regard to pH, LF1T, growing at a range of 7.5–10.5, is slightly more alkaliphilic than the other three strains, which grow at a pH range of 6.5–9.0. None of the strains grew at pH 5.5 or lower and the observed differences between the strains were less pronounced compared to the parameter temperature.
Genomic characteristics
All three strains show similarities in genome size (6.9–8.0 Mb) and G + C content (53.7–56.2%). The genome size (7.1 Mb) and G + C content (54.3%) of R. obstinata LF1T also fit in the ranges obtained for the novel species. The number of putative open reading frames (ORFs) ranges from 5182 to 6274 (of which 2096–2640 are annotated as hypothetical proteins), which corresponds to 746–785 putative protein-coding genes per Mb. These values give a nearly identical coding density of 89.0 ± 0.3%. Genomic characteristics are summarised in Table 2. The observed genome sizes of the three strains are not only comparable to R. obstinata LF1T, but also to other closely related species, e.g. Rhodopirellula baltica (7.1 Mb) and Roseimaritima ulvae (8.1 Mb). The genome of Pla22T (6.9 Mb) is around 1 Mb smaller than those of Poly51T and Poly59T (both 7.9 Mb). R. obstinata LF1T contains two copies of the 16S rRNA gene, while the gene is present in single copy in the three other strains. The number of 67 tRNAs in LF1T is similar to Pla22T (74 tRNAs) and Poly59T (71 tRNAs), but much lower compared to Poly51T (120 tRNAs).
Conclusion
Based on our physiological, genomic and phylogenetic analysis the three strains represent each a novel species of the genus Rubripirellula, for which we present the here characterised strains as respective type strains.
Emended description of the genus Rubripirellula Bondoso et al. 2016
The description of the genus Rubripirellula is as given previously (Bondoso et al. 2015), with the following modification: Colonies are amaranth pink- to red-coloured.
Description of Rubripirellula amarantea sp. nov
Rubripirellula amarantea (a.ma.ran’te.a. N.L. fem. adj. amarantea of amaranth; corresponding to the amaranth colour of the cells).
Cells are round grain rice-shaped (length: 1.7 ± 0.3 µm, width: 0.9 ± 0.2 µm), form aggregates and divide by polar budding. Colonies have amaranth pink color. Cells of the type strain grow at ranges of 10–36 °C (optimum 33 °C) and at pH 6.0–8.5 (optimum 7.5). The genome of the type strain has a size of 6,945,823 bp and a G + C content of 53.7 ± 0.9%. The type strain genome (acc. no. SJPI00000000) and 16S rRNA gene sequence (acc. no. MK554581) are available from GenBank.
The type strain is Pla22T (DSM 102267T = LMG 29691T), isolated from polyethylene particles incubated in brackish water of the Warnow river estuary close to a wastewater treatment plant near Rostock, Germany.
Description of Rubripirellula tenax sp. nov
Rubripirellula tenax (te’nax. L. fem. adj. tenax holding fast, clinging; corresponding to the characteristic of the cells to be adhesive).
Cells are round grain-rice shaped (length: 1.4 ± 0.2 µm, width: 0.9 ± 0.1 µm), form aggregates and divide by polar budding. Pink amaranth coloured colonies are formed. The temperature optimum is 22–24 °C (growth observed from 10 to 28 °C). The type strain failed to grow at 30 °C or higher. The preferred pH is 8.0, but growth is also observed at pH 6.5–9.0. The type strain genome has a size of 7,988,747 and a G + C content of 56.2 ± 2.1%. The type strain genome (acc. no. SJPW00000000) and 16S rRNA gene sequence (acc. no. MK554552) are available from the GenBank database.
The type strain is Poly51T (DSM 103356T = VKM B-3438T), isolated from polystyrene particles incubated in the Baltic Sea in 2 m depth near Heiligendamm, Germany.
Description of Rubripirellula reticaptiva sp. nov
Rubripirellula reticaptiva (re.ti.cap.ti’va. L. n. rete a fishing net; L. fem. adj. captiva captive; N.L. fem. adj. reticaptiva corresponding to the capture of the cells in water).
Cells are round rice grain-shaped to round (1.5 ± 0.3 µm × 1.0 ± 0.2 µm), form aggregates and divide by polar budding. Colonies have a red colour. The preferred temperature and pH is 24 °C and 7.5–8.0, respectively, while growth is observed in the range of 10–33 °C and at pH 6.5–9.0. The type strain genome has a size of 7,852,560 and a G + C content of 54.8 ± 1.7%. The type strain genome (acc. no. SJPX00000000) and 16S rRNA gene sequence (acc. no. MK554553) are available from GenBank.
The type strain is Poly59T (DSM 103767T = LMG 29696T), isolated from polyethylene particles incubated in the Baltic Sea in 2 m depth near Heiligendamm, Germany.
References
Acehan D, Santarella-Mellwig R, Devos DP (2013) A bacterial tubulovesicular network. J Cell Sci 127:277–280
Bengtsson MM, Øvreås L (2010) Planctomycetes dominate biofilms on surfaces of the kelp Laminaria hyperborea. BMC Microbiol 10:261
Bengtsson MM, Sjøtun K, Lanzén A, Øvreås L (2012) Bacterial diversity in relation to secondary production and succession on surfaces of the kelp Laminaria hyperborea. ISME J 6:2188–2198
Boedeker C, Schuler M, Reintjes G, Jeske O, van Teeseling MC, Jogler M, Rast P, Borchert D, Devos DP, Kucklick M, Schaffer M, Kolter R, van Niftrik L, Engelmann S, Amann R, Rohde M, Engelhardt H, Jogler C (2017) Determining the bacterial cell biology of Planctomycetes. Nat Commun 8:14853
Bondoso J, Harder J, Lage OM (2013) rpoB gene as a novel molecular marker to infer phylogeny in Planctomycetales. Antonie Van Leeuwenhoek 104:477–488
Bondoso J, Balague V, Gasol JM, Lage OM (2014) Community composition of the Planctomycetes associated with different macroalgae. FEMS Microbiol Ecol 88:445–456
Bondoso J, Albuquerque L, Nobre MF, Lobo-da-Cunha A, da Costa MS, Lage OM (2015) Roseimaritima ulvae gen. nov., sp. nov. and Rubripirellula obstinata gen. nov., sp. nov. two novel planctomycetes isolated from the epiphytic community of macroalgae. Syst Appl Microbiol 38:8–15
Bondoso J, Godoy-Vitorino F, Balague V, Gasol JM, Harder J, Lage OM (2017) Epiphytic Planctomycetes communities associated with three main groups of macroalgae. FEMS Microbiol Ecol 93(3):fiw255
Cayrou C, Raoult D, Drancourt M (2010) Broad-spectrum antibiotic resistance of Planctomycetes organisms determined by Etest. J Antimicrob Chemother 65:2119–2122
Devos DP (2014) PVC bacteria: variation of, but not exception to, the Gram-negative cell plan. Trends Microbiol 22:14–20
Devos DP, Reynaud EG (2010) Evolution. Intermediate steps. Science 330:1187–1188
Devos DP, Jogler C, Fuerst JA (2013) The 1st EMBO workshop on PVC bacteria-Planctomycetes-Verrucomicrobia-Chlamydiae superphylum: exceptions to the bacterial definition? Antonie Van Leeuwenhoek 104:443–449
Frank O, Michael V, Pauker O, Boedeker C, Jogler C, Rohde M, Petersen J (2014) Plasmid curing and the loss of grip—the 65-kb replicon of Phaeobacter inhibens DSM 17395 is required for biofilm formation, motility and the colonization of marine algae. Syst Appl Microbiol 38:120–127
Fuerst JA, Sagulenko E (2011) Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol 9:403–413
Godinho O, Calisto R, Ovreas L, Quinteira S, Lage OM (2019) Antibiotic susceptibility of marine Planctomycetes. Antonie Van Leeuwenhoek 112:1273–1280
Graca AP, Calisto R, Lage OM (2016) Planctomycetes as novel source of bioactive molecules. Front Microbiol 7:1241
Jeske O, Jogler M, Petersen J, Sikorski J, Jogler C (2013) From genome mining to phenotypic microarrays: planctomycetes as source for novel bioactive molecules. Antonie Van Leeuwenhoek 104:551–567
Jeske O, Schüler M, Schumann P, Schneider A, Boedeker C, Jogler M, Bollschweiler D, Rohde M, Mayer C, Engelhardt H, Spring S, Jogler C (2015) Planctomycetes do possess a peptidoglycan cell wall. Nat Commun 6:7116
Jeske O, Surup F, Ketteniß M, Rast P, Förster B, Jogler M, Wink J, Jogler C (2016) Developing techniques for the utilization of Planctomycetes as producers of bioactive molecules. Front Microbiol 7:1242
Jogler C, Glöckner FO, Kolter R (2011) Characterization of Planctomyces limnophilus and development of genetic tools for its manipulation establish it as a model species for the phylum Planctomycetes. Appl Environ Microbiol 77:5826–5829
Jogler C, Waldmann J, Huang X, Jogler M, Glöckner FO, Mascher T, Kolter R (2012) Identification of proteins likely to be involved in morphogenesis, cell division, and signal transduction in Planctomycetes by comparative genomics. J Bacteriol 194:6419–6430
Kallscheuer N, Jogler M, Wiegand S, Peeters S, Heuer A, Boedeker C, Jetten M, Rohde M, Jogler C (2019) Rubinisphaera italica sp. nov. isolated from a hydrothermal area in the Tyrrhenian Sea close to the volcanic island Panarea. Antonie van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01329-w
Kim M, Oh HS, Park SC, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351
Kohn T, Heuer A, Jogler M, Vollmers J, Boedeker C, Bunk B, Rast P, Borchert D, Glöckner I, Freese HM, Klenk HP, Overmann J, Kaster AK, Wiegand S, Rohde M, Jogler C (2016) Fuerstia marisgermanicae gen. nov., sp. nov., an unusual member of the phylum Planctomycetes from the German Wadden Sea. Front Microbiol 7:2079
Kohn T, Wiegand S, Boedeker C, Rast P, Heuer A, Jetten MSM, Schüler M, Becker S, Rohde C, Müller R-W, Brümmer F, Rohde M, Engelhardt H, Jogler M, Jogler C (2019) Planctopirus ephydatiae, a novel Planctomycete isolated from a freshwater sponge. Syst Appl Microbiol. https://doi.org/10.1016/j.syapm.2019.126022
König E, Schlesner H, Hirsch P (1984) Cell wall studies on budding bacteria of the Planctomyces/Pasteuria group and on a Prosthecomicrobium sp. Arch Microbiol 138:200–205
Lachnit T, Fischer M, Kunzel S, Baines JF, Harder T (2013) Compounds associated with algal surfaces mediate epiphytic colonization of the marine macroalga Fucus vesiculosus. FEMS Microbiol Ecol 84:411–420
Lage OM, Bondoso J (2014) Planctomycetes and macroalgae, a striking association. Front Microbiol 5:267
Lage OM, Bondoso J, Lobo-da-Cunha A (2013) Insights into the ultrastructural morphology of novel Planctomycetes. Antonie Van Leeuwenhoek 104:467–476
Lee I, Ouk Kim Y, Park SC, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103
Lindsay MR, Webb RI, Fuerst JA (1997) Pirellulosomes: a new type of membrane-bounded cell compartment in planctomycete bacteria of the genus Pirellula. Microbiology 143:739–748
Luo C, Rodriguez RL, Konstantinidis KT (2014) MyTaxa: an advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res 42:e73
Oberbeckmann S, Kreikemeyer B, Labrenz M (2018) Environmental factors support the formation of specific bacterial assemblages on microplastics. Front Microbiol 8:2709
Pilhofer M, Rappl K, Eckl C, Bauer AP, Ludwig W, Schleifer KH, Petroni G (2008) Characterization and evolution of cell division and cell wall synthesis genes in the bacterial phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and phylogenetic comparison with rRNA genes. J Bacteriol 190:3192–3202
Pruesse E, Peplies J, Glöckner FO (2012) SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829
Qin Q-L, Xie B-B, Zhang X-Y, Chen X-L, Zhou B-C, Zhou J, Oren A, Zhang Y-Z (2014) A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol 196:2210–2215
Rast P, Glöckner I, Boedeker C, Jeske O, Wiegand S, Reinhardt R, Schumann P, Rohde M, Spring S, Glöckner FO (2017) Three novel species with peptidoglycan cell walls form the new genus Lacunisphaera gen. nov. in the family Opitutaceae of the verrucomicrobial subdivision 4. Front Microbiol 8:202
Rodriguez-R LM, Konstantinidis KT (2016) The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints, 1900v1
Santarella-Mellwig R, Pruggnaller S, Roos N, Mattaj IW, Devos DP (2013) Three-dimensional reconstruction of bacteria with a complex endomembrane system. PLoS Biol 11:e1001565
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539
Spring S, Bunk B, Sproer C, Schumann P, Rohde M, Tindall BJ, Klenk HP (2016) Characterization of the first cultured representative of Verrucomicrobia subdivision 5 indicates the proposal of a novel phylum. ISME J 10:2801–2816
Stackebrandt E, Ebers J (2006) Taxonomic parameter revisited: tarnished gold standards. Microbiol Today 33:152–155
Staley JT, Fuerst JA, Giovannoni S, Schlesner H (1992) The order Planctomycetales and the genera Planctomyces, Pirellula, Gemmata, and Isosphaera. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer K-H (eds) The Prokaryotes. Springer, New York, pp 3710–3731
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
van Teeseling MC, Mesman RJ, Kuru E, Espaillat A, Cava F, Brun YV, Van Nieuwenhze MS, Kartal B, van Niftrik L (2015) Anammo× Planctomycetes have a peptidoglycan cell wall. Nat Commun 6:6878
Vollmers J, Frentrup M, Rast P, Jogler C, Kaster AK (2017) untangling genomes of novel Planctomycetal and Verrucomicrobial species from monterey bay kelp forest metagenomes by refined binning. Front Microbiol 8:472
Wagner M, Horn M (2006) The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol 17:241–249
Wiegand S, Jogler M, Jogler C (2018) On the maverick Planctomycetes. FEMS Microbiol Rev 42:739–776
Wiegand S, Jogler M, Boedeker C, Pinto D, Vollmers J, Rivas-Marín E, Kohn T, Peeters SH, Heuer A, Rast P, Oberbeckmann S, Bunk B, Jeske O, Meyerdierks A, Storesund JE, Kallscheuer N, Lücker S, Lage OM, Pohl T, Merkel BJ, Hornburger P, Müller R-W, Brümmer F, Labrenz M, Spormann AM, Op den Camp HJM, Overmann J, Amann R, Jetten MSM, Mascher T, Medema MH, Devos DP, Kaster A-K, Øvreås L, Rohde M, Galperin MY, Jogler C (2019) Cultivation and functional characterization of 79 planctomycetes uncovers their unique biology. Nat Microbiol. https://doi.org/10.1038/s41564-019-0588-1
Will SE, Henke P, Boedeker C, Huang S, Brinkmann H, Rohde M, Jarek M, Friedl T, Seufert S, Schumacher M (2018) Day and night: metabolic profiles and evolutionary relationships of six axenic non-marine cyanobacteria. Gen Biol Evol 11:270–294
Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer KH, Whitman WB, Euzeby J, Amann R, Rossello-Mora R (2014) Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12:635–645
Acknowledgements
Part of this research was funded by the Deutsche Forschungsgemeinschaft (DFG) Grants No. KA 4967/1-1 and JO 893/4-1, Grant ALWOP.308 of the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO), SIAM (Soehngen Institute for Anaerobic Microbiology) Grant No. 024002002 and the Radboud Excellence fellowship. We thank Ina Schleicher for skillful technical assistance and our collaborators Sonja Oberbeckmann and Matthias Labrenz (IOW Warnemünde, Germany) for sampling support. We would also like to thank Brian Tindall and Regine Fähnrich as well as the BCCM/LMG Bacteria collection for on-going support during strain deposition.
Author information
Authors and Affiliations
Contributions
NK wrote the manuscript, analyzed the data and prepared the figures; SW and MJ performed the genomic and phylogenetic analysis; AH isolated the strains and performed the initial cultivation and strain deposition; SHP and CB performed the light microscopic analysis; MSMJ contributed to text preparation and revised the manuscript; MR performed the electron microscopic analysis; CJ and MJ took the samples in the Baltic Sea and the river Warnow and CJ supervised the study. All authors read and approved the final version of the manuscript
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethicals statement
This article does not contain any studies with animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kallscheuer, N., Jogler, M., Wiegand, S. et al. Three novel Rubripirellula species isolated from plastic particles submerged in the Baltic Sea and the estuary of the river Warnow in northern Germany. Antonie van Leeuwenhoek 113, 1767–1778 (2020). https://doi.org/10.1007/s10482-019-01368-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-019-01368-3