Abstract
The phaZ gene of Pseudomonas resinovorans codes for a poly(hydroxyalkanoates) (PHA) depolymerase. Two phaZ mutants of Pseudomonas resinovorans NRRL B-2649, FOAC001 and FOAC002, were constructed by an in vitro transposition procedure followed by chromosomal integration via homologous recombination. A detailed mapping of the transposon insertion sites and an analysis of the resultant sequences showed that putative fusion polypeptides PhaZFOAC001 (239 amino-acid residues) and PhaZFOAC002 (85 amino-acid residues) could result from the mutant phaZ genes of FOAC001 and FOAC002, respectively. In vivo PHA degradation data indicated that PhaZFOAC001 might still retain a partial PHA depolymerization activity, while PhaZFOAC002 is completely devoid of this function. The cell yields and PHA contents of B-2649, FOAC001, and FOAC002 were similar when the cells were grown either under a limiting nitrogen-source (low-N) condition for up to 5 days or in excess N-source (high-N) for 3 days. A dramatic decrease in PHA content was observed in the PhaZ-active B-2649 and FOAC001 cells during prolonged cell growth (5 days) in high-N medium or in response to a shift-up in nitrogen-source. The repeat-unit compositions of the PHAs from FOAC001 and FOAC002 contained slightly lower amounts of β-hydroxyoctanoate and higher β-hydroxytetradecenoate than that of the wild-type B-2649 when grown under a high-N condition. While the molecular masses of the PHAs from FOAC001 and FOAC002 did not vary under any conditions used in this study, those of the wild-type B-2649 were markedly increased in cells either grown for 5 days under a high-N condition or subjected to a nitrogen-source shift-up. These phaZ mutants thus provide a valuable system to study the influence of PHA depolymerase on the accumulation and properties of medium-chain-length PHA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Poly(hydroxyalkanoates) (PHAs) are carbon and energy reserve materials synthesized by many microorganisms (Steinbüchel 1991). These biodegradable polyesters are potentially important substitutes for the hard-to-degrade plastics and elastomeric materials produced from petroleum-based feedstock (Anderson and Dawes 1990; Poirier et al. 1995). One class of PHAs, those of medium-chain-length (mcl), is synthesized chiefly by Pseudomonas species belonging to the rRNA–DNA homology group I. mcl-PHA contains as its repeat-unit monomers only β-hydroxyalkanoates having 6–14 carbon-atom chain length (Brandl et al. 1988; De Smet et al. 1983; Lageveen et al. 1988). These PHAs are potentially useful in the formulation of adhesives and elastomers (Anderson and Dawes 1990). However, an improvement in their economics of production and their performance characteristics is needed to render these PHAs more attractive for commercial application.
PHA depolymerases are a group of enzymes responsible for the specific degradation of short-chain-length (scl) or mcl-PHAs (see review by Jendrossek and Handrick, 2002). The PHA depolymerase enzyme (PhaZ) is coded for by phaZ in the mcl-PHA biosynthesis gene locus of Pseudomonas (Rehm and Steinbuchel 1999; Solaiman et al. 2000; Solaiman 2002). Inactivation of phaZ led to defective mobilization of intracellularly accumulated PHA in a mutant of Pseudomonas oleovorans (GPo500) (Huisman et al. 1991, 1992). Ruiz et al. (2001) related the failure of mutant GPo500 to mobilize its PHA reserve to its reduced survivability in river water microcosms. Interestingly, inactivation of phaZ in GPo500 did not lead to a net increase in the maximal accumulation level of the polymer (Huisman et al. 1992). This earlier study also did not find any significant differences in the physicochemical properties of the mcl-PHAs produced by P. oleovorans GPo1 and its GPo500 mutant. Since the genetic lesion of GPo500 had not been reported, we decided to construct additional phaZ mutants of P. resinovorans for a more detailed genetic and physiological study in the context of their effects on mcl-PHA biosynthesis. Here, we report the construction and characterization of two transposon-insertion phaZ mutants of P. resinovorans. We also describe PHA production characteristics of these mutants and the wild type strain under either a limiting or an excess nitrogen-source culturing condition. In addition, the physicochemical properties of the mcl-PHAs isolated from the three strains are reported. The results provide a better understanding of the influence of phaZ inactivation on the production and properties of mcl-PHA in P. resinovorans.
Materials and methods
Bacteria and culture conditions
P. resinovorans NRRL B-2649 (NCAUR-ARS-USDA's Microbial Culture Collection, Peoria, Ill., USA) and Escherichia coli DH5α (Invitrogen, Carlsbad, Calif., USA) were grown in either Luria-Bertani (LB) medium (1% w/v tryptone, 0.5% w/v yeast extract, 0.5% NaCl) or tryptic soy broth (Difco, Detroit, Mich., USA) at 30 °C (for P. resinovorans) or 37 °C (for E. coli) with shaking (200–250 rpm). For solid media, agar was added to a final concentration of 1.2–1.5% (w/v). Ampicillin (Ap, 50–100 μg/ml), carbenicillin (Cb, 50 μg/ml) and kanamycin (Km, 30–50 μg/ml) were added to the media as needed.
Molecular biology procedures
DNA restriction and modifying enzymes, and thermally stable DNA polymerases were variously purchased from Invitrogen, New England Biolabs (Beverly, Mass. ,USA) and Novagen (Madison, Wis., USA), and were used according to the procedures suggested by the vendors. Rapid, small-scale plasmid screening was carried out as described by Sambrook et al. (1989). Large-scale plasmid preparations were made either according to the method of Sambrook et al. (1989) or by using a Plasmid Midi Kit according to the manufacturer's protocol (Qiagen, Valencia, Calif., USA). DNA restriction fragments were analyzed by agarose gel electrophoresis in TBE buffer system (0.089 M Tris base, 0.089 M boric acid, 0.002 M Na-EDTA).
Electroporation
Plasmid DNA was introduced into P. resinovorans by an electroporation protocol (Solaiman 1998). Electroporation was carried out with a 0.1-cm gap-width cuvette and the settings of 2.5 kV, 25 μF and 200 Ω on a Gene Pulser II electroporator equipped with a Pulse Controller Plus module (Bio-Rad Laboratories, Hercules, Calif., USA). After electroporation, cells were allowed to recover in SOC medium (Hanahan 1983) for 2 h at 30 °C with 250 rpm shaking. The transformants were selected on LB solid-medium containing Km (30 μg/ml).
PCR cloning, transposon insertion, and nucleotide sequencing
phaZ was cloned by PCR from the genomic DNA of P. resinovorans NRRL B-2649 by using the primer-pair PHAZ-L (5′-CAATAGGGCTCCGCGCATGCC-3′) and PHAZ-R (5′-GTGTTAACCGCTCCGTAGCGGC-3′). The desired PCR product (876 bp) was gel-isolated by using the spin columns of the TOPO XL PCR Cloning Kit (Invitrogen). Purified phaZ was cloned into the EcoRV site of a pETBlue-1 vector by using the Perfectly Blunt Cloning Kit (Novagen). The resultant recombinant plasmids, arbitrarily designated as pETZ-a and pETZ-b depending on the orientation of the DNA insert, were isolated and separately subjected to a transposon-insertion reaction using the EZ::TN <KAN-2>Insertion Kit (Epicentre, Madison, Wis., USA). The transposon-insertion reaction mixture was used to transform E. coli DH5α cells. Transformants were selected on LB medium containing Km. The plasmids were screened on the kanamycin resistant clones to identify those containing a transposon insertion in phaZ. The selected recombinant plasmids containing a transposon-interrupted phaZ were used to electrotransform P. resinovorans NRRL B-2649.
To map the site of transposon insertion, the pertinent chromosomal DNA fragment was amplified from the P. resinovorans transposon mutants by using the primer-pair SQPRII170A (5′-CAAGACCAAGAAAGCCAG-3′)/PHAZ-R. The site of transposon insertion was mapped by sequencing the amplified fragment using the ABI PRISM BigDyeTerminators v3.0 Cycle Sequencing Kit (Applied Biosystems, Foster City, Calif., USA). Sequence data were collected and analyzed on an ABI PRISM 310 Genetic Analyzer or an ABI PRISM 3700 DNA Analyzer (Applied Biosystems).
Electron microscopy
Bacteria in culture samples were fixed in situ by the addition of glutaraldehyde (2.5% v/v). The cells were pelleted and treated in 2% osmium tetraoxide solution prior to being embedded in plastic. Embedding was carried out by sequential immersion in propylene oxide, a 1:1 (v/v) mixture of propylene oxide/epoxy resin, and finally in epoxy resin and cured for 2 days at 55 °C. Thin-sections were cut and stained sequentially with 2% uranyl acetate solution and lead citrate solution. The samples were viewed with an electron microscope (Philips/FEI model CM12; Hillsboro, Ore., USA).
Isolation and characterization of PHA
Ten ml of an overnight culture was used to inoculate 1 l of medium E* (Brandl et al. 1988) in 2-l Erlenmeyer flasks. For culturing under an excess nitrogen source condition, the concentration of (NH4)2HPO4 in the medium was 6.6 g/l (equivalent to 50 mM). The cultures were grown in a shaker-incubator (30 °C, 250 rpm) for various durations depending on the experiment. In nitrogen-source shift-up experiments in which the cultures were spiked with additional nitrogen source, an appropriate volume of a filter-sterilized (NH4)2HPO4 stock solution (4.4 g/12 ml) was added to the cultures at the specified time. Cells were harvested by centrifugation and lyophilized to dryness. The weights of the lyophilized cells constituted the cell dry weights (CDWs). PHA was extracted by vigorously shaking the dry cell pellets with excess chloroform in a shaker-incubator overnight at room temperature. After removing the cell debris by filtration through Whatman no.1 filter paper, the clear chloroform extract was rotoevaporated to a syrupy consistency. The weight of the syrupy materials constituted the crude PHA-yield. Purified PHA was obtained by re-dissolving the crude PHA in a minimal volume of chloroform, and the solution was added dropwise into a beaker containing pre-chilled methanol under constant stirring. The resultant PHA precipitates were collected and dried under vacuum overnight, and stored under nitrogen atmosphere until use. The repeat-unit composition of PHA was determined by gas chromatography/mass spectrometric (GC/MS) analysis according to a previously described method (Ashby et al. 2000). The molecular masses of the polymers were determined by gel permeation chromatography as described by Cromwick et al. (1996).
Results
Construction and verification of the phaZ::Tn<Kan-2>mutants of P. resinovorans.
The strategy for generating the mutants was to first introduce transposon-inserted phaZ carried on a non-replicating plasmid vector into the target host, P. resinovorans. Once inside the host cell, the mutant phaZ could then replace the native gene through a homologous recombination mechanism. Selection of the transformed cells against the kanamycin-resistance marker of the transposon allowed identification of recombinant bacteria. Using this strategy, phaZ was first cloned into an E. coli expression vector, pETBlue-1, which is not known to replicate in P. resinovorans. A synthetic transposon, EZ::TN<KAN-2>(Epicentre), was inserted into phaZ of the plasmid by a transposase-mediated in vitro reaction (Goryshin and Reznikoff, 1998). Two recombinant plasmids, each containing a transposon-inserted mutant phaZ oriented in the opposite direction, were selected for electroporation into P. resinovorans NRRL B-2649. As expected in the low-frequency homologous recombination event, only one to four kanamycin-resistant P. resinovorans colonies appeared on the selection agar-medium plates per experiment. Two recombinant P. resinovorans clones, designated as strains FOAC001 and FOAC002, were selected for further study.
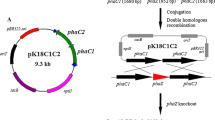
The insertion site and orientation of the transposon in each P. resinovorans phaZ mutant were mapped by sequence analysis. The chromosomally integrated mutant phaZ genes of FOAC001 and FOAC002 were amplified by PCR using the primer-pairs SQPRII170A/PHAZ-R. The ~2.4-kb amplified fragments were purified by gel electrophoresis and used as templates in sequencing reactions. Primers KAN-2 FP1 (Epicentre; 5′-ACCTACAACAAAGCTCTCATCAACC-3′) and KAN-2 RP1 (Epicentre; 5′-GCAATGTAACATCAGAGATTTTGAG-3′) were used in the reactions to read outwardly from the transposon. Second-strand sequencing was done using primers SQPRII170A and PHAZ-R. The results show that, in strain FOAC001, the transposon (designated as Tn<kan-2>) integrated into phaZ after nucleotide 664 (Fig. 1A). The orientation of the kanamycin-resistance determinant (KmR) of transposon in FOAC001 is opposite to that of phaZ. An open-reading-frame (ORF) analysis showed that a part of the contiguous transposon sequence had added 18 amino-acid residues to the truncated PhaZ to form a putative fusion protein, PhaZFOAC001, of 239 amino acids (Fig. 1B). For comparison, the complete P. resinovorans PhaZ contains 285 amino acids. For mutant strain FOAC002, integration of the transposon occurred after nucleotide 163 of phaZ (Fig. 1A). Unlike the case with FOAC001, the orientation of the KmR in FOAC002 is the same as that of phaZ. An ORF analysis indicated that an 85-amino-acid putative fusion polypeptide, PhaZFOAC002, consisting of truncated PhaZ(54 amino acids) and the transposon sequence (31 amino acids) could occur in FOAC002 (Fig. 1B). Short direct-repeat sequences (9 bases) of phaZ were found flanking the transposon inserts in both mutants. This target-site duplication is characteristic of the transposon insertion mechanism.
The phaZ genes of wild-type and transposon-mutated Pseudomonas resinovorans. A The P. resinovorans pha locus, showing the site of transposon insertion in phaZ of FOAC001 and FOAC002. Solid filled arrows phaC1 and phaC2 (PHA synthase genes), left-hatched interrupted arrow phaZ (PHA depolymerase gene), open box Tn<kan-2>(EZ::TN<KAN-2>cassette), right-hatched arrow KmR (kanamycin-resistance gene). B Comparison of the putative gene products of the wild-type and transposon-inserted phaZ genes. Open box native PhaZ amino-acid sequence, right-hatched box predicted α/β-hydrolase fold of PhaZ, filled box putative lipase box, left-hatched box extraneous fusion-peptide sequence resulting from transposon insertion
Synthesis and properties of PHA
Synthesis and properties of PHA from the phaZ mutants were studied with cells grown on oleic acid as the carbon source. Table 1 shows that under the same growth conditions with a limiting (or low) nitrogen-source of 8.3 mM (NH4)2HPO4, both P. resinovorans FOAC001 and FOAC002 grew to a cell yield of 3.1–3.3 g CDW/l culture, which is close to the value obtained with the wild-type cells. The PHA contents of the three cell lines were in the range of 32.5–40.3%CDW and were thus not significantly different from each other. Furthermore, the length of cell cultivation did not significantly affect the cell densities and PHA contents of the three cell lines.
A similar experiment was carried out under an excess (or high) nitrogen-source condition (i.e., 50 mM (NH4)2HPO4) to evaluate its effect on cell growth and PHA yields. The results (Table 1) showed that excess nitrogen did not affect the cell growth of the three strains of P. resinovorans. Regardless of a cultivation time of 3 or 5 days, all three cell lines grown under this condition gave a cell yield of 2.6–3.7 g/l. Furthermore, all three strains at the end of a 3-day growth period contained comparable amounts of PHA, 35.1–42.8%CDW. At the end of a 5-day growth period, however, there was a dramatic decrease in the polymer content of the wild-type and the FOAC001 mutant (about 6%CDW), while FOAC002 retained its accumulated PHA (41.7%CDW).
The repeat-unit compositions of the PHAs extracted from cells grown under the various conditions described above were compared. The results (Table 2) showed that, in general, all of the isolated PHAs contained β-hydroxyoctanoate (C8:0), β-hydroxydecanoate (C10:0) and β-hydroxytetradecenoate (C14:1) as their major monomers. The data further showed that cells grown for 3 or 5 days under the limiting nitrogen-source condition yielded PHAs with similar repeat-unit compositions. Furthermore, the PHA compositions were very similar for the three cell lines grown for 3 days under an excess nitrogen-source condition. After a 5 days of growth under this condition, however, the PHAs of the two mutants appeared to have a slightly lower C8:0 content and a higher C14:1 content than that of wild-type P. resinovorans (Table 2).
The molecular masses of the PHAs obtained from these cells were determined by gel-permeation chromatography. There was a slight but noticeable time-dependent increase in the molecular masses of PHAs from the wild-type and the FOAC001 mutant grown under either low- or high-nitrogen source conditions (Table 3). The FOAC002 mutant, however, did not exhibit such behavior. The PHAs of the 5-day-old wild-type cells grown under the limiting nitrogen-source condition had markedly higher molecular masses than those of the other test samples (Table 3). Under this condition, the number-average molecular weight of the wild-type PHA was 2.5–3 times that of the PHAs from the FOAC001 and FOAC002 mutants (Table 3).
Using the differential scanning calorimetric method, the melting temperature (Tm) and glass-transition temperature (Tg) of each PHA sample was measured . The results showed that all of the samples exhibited a Tg value in the range of −50.6 to −41.9 °C (data not shown). The Tm values, however, were not readily discernable from the calorimetric tracings, indicating the amorphous behavior of the isolated polymers.
PHA degradation under nitrogen-source shift-up
PHA is purportedly the carbon and energy reserve material of the many microorganisms that produce the polymer. To further understand its roles in cell maintenance, the content and properties of PHA in the wild-type were compared with those of the phaZ mutants of P. resinovornas on nitrogen-source shift-up. Accordingly, cells of the three strains were grown for 3 days under the limiting N-source conditions. This allowed the cells to accumulate PHAs of similar properties to similar levels (Tables 1, 2, 3). A fresh dose (8.3 mM) of (NH4)2HPO4 was then added to the cultures, and the cells were grown for an additional 2 days before they were harvested and processed for PHA isolation and characterization. The results showed that the three cell lines yielded similar cell yields (Table 4). Data in Table 4 further show that wild-type P. resinovorans lost virtually all of its accumulated PHA. A marked decrease in PHA content was also observed with the FOAC001 mutant. P. resinovorans FOAC002 mutant, however, retained all the accumulated PHA under the nitrogen-source shift-up (Table 4). Pre- (3-day growth) and post- (5-day growth) shift-up cells were examined by transmission electron microscopy to visualize the state of the PHA intracellular granules. The series of micrographs in Fig. 2 clearly show that a copious number of large PHA granules remained un-degraded in the post-shift-up 5-day FOAC002 cells. By contrast, there were markedly fewer small PHA granules in the 5-day wild-type and FOAC001 phaZ mutant.
Transmission electron micrographs of P. resinovorans NRRL B-2649 (left), FOAC001 phaZ transposon-mutant (center), and FOAC002 phaZ transposon-mutant (right). Cells were first grown in E* medium ([(NH4)2HPO4]=8.3 mM) for 3 days, and an aliquot of each culture was removed for transmission electron microscopy (top). Additional (NH4)2HPO4 (7.7 mM) was then added to each culture, and cultivation was continued for 2 days (bottom)
The repeat-unit compositions of the PHAs obtained from the post-shift-up cells were determined. Table 5 shows that the compositions of the polymers were not significantly different from each other. In all cases, the three major monomers were β-hydroxyoctanoate (C8:0), β-hydroxydecanoate (C10:0) and β-hydroxytetradecenoate (C14:1). Furthermore, the composition profiles were very similar to those obtained with cells grown in low (limiting) nitrogen-source conditions (Table 2). Whereas a slightly lower C8:0 content and a higher C14:1 content were previously observed in the PHAs of the 5-day cells grown in excess nitrogen-source (Table 2), this trend was not apparent in PHAs of the nitrogen-source shift-up cells (Table 5).
The molecular masses of the PHAs in this nitrogen-source shift-up experiment were determined and are presented in Table 5. The results showed that the FOAC001 and FOAC002 mutants contained PHAs that have a number-average molecular weight (Mn) of about 40×103 g/mol. The apparent Mn value of the polymer of the wild-type strain was determined as (271±9)×103 g/mol (Table 5). The Tgs of the PHAs from FOAC001 and FOAC002 strains range from −43.8 to −40.9 °C. The differential scanning calorimetric measurement failed to yield a Tg for the wild-type polymer and the Tms for the PHAs of the three cell lines.
Discussion
In this study, two strains of P. resinovorans phaZ transposon-mutants, FOAC001 and FOAC002, were constructed. Unlike the well-studied phaZ mutant of P. oleovorans (GPo500), the precise genetic lesion of which was never reported, we have mapped the transposon-insertion site of phaZ in P. resinovorans FOAC001 and FOAC002 (Fig. 1A). An ORF analysis showed that the mutated phaZ of FOAC001 could putatively code for a PhaZFOAC001 fusion protein (Fig. 1B). A conserved domain analysis had shown that the PHA depolymerase (PhaZPre) encoded by the P. resinovorans phaZ contains an α/β-hydrolase fold spanning amino acids 55–266. A conserved lipase-box sequence (Huisman et al. 1991) was located in amino acids 96–105 of PhaZPre. In PhaZFOAC001, nearly 80% of the α/β-hydrolase fold and the entire lipase box identified in PhaZPre were retained. An additional 17-amino-acid fusion peptide was created at the carboxy-terminus by an in-frame part of the inserted transposon sequence. The putative gene product (PhaZFOAC002, 85 amino acids) of the mutated phaZ in FOAC002 consists of 54 amino acid residues of PhaZPre at the amino-terminal end and a 31-amino-acid fusion peptide at the carboxy-terminus. The PhaZFOAC002 polypeptide had lost the entire α/β-hydrolase fold of the native PhaZPre (Fig. 1B). The results of the structural analysis provide a plausible explanation for the ability of strain FOAC001 to still degrade intracellularly accumulated PHA (Table 4). The putative PhaZFOAC001 of FOAC001 not only contains 78% of the amino-terminal portion of the native PhaZPre enzyme, but also retains 80% of the predicted α/β-hydrolase fold. PhaZFOAC001 is thus expected to retain at least a partial PHA depolymerization activity. The data in Table 4 indeed show that, while there was a near-complete depletion of PHA (3.0% CDW) in the wild-type strain, FOAC001 still retained a comparatively higher average PHA content (13.3%CDW). An examination of the putative PhaZFOAC002 protein sequence of strain FOAC002, on the other hand, clearly showed that the protein is unlikely to exhibit any PHA depolymerization activity. Definitive conclusions regarding the activities of PhaZFOAC001 and PhaZFOAC001 await further in vitro study (Foster et al. 1994).
We had expected that inactivation of phaZ would lead to decreased degradation of mcl-PHAs and thus to a concomitant increased accumulation of the polymer. Experimental results, however, showed that the PHA contents of P. resinovorans FOAC001 and FOAC002 grown under nitrogen-source limiting conditions were similar to that of the wild-type cells (Table 1). Furthermore, extending the cultivation time from 3 days to 5 days did not affect the results. Huisman et al. (1992) reported that both P. oleovorans GPo1 and its GPo500 phaZ mutant accumulated PHA to a similar level of 40%CDW after a 1-day cultivation in a nitrogen-source-limiting medium. Their data, however, further showed that while the accumulated PHA in GPo1 was degraded following an additional day of cultivation, that in the GPo500 strain remained essentially unchanged. Although the reason for the discrepancy between our results and those of Huisman et al. (1992) is not clear, one likely explanation is that the organisms or growth conditions used in their experiments somehow continued to mandate metabolic activities that required the expenditure of carbon or energy source. For example, P. oleovorans GPo1 under a duress condition might have a high carbon or energy demand for maintaining a cell survival mechanism similar to that described in the study of Ruiz et al.(2001). As a result, the wild-type GPo1 strain would have depleted the accumulated PHA via the action of its active PHA depolymerase to meet the carbon or energy needs; the phaZ-mutant GPo500 was incapable of doing so and thus retained its PHA content, leading to the observation of Huisman et al. (1992). The results of our study with P. resinovorans grown under an excess nitrogen-source condition support this contention (Table 1). Upon prolonged cultivation (5 days) under this condition, P. resinovorans wild-type and FOAC001 mutant apparently consumed the accumulated PHA reserves to support some undefined cellular activities, leading to a drastic decrease in their PHA contents (Table 1). The FOAC002 mutant strain apparently lacked the depolymerase activity to carry out similar PHA degradation. The nitrogen-source shift-up experiments lend further support for this notion. Data in Table 4 as well as the transmission electron micrographs in Fig. 2 clearly show that when nitrogen limitation was relieved, the wild-type and the FOAC001 mutant, but not the FOAC002 strain, depleted the previously accumulated PHA.
Lageveen et al. (1988) observed that the synthesis of mcl-PHA by P. oleovorans grown on hydrocarbons only occurred under a nitrogen-limiting growth condition. By contrast, our results (Table 1) showed that mcl-PHA was accumulated by P. resinovorans grown in medium containing oleic acid either under the limiting or excess nitrogen-source condition. Brandl et al. (1988) also showed that the PHA contents of P. oleovorans grown with octanoate as a carbon source were not significantly changed by NH4 + concentrations of 0–50 mM. It seems that the effect of nitrogen limitation on PHA accumulation might be dependent on the type of carbon source used. Thus, the accumulation of PHA from a fatty-acid substrate, such as oleic acid or octanoate, is not as sensitive to nitrogen-source modulation as PHA accumulation from hydrocarbons. The results of our repeat-unit composition analyses, however, did show that under the excess nitrogen-source condition, the mcl-PHAs of the three strains exhibited a slight but noticeable difference in their compositions. Compared to wild-type P. resinovorans grown under this condition on oleic-acid, FOAC001 and FOAC002 phaZ mutants accumulated mcl-PHAs with a slightly lower β-hydroxyoctanoate (C8:0) and higher β-hydroxytetradecenoate (C14:1) content (Table 2). Although the reason for this phenomenon is not clear, it seems to indicate that under high nitrogen-source condition, the preferred precursor pool for the synthesis of mcl-PHA might have shifted from the fatty-acid de novo biosynthesis pathway (which provides the C8:0 monomer) to the fatty-acid β-oxidation degradation pathway (which supplies the C14:1 repeat unit).
The effect of the insertion of a transposon into phaZ on the molecular weight of the PHA is most intriguing. High-molecular-weight mcl-PHAs were obtained from wild-type P. resinovorans grown either under a limited nitrogen-source condition for 5 days (Table 3) or on nitrogen-source shift-up (Table 5). Under either condition, FOAC001 and FOAC002 mutants did not accumulate high-molecular-weight polymer. Huisman et al. (1992) did not observe this phenomenon when comparing wild-type P. oleovorans GPo1 and the GPo500 phaZ mutant, possibly because they had isolated and characterized the polymers at a much earlier time-point in cell growth than the 5-day cultivation time used in the present study. To explain our observation that the phaZ mutants were incapable of accumulating the high-molecular-weight PHAs, it was tempting to speculate that the transposon insertion had somehow also inactivated the downstream phaC2 gene. Such inactivation should create a less active PHA polymerization environment and thus result in the synthesis of a lower molecular weight PHA. However, this explanation is refuted by a previously reported finding that a less active PHA synthesis environment in fact promotes accumulation of the higher-molecular weight polymers (Brandl et al. 1988). Thus, our results might in fact support the notion that PHA biosynthesis is a dynamic process which intricately involves both polymerization and depolymerization activities (Doi et al. 1990). This cycle may have been somehow disrupted in the transposon-insertion phaZ mutants, leading to the observations described herein.
Construction of the two phaZ mutants in this study has allowed a more in-depth examination of the roles of PHA depolymerase in PHA metabolism. More importantly, it has demonstrated that the chemical and physical properties of the polymers may be affected and thus manipulated by modulation of phaZ. Further study should allow not only a better understanding of the physiological functions of the mcl-PHA depolymerase, but also the development of a means to control and synthesize tailored polymers to suit specific applications.
References
Anderson AJ, Dawes EA (1990) Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev 54:450–472
Ashby RD, Foglia TA, Solaiman DKY, Liu C-K, Nunez A, Eggink G (2000) Viscoelastic properties of linseed oil-based medium chain length poly(hydroxyalkanoate) films: effects of epoxidation and curing. Int J Biol Macromol 27:355–361
Brandl H, Gross RA, Lenz RW, Fuller RC (1988) Pseudomonas oleovorans as a source of poly(β-hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl Environ Microbiol 54:1977–1982
Cromwick A-M, Foglia T, Lenz RW (1996) The microbial production of poly(hydroxyalkanoates) from tallow. Appl Microbiol Biotechnol 46:464–469
De Smet MJ, Eggink G, Witholt B, Kingma J, Wynberg H (1983) Characterization of intracellular inclusions formed by Pseudomonas oleovovans during growth on octane. J Bacteriol 154:870-878
Doi Y, Segawa A, Kawaguchi Y, Kunioka M (1990) Cyclic nature of poly(3-hydroxyalkanoate) metabolism in Alcaligenes eutrophus. FEMS Microbiol Lett 67:165–170
Foster LJR, Lenz RW, Fuller RC (1994) Quantitative determination of intracellular depolymerase activity in Pseudomonas oleovorans inclusions containing poly-3-hydroxyalkanoates with long alkyl substitutents. FEMS Microbiol Lett 118:279–282
Goryshin IY, Reznikoff WS (1998) Tn5 in vitro transposition. J Biol Chem 273:7367–7374
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580
Huisman GW, Wonink E, Meima R, Kazemier B, Terpstra P, Witholt B (1991) Metabolism of poly(3-hydroxyalkanoates) (PHAs) by Pseudomonas oleovorans. J Biol Chem 266:2191–2198
Huisman GW, Wonink E, de Koning G, Preusting H, Witholt B (1992) Synthesis of poly(3-hydroxyalkanoates) by mutant and recombinant Pseudomonas strains. Appl Microbiol Biotechnol 38:1–5
Jendrossek D, Handrick R (2002) Microbial degradation of polyhydroxyalkanoates. Annu. Rev. Microbiol. 56:403–432
Lageveen RG, Huisman GW, Preusting H, Ketelaar P, Eggink G, Witholt B (1988) Formation of polyesters by Pseudomonas oleovorans: Effect of substrates on formation and composition of poly-(R)-3-hydroxyalkanoates and poly-(R)-3-hydroxyalkenoates. Appl Environ Microbiol 54:2924–2932
Poirier Y, Nawrath C, Somerville C (1995) Production of polyhydroxyalkanoate, a family of biodegradable plastics and elastomers, in bacteria and plants. Bio/Technol 13:142–150
Rehm BHA, Steinbüchel A (1999) Biochemical and genetic analysis of PHA synthases and other proteins required for PHA synthesis. Int J Biol Macromol 25:3–19
Ruiz JA, Lopez NI, Fernandez RO, Mendez BS (2001) Polyhydroxyalkanoate degradation is associated with nucleotide accumulation and enhances stress resistance and survival of Pseudomonas oleovorans in natural water microcosms. Appl Environ Microbiol 67:225–230
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Solaiman DKY (1998) Genetic transformation of Pseudomonas oleovorans by electroporation. Biotechnol Techniques 12:829–832
Solaiman DKY (2002) Polymerase-chain-reaction-based detection of individual polyhydroxyalkanoate synthase phaC1 and phaC2 genes. Biotechnol Lett 24:245–250
Solaiman DKY, Ashby RD, Foglia TA (2000) Rapid and specific identification of medium-chain-length polyhydroxyalkanoate synthase gene by polymerase chain reaction. Appl Microbiol Biotechnol 53:690–694
Steinbüchel A (1991) Polyhydroxyalkanoic acids. In: Byrom D (ed) Biomaterials. Macmillan, New York, pp 123–213
Acknowledgements
The authors thank Dr. Peter Cooke of the Microscopic Imaging Group of Eastern Regional Research Center for acquiring the transmission electron micrographs, and Nicole Cross and Marshall Reed for technical assistance. Mention of brand or firm name does not constitute an endorsement by the U.S. Department of Agriculture over others of a similar nature not mentioned.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Solaiman, D.K.Y., Ashby, R.D. & Foglia, T.A. Effect of inactivation of poly(hydroxyalkanoates) depolymerase gene on the properties of poly(hydroxyalkanoates) in Pseudomonas resinovorans . Appl Microbiol Biotechnol 62, 536–543 (2003). https://doi.org/10.1007/s00253-003-1317-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-003-1317-4