Abstract
A medium-chain-length poly-3-hydroxyalkanote (MCL-PHA) depolymerase knockout mutant of Pseudomonas putida KT2440 was produced by double homologous recombination. A carbon-limited shake-flask study confirmed that depolymerase activity was eliminated. Lysis of both mutant and wild-type strains occurred under these conditions. In carbon-limited, fed-batch culture, the yield of unsaturated monomers from unsaturated substrate averaged only 0.62 mol mol−1 for the phaZ minus strain compared to 0.72 mol mol−1 for the wild type. The mutant strain also produced more CO2 and less residual biomass from the same amount of carbon substrate. However, most results indicated that elimination of PHA depolymerase activity had little impact on the overall yield of biomass and PHA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Poly-3-hydroxyalkanote (PHA) polyesters are synthesized by many bacteria and have attracted extensive commercial interest due to their biocompatibility and biodegradability [14, 18]. PHAs are classified as short-chain-length PHAs (SCL-PHAs) when the pendant group of monomer varies from 0 to 2 carbons, medium-chain-length PHAs (MCL-PHAs) when there are three or more carbons on the pendant group and SCL–MCL-PHAs when polymers consist of both SCL and MCL monomers [19, 22]. MCL-PHAs are of increasing interest because of their low crystallinity, high elasticity [13, 25] and the possibility of having different functional groups, such as alkenes [21], aromatic groups [12], halogen [9, 16, 20], esters [30] and phenoxy groups [27] on their side chains. Since more than 100 different MCL-PHA monomers can be incorporated, MCL-PHAs can exhibit a wide variety of properties with many possible applications including coatings, medical implants, drug delivery, water-based latex paints and others [39, 40]. PHAs are more expensive to produce than conventional plastics with expenditures almost evenly divided between carbon source, fermentation process and separation process [37]. Metabolic engineering may be used to achieve higher PHA cellular content, more effective carbon source usage, and to obtain novel PHAs with valuable properties, thus increasing commercially viability. One such approach is the elimination of PhaZ activity.
The phaZ gene encoding PhaZ is located between phaC1 and phaC2 genes of the MCL-PHA metabolism gene cluster which consists of phaC1, phaZ, phaC2, phaD, phaF and phaI genes in Pseudomonas putida KT2440 whose entire genome has been mapped [17, 24]. The PhaZ of P. putida KT2442, a spontaneous rif r mutant of P. putida KT2440 [2], is an intracellular MCL-PHA depolymerase and an MCL-PHA granule surface-associated protein [7, 8]. PhaZ is reported to play a crucial role in the turnover of MCL-PHAs under carbon starvation in P. putida KT2442 [5].
During MCL-PHA synthesis, both PHA synthases and depolymerases are found on the surface of the granules along with other “phasins”. This observation, work on scl-PHAs [10] and other data have led to the belief that synthesis and degradation may occur simultaneously [26]. Thus, it can be speculated that phaZ minus mutants should be more productive in terms of PHA synthesis. Reports have demonstrated increased production in phaZ-knockout mutants of P. putida strain U [1] and P. putida KT2442 [3], while others have found that neither a mutant of P. putida GPo1 lacking part of the phaZ gene [14] nor transposon-disrupted phaZ minus mutants of P. resinovorans [34] showed any substantial difference in PHA accumulation under conditions favorable to PHA synthesis. In particular, since deletion of phaZ has been shown to allow more efficient accumulation of “longer” medium-chain-length substrates such as 11-phenoxyundecanoic acid in P. fluorescens BM07 [4], we wished to examine the effects of eliminating PhaZ activity on accumulation of undecylenic acid. This substrate results in the formation of PHA with unsaturated (vinyl) side chains [38] which have many possible applications due to their chemical reactivity. To date, all studies of phaZ minus strains accumulating MCL-PHA have been conducted in flasks where physiological conditions cannot be well controlled. An understanding of the effects of phaZ on PHA accumulation under different physiological conditions is required if MCL-PHA is to be produced efficiently.
In the current study, a phaZ minus mutant of P. putida KT2440 was created as a way to prevent degradation of unsaturated MCL-PHA granules during commercial production of MCL-PHA. Fed-batch culture is typically used for commercial production, as it allows high product concentrations to be achieved. The carboxylic acids needed to achieve maximum accumulation of MCL-PHA inhibit growth at relatively low concentrations. For example, concentrations of sodium octanoate greater than 30 mM (5 g l−1) are toxic to P. putida GPo1 [11]. To avoid carboxylic acid toxicity and since P. putida KT2440, unlike GPo1, exhibits high PHA productivity and yield in carbon-limited high-density culture [23], single-stage, carbon-limited, fed-batch fermentations were used to compare the wild type and the MCL-PHA depolymerase knockout in terms of growth and unsaturated MCL-PHA production. To this end, the phaZ minus mutant was evaluated for its potential to more efficiently produce MCL-PHA under well-controlled PHA production conditions. This is the first report of the behavior of an MCL-PHA depolymerase minus mutant in fed-batch culture.
Materials and methods
Bacterial strains, plasmids, primers and chemicals
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli DH5αF′ was used for routine cloning work, E. coli S17-1 was employed to transfer constructed gene-deletion plasmids into P. putida KT2440 in conjugal experiments and P. putida KT2440 was the MCL-PHA producer whose phaZ gene was deleted to create the phaZ minus mutant. For genetic manipulations, E. coli and P. putida strains were grown in Luria–Bertani (LB) medium at 37 °C overnight. Ampicillin sodium (150 μg ml−1) or kanamycin sulfate (100 μg ml−1) were added as required. pBluescript II KS (+) was used to subclone phaC1 and phaC2 genes from genomic DNA of P. putida KT2440. Suicide plasmid pK18mobsacB was used to construct the plasmid for excising the phaZ gene from the P. putida KT2440 chromosome by double homologous recombination. pGEM®-T Easy was used to subclone the DNA fragment containing phaC1 and phaC2 genes from phaZ minus mutant to confirm the nucleotide sequence. Primers were synthesized by Life Technologies Corporation (USA) and are listed in Table 2. Nonanoic acid (NA) and undecylenic acid (UDA) were purchased from Sigma-Aldrich (Canada). Pseudomonas Isolation Agar was purchased from Thermo Fisher Scientific Inc. (Fisher Scientific, Canada).
Genetic manipulations
Restriction endonucleases, FastAP thermosensitive alkaline phosphatase and Pfu DNA polymerase from Thermo Fisher Scientific Inc. (Fisher Scientific, Canada), and 1-kb DNA Step Ladder, GoTaq Flexi DNA polymerase and T4 DNA ligase from Promega Corporation (Fisher Scientific, Canada) were used as recommended by the suppliers. Plasmid isolation, agarose gel electrophoresis, preparation and transformation of competent E. coli using calcium chloride and electroporation followed the procedures described by Sambrook and Russell [28]. Genomic DNA of P. putida strains were extracted using an E.Z.N.A.® Bacterial DNA kit (Omega Bio-tek Inc., USA), plasmids and PCR products were purified using a Gel/PCR DNA Fragment Extraction kit (IBI Scientific, Canada), and the DNA fragments from restriction endonuclease reactions were purified using an E.Z.N.A.® MicroElute DNA Clean-Up kit (Omega Bio-tek Inc.) according to the protocol provided by the manufacturer. The cloned DNA fragment was sequenced for confirmation (Genome Quebec Innovation Centre, Montreal, Canada).
Construction of phaZ gene-deletion plasmid pK18C1C2 and MCL-PHA depolymerase minus mutant of P. putida KT2440
The phaC1 gene with ribosome binding site was amplified from genomic DNA of P. putida KT2440 by PCR employing Pfu DNA polymerase and a pair of primers amC1-F/amC1-R. The PCR products of phaC1 were purified, digested with HindIII/XhoI, purified and inserted into purified HindIII/XhoI-hydrolyzed pBluescript II KS (+), resulting in plasmid pBlueC1. The phaC2 gene with ribosome binding site was amplified from genomic DNA of P. putida KT2440 by PCR employing Pfu DNA polymerase and a pair of primers delC2-F/delC2-R. The PCR products of phaC2 were digested with XhoI/KpnI and inserted into XhoI/KpnI-hydrolyzed pBlueC1, resulting in plasmid pBlueC1C2. A 3,624-bp DNA fragment containing the phaC1 and phaC2 genes was amplified from pBlueC1C2 by PCR employing Pfu DNA polymerase and a pair of primers amC1-F/C2-R1. The PCR products of phaC1phaC2 were digested with HindIII and inserted into HindIII/alkaline phosphatase-treated pK18mobsacB, resulting in phaZ gene-deletion plasmid pK18C1C2 as shown in Fig. 1a.
The phaZ minus mutant of P. putida KT2440 was created by a double homologous recombination as shown in Fig. 1b and as described by Simon et al. [32] and Schweizer and Hoang [31]. Three ml of both early-log-phase cultures of donor pK18C1C2-bearing E. coli S17-1 and recipient P. putida KT2440 grown in LB medium were mixed and centrifuged to harvest cells; the pellet was resuspended in 100 μl of LB medium, which was then dropped on a filter placed on an LB agar plate that was then incubated at 37 °C overnight to allow conjugation to occur. P. putida KT2440 transconjugants on the filter were selected on Pseudomonas Isolation Agar containing 100 μg ml−1 kanamycin (PIAKan100). Single colonies were subsequently streaked on LB agar containing 15 % sucrose (wt/vol), and incubated at 30 °C overnight. Sucrose-resistant colonies were screened for the appropriate deletion by colony PCR employing GoTaq Flexi DNA polymerase and a pair of primers amZ-F/amZ-R. To verify the nucleotide sequence of phaC1phaC2 cluster of the phaZ minus mutants, a DNA fragment containing phaC1phaC2 genes from genomic DNA of the mutants was amplified by PCR employing GoTaq Flexi DNA polymerase and a pair of primers amC1-F/C2-R1, ligated into pGEM®-T Easy resulting in pGC1∆ZC2, and sequenced using SP6, T7, C1seq 1–3 and C3seq 1–3 primers.
Shake flasks for verifying the function and evaluating the unsaturated MCL-PHA production of the MCL-PHA depolymerase minus mutant
A defined mineral salts (DMS) medium plus 5.0 g l−1 NA or 5.0 g l−1 UDA or a mixture of 2.5 g l−1 NA and 2.5 g l−1 UDA as the only carbon sources was used. The DMS medium contained per liter: 6.35 g Na2HPO4, 2.7 g KH2PO4, 0.39 g MgSO4, 0.5 g (NH4)2SO4 and 1 ml of trace element solution [35]. Cultures grown overnight in Difco nutrient broth (NB) were used as the seed cultures and transferred into Erlenmeyer flasks containing the DMS medium and fatty acid to achieve the initial optical densities OD600nm = 0.2.
Fermentation conditions
The carbon-limited fed-batch fermentations were performed at 30 °C with 3.0 l initial working volume in a Minifors 5 l stirred tank bioreactor (InforsHT, Bottmingen, Switzerland). The inoculum medium, the initial fermentation medium, seed culture preparation and fermentation system were set up and controlled with data acquisition conducted as described previously [38].
A mixture of NA + UDA (1:1 mol/mol) was fed based on the mass of the substrate reservoir. A total of 1 g l−1 of NA + UDA was added to the initial fermentation medium. The additional carbon substrate was exponentially fed according to Eq. 1.
where S t (g) is the total carbon source required to produce biomass X t (g) at time t (h), Y X/S (g g−1) is the yield of biomass from total carbon, X 0 is the initial biomass and μ is the desired specific growth rate (0.15 h−1 was used in this study).
Analytical procedures
Dry cell weights (DCW) were determined with lyophilized biomass. The biomass was obtained by centrifugation of 30–90 ml of culture at 12,000g for 10 min and a distilled water wash. Residual phosphate and ammonium concentrations in the supernatant of the centrifuged broth were determined as described previously [35]. Unsaturated MCL-PHA content and composition were determined by GC analysis after methylation using benzoic acid as the internal standard [36]. The unsaturated MCL-PHA standard for the GC analysis was prepared by repeated cycles of solvent extraction followed by precipitation in cold methanol as described previously [15]. Carbon dioxide (CO2) content in the exit gas was measured with an infrared CO2 monitor (Guardian Plus, Topac Inc. Hingham, MA, USA) and the data were acquired by a LabVIEW 6.1 program (National Instruments) to calculate the CO2 production rate (CPR, g l−1 h−1). The monitor was calibrated with air and a gas mixture containing a known amount of CO2 before each fermentation.
Results
Construction of MCL-PHA depolymerase minus mutants of P. putida KT2440
MCL-PHA depolymerase minus mutants of P. putida KT2440 were created successfully by excising the phaZ from the phaC1ZC2 gene cluster. This was achieved by double homologous recombination between phaC1 and phaC2 genes of the P. putida KT2440 chromosome and phaC1 and phaC2 genes of the introduced plasmid pK18C1C2 (see “Materials and methods” for construction details). The pK18C1C2 plasmid was conjugally transformed into P. putida KT2440 cells (Fig. 1). Candidate phaZ minus mutants were selected from sucrose-resistant colonies and screened by colony PCR using the primer pair, amZ-F/amZ-R. Colonies with successful recombinations would give rise to PCR products of 626-bp DNA fragment without phaZ. Unsuccessful recombination would give rise to a 1,426-bp DNA fragment with phaZ (852 bp). These 1,426-bp products were observed with a control colony of wild-type P. putida KT2440.
PCR products derived from 39 candidate colonies were analyzed by agarose gel electrophoresis. Seven colonies gave the expected DNA fragment size of about 0.6 kb. The other 32 colonies produced DNA products of about 1.4 kb, similar to the control (data not shown). The seven positive candidate colonies were then assessed further using their extracted genomic DNA as templates to amplify their phaC1phaC2 gene cluster by PCR. The primer pair amC1-F/C2-R1 was used for these particular PCR reactions. The PCR products were also cloned into pGEM-T Easy and subjected to sequencing for confirmation.
A comparison of genome-derived PCR products between candidate phaZ mutants and the wild-type P. putida KT2440 confirmed that phaZ was excised from the genome of P. putida KT2440 (Fig. 2). Using the primer pair amC1-F/C2-R1, mutant strains gave rise to a 3.6-kbp product (without phaZ), whereas control strains gave rise to a longer DNA product of about 4.4 kbp (with phaZ). Confirmatory PCR reactions employing the primer pair amZ-F/amZ-R and the same genomic DNA preparations showed the same results as above in the colony PCR screens (0.6 versus 1.4-kb DNA fragments). In addition, the sequencing results for the seven candidate mutants showed that there were no changes in nucleotide composition or in lengths of the phaC1 and phaC2 as a consequence of the double homologous recombination process.
Analysis of PCR products of one representative of the MCL-PHA depolymerase minus mutant of P. putida KT2440 (a) and nucleotide sequence of its phaC1phaC2 gene cluster (b). The lanes in panel a are as follows: lane 1, 1-kb DNA Step Ladder; lanes 2–4, genomic DNA of P. putida KT2440, P. putida DZ18 and plasmid pK18C1C2, respectively, used as templates and employing the primer pair amC1-F/C2-R1; lanes 5–7, genomic DNA of P. putida KT2440, P. putida DZ18 and plasmid pK18C1C2, respectively, used as templates and employing the primer pair amZ-F/amZ-R. In panel b, coding regions of phaC1 and phaC2 are boxed with start and stop codons. Residual nucleotides of phaZ gene are highlighted by gray shading. Native linker nucleotides between phaC1, phaZ and phaC2 genes are bolded and italicized. The genetically added Xho I restriction site is indicated by the strike-through. Nucleotide sequences used to design the primers amC1-F, amC1-R, delC2-F, delC2-R, C2-R1, amZ-F and amZ-R are underlined
These results indicated that the MCL-PHA depolymerase minus mutants were successfully created from double homologous recombination in which the coding region of the phaZ was excised from the genome of P. putida KT2440. One of these seven mutants was selected and designated as P. putida DZ18 for further study.
Functional verification of the MCL-PHA depolymerase in P. putida DZ18
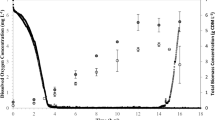
Pseudomonas putida DZ18 and the wild type were grown on NB medium for 20 h and then transferred into baffled shake flasks containing 300 ml DMS medium with 5 g l−1 NA to verify the activity of the MCL-PHA depolymerase in vivo. This was achieved by observing cell growth and MCL-PHA content under nitrogen-limited conditions (Fig. 3). The yield of dry biomass is approximately 5.44 g g−1 NH4 [36]. Since, the medium initially contained only 0.5 g (NH4)2SO4, there was only enough nitrogen to support 0.76 g l−1 of biomass before the N-source was exhausted.
Dry biomass (a), MCL-PHA content (b) and MCL-PHA concentration (c) of wild-type (circle) and MCL-PHA depolymerase minus (triangle) P. putida KT2440. Strains were cultivated at the initial OD600nm ~ 0.2 and shaken at 200 rpm, at 30 °C. 20 ml of cultures were taken at each point of time to analyze biomass and MCL-PHA accumulation. Dry residual biomass (RB) was calculated by subtracting accumulated MCL-PHA from dry total biomass (TB). Data represent the average of the two independent experiments
Both the wild-type P. putida KT2440 and DZ18 grew to a maximum of 2.50 g l−1 dry biomass. The biomass concentration of both strains declined after the carbon and energy source was exhausted, but the rate of decrease was more rapid in the wild type. The % PHA was constant in the PhaZ minus, while it decreased rapidly in the wild type. Moreover, a decrease in residual biomass (total biomass minus PHA) concentration indicated that lysis occurred in both strains during carbon limitation. This result showed that the function of PhaZ in P. putida DZ18 was successfully removed through the excision of phaZ gene from P. putida KT2440 genomic DNA.
Production of unsaturated MCL-PHA by P. putida DZ18 in shake flasks
Pseudomonas putida KT2440 and P. putida DZ18 grown on NB medium for about 20 h were transferred into 500-ml shake flasks containing 100 ml of DMS medium with 5 g l−1 UDA alone or a mixture of NA + UDA (i.e., 2.5 g l−1 of each) to investigate the accumulation of unsaturated MCL-PHA. There was no apparent difference in either PHA content or composition between the wild-type and phaZ minus strains when grown on undecylenic acid alone (Table 3). When a mixture of nonanoic and undecylenic acids was fed, the mutant strain accumulated about 4 mol % more unsaturated subunits in its MCL-PHA. This difference was entirely due to an increase in C9:1 as the % of C11:1 subunits was almost identical.
Effect of phaZ knockout on production of unsaturated MCL-PHA in single-stage, carbon-limited, fed-batch fermentation
Single-stage, carbon-limited, fed-batch fermentations were performed to investigate the effects of the phaZ knockout on cell growth and accumulation of unsaturated MCL-PHA under the carbon-limited conditions. The pattern of growth was not greatly affected by the absence of phaZ (Fig. 4a, b). The overall carbon balance (assuming a residual biomass carbon content of 50 % by weight) was not affected (Fig. 4c). As in the shake flask experiment, much less C9:1 accumulated in the wild-type than in the mutant strain. However, under these conditions, the wild type accumulated more unsaturated subunits. The yield of unsaturated monomers from unsaturated substrate averaged only 0.62 mol mol−1 for the phaZ minus strain compared to 0.72 mol mol−1 for the wild type (Fig. 4d). The other notable difference between the two strains was a marked increase in CO2 production in the mutant strain (Fig. 5).
Dry biomass (a), MCL-PHA concentration (b), carbon balance (c) and yield of unsaturated MCL-PHA produced from UDA-fed (d) the wild-type and the MCL-PHA depolymerase minus P. putida DZ18 from four different batches of the single-stage, carbon-limited, fed-batch fermentations. Dry residual biomass (RB) was calculated by subtracting the accumulated MCL-PHA from the dry total biomass (TB)
Discussion
MCL-carboxylic acids are the most effective substrates for production of MCl-PHAs as they can be incorporated in very high yield. However, since MCL-carboxylic acids are toxic at relatively low concentrations, well-controlled feeding methods are required for their use in high-density fermentations. Carbon limitation is a simple and effective method of achieving this control in the production of MCL-PHA with P. putida KT2440, but carbon limitation may induce lysis. Lysis did occur during carbon and energy starvation of both the wild-type and phaZ minus strains (Fig. 3). Lysis releases surface-active compounds that cause foaming upon aeration. Foaming drastically increases fermentor liquid volume, eventually causing fermentation termination and decreasing overall productivity. The fact that lysis occurs under such conditions demonstrates the importance of a feeding profile that limits accumulation of toxic fatty acids while supplying sufficient carbon and energy to avoid carbon starvation but still allowing PHA synthesis.
Differences in physiology between the wild-type and mutant strains were detected. For example, in fed-batch culture the mutant strain accumulated less unsaturated PHA from the same molar amount of unsaturated substrate taken up. Previous examinations of phaZ minus P. putida have demonstrated that phaZ deletion can interfere in the transcription of phaC1 with a corresponding up-regulation of phaC2 [3]. However, differences in the MCL-PHA composition have been shown to be associated with different levels of phaZ expression and not due to a mutation polar effect [4]. Other variations in the flux of carbon were noted. Although 5 % of the carbon fed was not accounted for (Fig. 4c has a 0.95 slope), the straight line fit demonstrates that data from the four fed-batch fermentations were consistent throughout each individual fermentation. The increased production of CO2 by the phaZ minus strain corresponded to a decrease in residual biomass indicating a significant change in the mutant strain’s physiology not directly related to either the depolymerase or disruption of transcription of phaCs.
MCL-PHA synthesis is controlled by NADH/NAD ratios and carbon substrate availability [26]. In our investigation, the phaZ minus mutant and the wild type produced similar amounts of biomass and PHA during carbon and energy limitation (exponential fed-batch fermentation). The only decrease in MCL-PHA content occurred during carbon and energy starvation of the wild-type strain. However, it has been demonstrated that both MCL-PHA depolymerase and acyl-CoA synthase are present on the surface of MCL-PHA inclusions, facilitating rapid turnover. Acyl substituents released from the granule can be reactivated in a form suitable for polymerization by binding to CoA, which requires ATP. It has also been shown that conditions leading to increased MCL-PHA synthesis, such as the presence of MCL-fatty acids, are associated with increased transcription of phaZ [6]. Thus, although our data do not show any significant advantage in PHA yield by elimination of the depolymerase, there is likely some value in achieving its elimination. A method of eliminating PhaZ activity is needed that retains transcription rates of both phaC1 and phaC2. However, since elimination of PHA depolymerase activity had little effect on PHA productivity, its elimination is not required for commercial production by carbon-limited fed-batch culture of P putida KT2440.
References
Arias S, Bassas-Galia M, Molinari G, Timmis KN (2013) Tight coupling of polymerization and depolymerization of polyhydroxyalkanoates ensures efficient management of carbon resources in Pseudomonas putida. Microb Biotechnol 6(5):551–563. doi:10.1111/1751-7915.12040
Bagdasarian M, Timmis KN (1982) Host-vector systems for gene cloning in Pseudomonas. Curr Topics Microbiol Immunol 96:47–67
Cai L, Yuan MQ, Liu F, Jian J, Chen GQ (2009) Enhanced production of medium-chain-length polyhydroxyalkanoates (PHA) by PHA depolymerase knockout mutant of Pseudomonas putida KT2442. Bioresour Technol 100(7):2265–2270. doi:10.1016/j.biortech.2008.11.020
Choi MH, Xu J, Rho JK, Zhao XP, Yoon SC (2010) Enhanced production of longer side chain polyhydroxyalkanoic acid with ω-aromatic group substitution in phaZ-disrupted Pseudomonas fluorescens BM07 mutant through unrelated carbon source cometabolism and salicylic acid β-oxidation inhibition. Bioresour Technol 101(12):4540–4548. doi:10.1016/j.biortech.2010.01.082
de Eugenio LI, Escapa IF, Morales V, Dinjaski N, Galán B, García JL, Prieto MA (2010) The turnover of medium-chain-length polyhydroxyalkanoates in Pseudomonas putida KT2442 and the fundamental role of PhaZ depolymerase for the metabolic balance. Environ Microbiol 12(1):207–221. doi:10.1111/j.1462-2920.2009.02061.x
de Eugenio LI, Galán B, Escapa IF, Maestro B, Sanz JM, García JL, Prieto MA (2010) The PhaD regulator controls the simultaneous expression of the pha genes involved in polyhydroxyalkanoate metabolism and turnover in Pseudomonas putida KT2442. Environ Microbiol 12(6):1591–1603. doi:10.1111/j.1462-2920.2010.02199.x
de Eugenio LI, García JL, García P, Prieto MA, Sanz JM (2008) Comparative analysis of the physiological and structural properties of a medium-chain-length polyhydroxyalkanoate depolymerase from Pseudomonas putida KT2442. Eng Life Sci 8(3):260–267. doi:10.1002/elsc.200700057
de Eugenio LI, García P, Luengo JM, Sanz JM, San Román J, García JL, Prieto MA (2007) Biochemical evidence that phaZ gene encodes a specific intracellular medium-chain-length polyhydroxyalkanoate depolymerase in Pseudomonas putida KT2442. Characterization of a paradigmatic enzyme. J Biol Chem 282(7):4951–4962. doi:10.1074/jbc.M608119200
Doi Y, Abe C (1990) Biosynthesis and characterization of a new bacterial copolyester of 3-hydroxyalkanoates and 3-hydroxy-ω-chloroalkanoate. Macromolecules 23(15):3705–3707. doi:10.1021/ma00217a027
Doi Y, Segawa A, Kawaguchi Y, Kunioka M (1990) Cyclic nature of poly(3-hydroxyalkanoate) metabolism in Alcaligenes eutrophus. FEMS Microbiol Lett 67(1–2):165–170. doi:10.1111/j.1574-6968.1990.tb13856.x
Elbahloul Y, Steinbüchel A (2009) Large-scale production of poly(3-hydroxyoctanoic acid) by Pseudomonas putida GPo1 and a simplified downstream process. Appl Environ Microbiol 75(3):643–651. doi:10.1128/AEM.01869-08
Fritzsche K, Lenz RW, Fuller RC (1990) An unusual bacterial polyester with a phenyl pendant group. Die Makromolekulare Chemie 191(8):1957–1965. doi:10.1002/macp.1990.021910821
Gross RA, DeMello C, Lenz RW, Brandl H, Fuller RC (1989) The biosynthesis and characterization of poly(β-hydroxyalkanoates) produced by Pseudomonas oleovorans. Macromolecules 22(3):1106–1115. doi:10.1021/ma00193a018
Huisman GW, Wonink E, de Koning G, Preusting H, Witholt B (1992) Synthesis of poly(3-hydroxyalkanoates) by mutant and recombinant Pseudomonas strains. Appl Microbiol Biotechnol 38(1):1–5. doi:10.1007/BF00169409
Jiang X, Ramsay JA, Ramsay BA (2006) Acetone extraction of MCL-PHA from Pseudomonas putida KT2440. J Microbiol Methods 67(2):212–219. doi:10.1016/j.mimet.2006.03.015
Kim OY, Gross RA, Hammar WJ, Newmark RA (1996) Microbial synthesis of poly(β-hydroxyalkanoates) containing fluorinated side-chain substituents. Macromolecules 29(13):4572–4581. doi:10.1021/ma960059j
Kim TK, Shin HD, Seo MC, Lee JN, Lee YH (2003) Molecular structure of PCR cloned PHA synthase genes of Pseudomonas putida KT2440 and its utilization for medium-chain-length polyhydroxyalkanoate production. J Microbiol Biotech 13(2):182–190
Kim YB, Kim DY, Rhee YH (1999) PHAs produced by Pseudomonas putida and Pseudomonas oleovorans grown with n-alkanoic acids containing aromatic groups. Macromolecules 32(19):6058–6064. doi:10.1021/ma981904w
Kim YB, Lenz RW (2001) Polyesters from microorganisms. In: Babel W, Steinbüchel A (eds) Advances in biochemical engineering/biotechnology: biopolyesters, vol 71. Springer, Berlin, Heidelberg, Germany, pp 51–79. doi:10.1007/3-540-40021-4_2
Kim YB, Lenz RW, Fuller RC (1992) Poly(β-hydroxyalkanoate) copolymers containing brominated repeating units produced by Pseudomonas oleovorans. Macromolecules 25(7):1852–1857. doi:10.1021/ma00033a002
Lageveen RG, Huisman GW, Preusting H, Ketelaar P, Eggink G, Witholt B (1988) Formation of polyesters by Pseudomonas oleovorans: effect of substrates on formation and composition of poly-(R)-3-hydroxyalkanoates and poly-(R)-3-hydroxyalkenoates. Appl Environ Microbiol 54(12):2924–2932
Lenz RW, Kim YB, Fuller RC (1992) Production of unusual bacterial polyesters by Pseudomonas oleovorans through cometabolism. FEMS Microbiol Rev 103(2–4):207–214. doi:10.1111/j.1574-6968.1992.tb05839.x
Maclean H, Ramsay J, Ramsay B (2008) Declining exponential feeding as a strategy for the high-density production of medium-chain-length poly-3-hydroxyalkanoates. Can J Chem 86:564–569
Nelson KE, Weinel C, Paulsen IT, Dodson RJ, Hilbert H, Martinsdos Santos VA, Fouts DE, Gill SR, Pop M, Holmes M et al (2002) Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol 4(12):799–808. doi:10.1046/j.1462-2920.2002.00366.x
Preusting H, Nijenhuis A, Witholt B (1990) Physical characteristics of poly(3-hydroxyalkanoates) and poly(3-hydroxyalkenoates) produced by Pseudomonas oleovorans grown on aliphatic hydrocarbons. Macromolecules 23(19):4220–4224. doi:10.1021/ma00221a007
Ren Q, de Roo G, Ruth K, Witholt B, Zinn M, Thöny-Meyer L (2009) Simultaneous accumulation and degradation of polyhydroxyalkanoates: futile cycle or clever regulation? Biomacromolecules 10(4):916–922. doi:10.1021/bm801431c
Ritter H, von Spee AG (1994) Bacterial production of polyesters bearing phenoxygroups in the side chains: poly(3-hydroxy-5-phenoxypentanoate-co-3-hydroxy-9-phenoxynonanoate) from Pseudomonas oleovorans. Macromol Chem Phys 195(5):1665–1672. doi:10.1002/macp.1994.021950517
Sambrook J, David WR (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145(1):69–73. doi:10.1016/0378-1119(94)90324-7
Scholz C, Fuller RC, Lenz RW (1994) Production of poly(β-hydroxyalkanoates) with β-substituents containing terminal ester groups by Pseudomonas oleovorans. Macromol Chem Phys 195(4):1405–1421. doi:10.1002/macp.1994.021950424
Schweizer HP, Hoang TT (1995) An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158(1):15–22. doi:10.1016/0378-1119(95)00055-B
Simon R, O’Connell M, Labes M, Puhler A (1986) Plasmid vector for the genetic analysis and manipulation of rhizobia and other Gram-negative bacteria. Methods Enzymol 118:640–659. doi:10.1016/0076-6879(86)18106-7
Simon R, Priefer U, Puehler A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nature Biotechnol 1(9):784–791. doi:10.1038/nbt1183-784
Solaiman DKY, Ashby RD, Foglia TA (2003) Effect of inactivation of poly (hydroxyalkanoates) depolymerase gene on the properties of poly (hydroxyalkanoates) in Pseudomonas resinovorans. Appl Microbiol Biotechnol 62(5–6):536–543. doi:10.1007/s00253-003-1317-4
Sun Z, Ramsay JA, Guay M, Ramsay BA (2006) Automated feeding strategies for high-cell-density fed-batch cultivation of Pseudomonas putida KT2440. Appl Microbiol Biotechnol 71(4):423–431. doi:10.1007/s00253-005-0191-7
Sun Z, Ramsay JA, Guay M, Ramsay BA (2007) Carbon-limited fed-batch production of medium-chain-length polyhydroxyalkanoates from nonanoic acid by Pseudomonas putida KT2440. Appl Microbiol Biotechnol 74(1):69–77. doi:10.1007/s00253-006-0655-4
Sun Z, Ramsay JA, Guay M, Ramsay BA (2007) Fermentation process development for the production of medium-chain-length poly-3-hydroxyalkanoates. Appl Microbiol Biotechnol 75(3):475–485. doi:10.1007/s00253-007-0857-4
Sun Z, Ramsay JA, Guay M, Ramsay BA (2009) Fed-batch production of unsaturated medium-chain-length polyhydroxyalkanoates with controlled composition by Pseudomonas putida KT2440. Appl Microbiol Biotechnol 82(4):657–662. doi:10.1007/s00253-008-1785-7
Witholt B, Kessler B (1999) Perspectives of medium-chain-length poly(hydroxyaIkanoates), a versatile set of bacterial bioplastics. Curr Opin Biotechnol 10(3):279–285. doi:10.1016/S0958-1669(99)80049-4
Zinn M, Witholt B, Egli T (2001) Occurrence, synthesis and medical application of bacterial polyhydroxyalkanoate. Adv Drug Deliv Rev 53(1):5–21. doi:10.1016/S0169-409X(01)00218-6
Acknowledgments
The authors gratefully acknowledge the provision of plasmid pEX18Tc, pK18mobsacB and E. coli S17-1 by Professor Keith Poole at the Department of Biomedical and Molecular Sciences, Queen’s University, Kingston, Ontario, Canada, and the financial support of the Ontario Centres of Excellence.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vo, M.T., Ko, K. & Ramsay, B. Carbon-limited fed-batch production of medium-chain-length polyhydroxyalkanoates by a phaZ-knockout strain of Pseudomonas putida KT2440. J Ind Microbiol Biotechnol 42, 637–646 (2015). https://doi.org/10.1007/s10295-014-1574-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-014-1574-5