Abstract
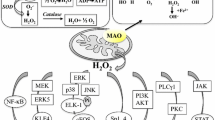
Type B monoamine oxidase (MAO-B) in glial cells has been considered to be associated with neuronal death in Parkinson’s disease. MAO-B inhibitors, rasagiline and selegiline [(−)deprenyl], protect neurons in animal and cellular models of neurodegeneration. However, the role of MAO-B itself in the regulation of cell death processing remains elusive, whereas type A MAO (MAO-A) mediates the induction of anti-apoptotic Bcl-2 genes by rasagiline and selegiline. In this paper, the involvement of MAOs in the induction of neuroprotective genes by MAO inhibitors was investigated in human glioblastoma U118MG cells expressing mainly MAO-B. Selegiline significantly increased Mao-B, which was suppressed by Mao-A knockdown with short interfering (si)RNA, whereas rasagiline less markedly increased Mao-B, which was not affected by Mao-A knockdown. Mao-A mRNA was also markedly increased by rasagiline and selegiline, and Mao-B knockdown significantly enhanced the induction by selegiline, but not by rasagiline. Mao-B knockdown also significantly increased mRNA levels of Bcl-2, brain-derived neurotrophic factor (BDNF) and glial cell line-derived neurotrophic factor (GDNF). Selegiline synergistically enhanced the expression of these genes in Mao-B knockdown cells, but Mao-A knockdown suppressed the increase. Rasagiline increased BDNF and GDNF, which Mao-B and Mao-A knockdown inhibited. These results show that MAO-B might function as a repressor and MAO-A as a mediator in the constitutional expression of pro-survival genes, and that MAO-B and MAO-A might regulate different signal pathways for rasagiline and selegiline to induce neuroprotective genes. The novel role of glial MAOs in the regulation of gene expression is discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Monoamine oxidase (MAO, amine: oxygen oxidoreductase, EC 1.4.3.4) catalyzes oxidative catabolism of monoamines and regulates the homeostasis of neurotransmitter monoamines in the brain. MAO is classified as type A and B (MAO-A, MAO-B), depending on the selective affinity to substrates and the sensitivity to distinct inhibitors (Shih et al. 1999). MAO-A and MAO-B are coded by different genes located on the X chromosome (Xp11.23) and are composed of different protein species, but they share many similar properties, such as 70% sequence identity and covalent FAD as the prosthetic group. MAO-A is expressed in catecholaminergic neurons and MAO-B in serotonergic and histaminergic neurons and glial cells, suggesting their different role in brain functions. MAO-B expression increases age dependently in the brain, suggesting the association of MAO-B with neuronal loss in age-related disorders, such as Parkinson’s disease by increased production of reactive oxygen species (ROS) (Jenner and Olanow 1996). However, age-dependent MAO-B increase was absent in the substantia nigra of control subjects (Saura et al. 1997), and MAO-B knockdown could not protect mice from ischemic injury, suggesting that MAO-B might be not directly involved in neuronal loss (Holschneider et al. 1999). More direct evidences are required to support the association of MAO-B itself with the pathogenesis of Parkinson’s disease (Damier et al. 1996).
Irreversible MAO-B inhibitors, selegiline [(−)deprenyl, (2R)-N-methyl-1-phenyl-N-pro-2-ynyl-propan-2-amine] and rasagiline [(1R)-N-prop-2-ynyl-2,3-dihydro-1H-amine] protect neurons from cell death in animal and cellular models of neurodegeneration, which is independent of MAO inhibition (Magyar 2011; Naoi et al. 2012, 2013a; Youdim and Bakhle 2006; Youdim et al. 2006). Rasagiline and selegiline directly suppress mitochondrial apoptotic signaling (Naoi et al. 2013b; Wu et al. 2015, 2016) and induce anti-apoptotic Bcl-2 and pro-survival neurotrophic factors and antioxidant enzymes (Maruyama et al. 2004; Weinreb et al. 2007; Maruyama and Naoi 2010). The role of MAO in neuroprotection by MAO-B inhibitors has been investigated using MAO-knockdown animals and cellular models treated with short interfering RNA (siRNA), microRNA (miRNA) and antisense oligonucleotides. In the cortex, striatum and corpus callosum of MAO-B-knockout mice, selegiline lost almost all the specific binding and the remaining 2–4% of 3H-selegiline was bound to MAO-A, suggesting that the selegiline still targets MAO in the brain (Ekblom et al. 1998). MAO-A expression was knocked down with siRNA against MAO-A (siMao-A), which inhibited rasagiline-induced increase in the mRNA and protein of Bcl-2 and MAO-A, and the enzymatic activity of MAO-A itself (Inaba-Hasegawa et al. 2012, 2013).
Glial cells, especially microglia and astrocytes, play a vital role in the disease progress of neurodegenerative disorders (Mosley et al. 2006). Microglia and astrocytes promote neuronal survival through removal of toxic molecules (α-synuclein, glutamate, potassium and calcium) in the extracellular space and control synaptogenesis and plasticity by release of trophic factors [nerve growth factor (NGF), neurotrophin-3 (NT-3) and basic fibroblast growth factor (bFGF)] and antioxidant glutathione (Rappold and Tieu 2010). However, it is not well documented whether MAO-B is associated with gene induction by the MAO-B inhibitors in glial cells.
This paper presents the results on the involvement of MAO-B in the constitutive expression of genes coding MAO, Bcl-2 and neurotrophic factors in human U118MG cell line composed of a mixture of neuroblastoma and astrocyte cells. Gene induction by rasagiline, selegiline and an MAO-A inhibitor, clorgyline [3-(2,4-dichlorophenoxy)-N-methyl-N-prop-2-ynyl-propan-1-amine], was also investigated in MAO-knockdown cells, by use of siRNA against Mao-B and Mao-A (siMao-B, siMao-A), respectively. The role of MAO observed in glial U118MG cells was discussed by comparison with those in neuronal SH-SY5Y cells. Regulation of neuroprotective genes by MAOs and the inhibitors is discussed with respect to the role of glial cells in neuroprotection for Parkinson’s disease and other neurodegenerative disorders.
Materials and methods
Materials
Professor J. Knoll (Semmellweis University, Budapest, Hungary), TEVA (Netanya, Israel) and May-Baker (Dagenham, UK) kindly donated selegiline, rasagiline and clorgyline, respectively. Kynuramine was purchased from Sigma (St. Louis, MO, USA), and Dulbecco’s modified Eagle’s medium (DMEM) and other chemicals were from Wako (Osaka, Japan).
Culture of U118MG cells and wild and MAO-B-overexpressed SH-SY5Y cells
U118MG cell line was purchased from American Type Culture Collection (Manassas, VA, USA) and cultured in DMEM containing 10% fetal calf serum (PBS, ATCC 30-2020), 4 mM l-glutamine, 100 IU/ml penicillin and 100 µg/ml streptomycin. SH-SY5Y cells were cultured in Cosmedium-001 tissue culture medium (CosmoBio, Tokyo, Japan) supplemented by 5% fetal calf serum in 95% air–5% CO2. Stable clone overexpressing MAO-B was established in SH-SY5Y cells (MAOB-SH cells) by DNA transfection of MAO-B gene, as reported (Yi et al. 2006).
MAO-B and MAO-A knockdown with siRNA
Expression of MAO-B and MAO-A was knocked down in U118MG and SH-SY5Y cells by the use of siRNA targeting Mao-B and Mao-A mRNA (sc-35849, sc-35847, Santa Cruz Biotechnology, Dallas, Texas, USA), as reported previously (Inaba-Hasegawa et al. 2012, 2013). The cells (1.5 × 105 cells/well) were cultured in the six-well culture flask for 24 h and treated with siRNA for 48 h for the following experiments.
Quantitation of mRNA of MAO-A, MAO-B, Bcl-2 and neurotrophic factors
Induction of genes was quantitatively determined by the measurement of mRNA using the real-time reverse transcription-polymerase chain reaction (RT-PCR) method. U118MG and SH-SY5Y cells (3 × 105 cells/well) pretreated without or with siRNA against MAO were cultured in the six-well culture flask and then treated with MAO inhibitors (10−6–10−10 M) for 24 h. The mRNA levels of MAOs, Bcl-2 and neurotrophic factors were quantitatively determined by the real-time RT-PCR method. The cells were gathered, washed with phosphate-buffered saline and the total RNA was extracted by use of Trizol plus RNA purification kit according to the manufacture’s protocol (Invitrogen, San Diego, CA, USA). The cDNA was generated by reverse transcription of the total RNA (100 ng) using PrimeScript RT reagent kit (TaKaRa Bio, Ohtsu, Japan), and the cDNA fragments were amplified using the PCR primers and SYBR Premix EX Taq™ II (TaKaRa Bio). The used primers were as follows: Bcl-2, HA032558; Mao-A, HA071085; Mao-B, HA032947; BDNF, HA10999); NGF, HA115875; NT-3v1 and NT-3v2, HA1500423 and HA15558040; GDNF, HA003973; and β-action, HA067803-F/R.
The relative concentrations of PCR products were determined from the standard curve prepared by real-time PCR of serial dilutions of template cDNAs. The relative mRNA levels were quantitatively determined from the cycle threshold (C t) of the PCR reaction analyzed by Real time system software (TaKaRa Bio) and the standard curve. The mRNA level was normalized by comparison of β-actin levels in each sample and expressed as the percent of those in control cells treated without siRNA (non-treated cells).
Quantitative measurement of MAO activity in the mitochondrial fraction
MAO activity in the mitochondrial fraction was determined with kynuramine as a substrate by a microassay method modified from the method of Kraml (1965). The mitochondrial fraction (0.1–25 µg protein) was reacted with 1 mM kynuramine in 10 mM sodium phosphate buffer, pH 7.4 (the total volume, 100 µl) at 37 °C for 1 h, then 50 µl of 10% ZnSO4, 10 µl of 1 M NaOH and 40 µl of distilled water were added. After heating at 100 °C for 2 min, the sample was centrifuged at 1000g for 10 min and the supernatant (50 µl) was mixed with 50 µl NaOH in a 96-well microplate and the fluorescence at 450 nm was measured with excitation at 340 nm, using a Corona MTP-600F microplate fluorometer (Corona Electrics, Hitachinaka, Japan). MAO activity was expressed as pmoles of produced 4-hydroxyquinone per minute per microgram protein. MAO-A and MAO-B activity were differentiated by 20 min pre-treatment with 10 µM selegiline or clorgyline, respectively.
Quantitation of Bcl-2 protein
Bcl-2 protein was quantitatively determined by Western blot analysis, as reported (Inaba-Hasegawa et al. 2012). The cells (3 × 105 cells/well) were cultured for 24 h in a six-well poly-l-lysine-coated culture flask (Iwaki, Asahi Glass, Tokyo, Japan). The cells were treated with 10−5–10−11 M MAO inhibitors for 24 h, gathered, washed with phosphate-buffered saline (PBS) and suspended in RIPA buffer [10 mM Tris–HCl buffer, pH 7.5, containing 1% NP-40, 0.1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 150 mM NaCl and 1 mM EDTA 2 Na] and a protease inhibitor cocktail (Roche, Mannheim, Germany). The lyzed protein was separated by SDS-PAGE using 10–20% gradient polyacrylamide gel (Bio-Rad Lab., Hercules, VA, USA) and electroblotted onto a PVDF membrane (Amersham Hybond-P, GE Healthcare, Buchinghamshire, UK). After blockage with 5% nonfat milk in 10 mM Tris–HCl buffer, pH 7.5, containing 0.1% Tween 20 and 150 mM NaCl, the membrane was incubated overnight at 4 °C with anti-human Bcl-2 antibody (500× dilution, Santa Cruz Biotechnology, Santa Cruz, CA, USA) or anti-β-actin antibody for control (5000× dilution, Sigma). The membrane was incubated further with horseradish peroxidase (HRP)-linked anti-mouse IgG at room temperature. The immunoblots were visualized by use of Amersham ECL plus western blotting detection reagents (GE Healthcare) or Immunostar LD (Wako). The amounts of Bcl-2 protein were quantitatively determined using a Fujifilm LAS-4000 luminescent image analyzer (Tokyo, Japan). The protein amounts were quantified by densitometry, normalized against β-actin and expressed as the percent of those in non-treated cells.
Statistics
Experiments were repeated at least three to four times in triplicate or quadruplicate measurements, and the results were expressed as the mean and SD. Differences were statistically evaluated by analysis of variance (ANOVA) followed by Scheffe’s F test. A p value less than 0.05 was considered to be statistically significant.
Results
MAO-B in U118MG cells and effects of siRNA treatment on MAO levels
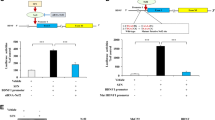
MAO protein in U118MG cells was detected as a single band with molecular mass of 520 kDa corresponding to MAO-B, but the amount was quite lower than MAO-A protein in wild SH-SY5Y cells (data not shown). MAO-B activity was the major part of the MAO activity in the mitochondrial fraction of U118MG cell: 15.5 ± 0.3 versus 1.5 ± 0.4 pol/min/mg mitochondrial protein, for MAO-B and MAO-A, respectively (Fig. 1a). In wild SH-SY5Y cells, MAO-A activity was 97.5% of the total MAO activity: 913 ± 57 out of 937 ± 44 pol/min/mg protein. In MAOB-SH cells MAO-B activity increased to 12 450 ± 1 020 pmol/min/mg protein from 31.3 ± 14.0 pmol/min/mg protein in wild SH-SY5Y cells, but MAO-A activity remained unchanged, 1 113 ± 43 pmol/min/mg protein.
The enzymatic activity of MAO-B and MAO-A in U118MG cells and wild and (Wild SH) and MAO-B overexpressed SH-SY5Y (MAOB-SH) cells. a MAO activity was expressed as pmol/min/mg of mitochondrial protein. b Mao mRNA levels in U118MG cells treated with siMao-B at 20, 40 and 60 nM or siMao-A at 20 nM. The mRNA levels were expressed as percent of the values in cells treated without siRNA (non-treated cells). c Mao mRNA levels in wild SH cells treated with 20 nM siMao-A and siMao-B. d Mao mRNA levels in MAOB-SH cells treated with 20 and 40 nM siMao-A and siMao-B. Column and bar represent the mean and SD. Asterisk and double asterisk statistically different from non-treated cells, p < 0.05 and 0.01, respectively
In U118MG cells, Mao-B mRNA level was deceased by treatment with 20–60 nM siMao-B, whereas siMao-B at 40 and 60 nM did not affect Mao-A expression and increased at 20 nM, 136 ± 18% of that in non-treated cells (Fig. 1b). Vice versa, siMao-A treatment downregulated Mao-A mRNA to 8% of non-treated cells, but did not affect Mao-B level. In SH-SY5Y cells, siMao-A (20 nM) suppressed only Mao-A expression, but siMao-B neither affected Mao-A nor Mao-B levels (Fig. 1c). In MAOB-SH cells, siMao-B significantly suppressed Mao-B expression, whereas siMao-B did not affect Mao-A levels (Fig. 1d). These results indicate that siRNA against Mao-B and Mao-A selectively decreased targeted Mao-B and Mao-A levels in these cells.
Selegiline and rasagiline increased Mao-B and Mao-A mRNA, and siMao-B synergistically increased selegiline-enhanced Mao-A expression
In SH-SY5Y cells, rasagiline and selegiline induced MAO-A mRNA, protein and enzyme activity, and the induction by rasagiline was mediated by MAO-A itself (Inaba-Hasegawa et al. 2013). The effects of rasagiline and selegiline on Mao-B levels were investigated in U118MG cells treated with 20 µM siMao-B, siMao-A, or non-specific siRNA (siNS) (Fig. 2). The effects of rasagiline and selegiline on Mao-A and Mao-B were not affected by siNS treatment, which was also the case with Bcl-2 and neurotrophic factors. Rasagiline at 10−7–10−10 M increased Mao-B mRNA in siMao-A- and siNS-treated cells virtually by the same extent, except at 10−9 M of siMao-A. Selegiline upregulated Mao-B mRNA in siNS-treated cells, which was suppressed by siMao-A, suggesting that MAO-A might mediate Mao-B mRNA increase by selegiline, but not by rasagiline in U118MG cells (Fig. 2a).
Effects of rasagiline, selegiline and clorgyline on Mao-B and Mao-A mRNA in siMao-B-, siMao-A- and siNS-treated U118MG cells. Cells were treated with 20 nM siMao-B, siMao-A or siNS and then with 10−6–10−10 M MAO inhibitors. a, b Mao-B and Mao-A mRNA levels in cells treated with siRNA and then rasagiline and selegiline. c Mao-A and Mao-B mRNA levels in cells treated with siMao-B and then with clorgyline. The mRNA levels were expressed as percent of the values in non-treated cells. Column and bar represent the mean and SD. Asterisk and double asterisk statistically different from the corresponding siRNA-treated cells without MAO inhibitor [(−)], p < 0.05 and 0.01, respectively
Effects of rasagiline and selegiline on Mao-A mRNA levels were also examined. In siNS-treated cells, rasagiline at 10−6–10−10 M and selegiline at 10−7 and 10−8 M increased Mao-A mRNA levels (Fig. 2b). In Mao-B-knockdown cells, selegiline at 10−6–10−10 M increased Mao-A mRNA synergistically, but rasagiline did not affect the levels, suggesting that MAO-B might suppress selegiline-dependent Mao-A induction (Fig. 2b).
The effects of clorgyline, an MAO-A inhibitor, on Mao-A and Mao-B expression were investigated. Clorgyline at 10−8–10−10 and 10−6–10−8 M increased Mao-A and Mao-B mRNA levels in siNS-treated cells (Fig. 2c). In Mao-B-knockdown cells, clorgyline did not change Mao-A levels, but increased Mao-B levels, which were significantly downregulated by siMao-B.
Rasagiline and selegiline increased anti-apoptotic Bcl-2 in U118MG cells
Rasagiline and selegiline increased Bcl-2 mRNA and protein in SH-SY5Y cells by mediation of MAO-A (Inaba-Hasegawa et al. 2012). In U118MG cells, the effects of rasagiline and selegiline on Bcl-2 protein levels were examined. Rasagiline at 10−6–10−10 M and selegiline at 10−5–10−7 and 10−10 M increased Bcl-2 protein (Fig. 3a, b). Treatment with 20 and 40 nM siMao-B increased Bcl-2 mRNA levels significantly, suggesting the suppression of the constitutional Bcl-2 levels by MAO-B (Fig. 3c). In cells treated with 20 nM siMao-B, the Bcl-2 mRNA levels were increased to 147 ± 13% of those in non-treated cells, whereas siMao-A treatment reduced those to 60 ± 10%.
Effects of rasagiline, selegiline, and siMao-B- and siMao-A-treatment on Bcl-2 protein and mRNA levels in U118MG cells. Cells were treated with rasagiline and selegiline at 10−5–10−11 M and subjected to quantitative analysis for Bcl-2. a, b Western blot analysis and quantitative results for Bcl-2 protein levels. The value was expressed as the percent of that in non-treated cells. c Bcl-2 mRNA in U118MG cells treated with 20, 40 and 60 nM siMao-B, 20 nM siMao-A or 20 nM siNS. Bcl-2 mRNA level was represented as percent of the value in non-treated control cells. d Bcl-2 mRNA levels in cells pretreated with 20 nM siMao-B, siMao-A or siNS, and then with 10−6–10−10 M rasagiline or selegiline. The value was expressed as percent of that in non-treated cells. Column and bar represent the mean and SD. Asterisk and double asterisk significantly different from corresponding siRNA-treated cells without MAO inhibitor, p < 0.01 and 0.05, respectively
In cells pretreated with 20 nM siNS, rasagiline and selegiline at 10−7–10−10 and 10−6–10−10 M markedly increased Bcl-2 mRNA. In siMao-B-treated cells, selegiline further increased Bcl-2 mRNA levels, but rasagiline did not. In siMao-A-treated cells rasagiline- and selegiline-dependent Bcl-2 expression was suppressed, except at 10−7 and 10−10 M of selegiline (Fig. 3d). These results indicate again that MAO-B negatively regulated Bcl-2 expression by selegiline, and MAO-A positively that by selegiline and rasagiline in U118MG cells.
Mao-B knockdown increased the mRNA levels of neurotrophic factors
In U118MG cells, treatment with 20 and 40 nM siMao-B significantly increased the mRNA levels of BDNF, NGF, NT-3v1, NT-3v2 and GDNF, whereas siNS at 20 nM was not affected, except for the downregulation of GDNF expression (Fig. 4a). The treatment with 60 nM siMao-B increased only NT-3 levels and did not affect BDNF, NGF and GDNF mRNA levels. On the other hand, the siMao-A treatment downregulated BDNF, NGF and GDNF mRNA levels (Fig. 4b). The constitutional expression of pro-survival neurotrophic factors was regulated by MAO-B and MAO-A in an opposite way.
Effects of Mao-B and Mao-A knockdown on mRNA levels of neurotrophic factors. a U118MG cells treated with 20, 40 and 60 nM siMao-B or 20 nM siNS, and the mRNA levels of neurotrophic factors were quantified by real-time RT-PCR method. The value was presented as percent of the each neurotrophic factor levels in non-treated cells. b U118MG cells treated with 20 nM siMao-A or siNS, and then BDNF, NGF and GDNF mRNA levels were quantified. Asterisk and double asterisk significantly different from non-treated cells, p < 0.05 and 0.01
Mao-B knockdown and selegiline synergistically enhanced BDNF and GDNF expression
Rasagiline and selegiline induced BDNF and GDNF protein and mRNA in SH-SY5Y cells (Maruyama and Naoi 2010). In U118MG cells, the effects of these MAO-B inhibitors on the expression of neurotrophic factors were investigated. In siNS-treated cells, rasagiline at 10−8–10−10 M and selegiline at 10−6–10−10 M increased BDNF mRNA significantly (Fig. 5a). As described above, siMao-B increased the basal expression of neurotrophic factors, whereas siMao-A inhibited that of BDNF, NGF and GDNF. In siMao-B-treatment cells, selegiline synergistically enhanced BDNF levels, but not in siMao-A-treated cells. Rasagiline did not increase BDNF levels in siMao-B- and siMao-A-treated cells, except at 10−7 M of siMao-A.
Effects of rasagiline and selegiline on BDNF and GDNF mRNA in siMao-B- or siMao-A-treated U118MG cells. Cells were treated with 20 nM siMao-B, siMao-A or siNS, and then 10−6–10−10 M rasagiline or selegiline. The mRNA levels were quantified, and the value was expressed as percent of that in non-treated cells. Asterisk and double asterisk significantly different from corresponding siRNA-treated cells without MAO inhibitor, p < 0.05 and 0.01, respectively
GDNF levels in siNS-treated ells were increased by rasagiline at 10−6–10−10 M, which was inhibited by siMao-A and siMao-B treatment. Selegiline also enhanced GDNF levels at 10−6–10−9 M in siNS-treated cells and also significantly increased them at 10−6–10−10 M in Mao-B-knockdown cells, but did not increase GDNF in siMao-A-treated cells (Fig. 5b).
Clorgyline increased BDNF and GDNF in Mao-B-knockdown U118MG cells
The effects of clorgyline on the expression of neurotrophic factors were investigated in U118MG cells treated with siMao-B and siNS (Fig. 6). In siNS-treated cells, clorgyline did not affect BDNF and GDNF mRNA levels, except BDNF increase at 10−8 M. In siMao-B-treated cells, clorgyline further upregulated BDNF and GDNF levels, suggesting the association of MAO-B with the induction (Fig. 6a). NGF and 3-NT levels were not affected by clorgyline in siNS-treated cells. siMao-B treatment increased the basal levels of NGF and 3-NT, but clorgyline did not further affect them (Fig. 6b).
Effects of clorgyline on mRNA of neurotrophic factors in siMao-B-treated U118MG cells. Cells were treated with 20 nM siMao-B or siNS and then with 10−6–10−10 M clorgyline. The results were expressed as percent of that in non-treated cells. Asterisk and double asterisk statistically different from the corresponding siRNA-treated cells without clorgyline, p < 0.05 and 0.01, respectively
In SH-SY5Y cells, MAO-A controversially regulated BDNF induction by rasagiline and selegiline
The association of MAO-A with BDNF induction by rasagiline and selegiline was examined in siMao-A-treated SH-SY5Y cells. Rasagiline at 10−6–10−8 and 10−10 M increased BDNF levels in siNS-treated cells, and siMao-A treatment inhibited the induction, whereas rasagiline increased NGF only at 10−6 M (Fig. 7a). Selegiline at 10−6–10−10 M increased BDNF in siNS-treated cells, and Mao-A-knockdown further enhanced BDNF, but did not increase NGF in siNS- and siMao-A-treated cells, except at 10−8 M (Fig. 7b). On the other hand, clorgyline did not affect BDNF and NGF mRNA levels in siMao-A- and siNS-treated SH-SY5Y cells, except BDNF increase at 10−7 M in siMao-A-treated cells (Fig. 7c). MAO-B overexpression in SH-SY5Y cells decreased the induction of Bcl-2, BDNF and GDNF gene by rasagiline and selegiline (data not shown).
Effects of Mao-A knockdown on BDNF and NGF induction by rasagiline, selegiline and clorgyline in wild SH-SY5Y cells. a–c Cells were treated with 20 nM siMao-A or siNS, and then with 10−5–10−10 M rasagiline, selegiline and clorgyline. The mRNA was expressed as percent of that in non-treated cells. Asterisk and double asterisk statistically different from the corresponding siRNA-treated cells without MAO inhibitor, p < 0.05 and 0.01, respectively
Discussion
In this paper, the role of MAO-B and MAO-A in the induction of anti-apoptotic and pro-survival genes in glial U118MG cells was confirmed, as summarized in Fig. 8. In the mouse model of Parkinson’s disease, where MAO-B expression was induced in astroglia at adulthood, MAO-B was directly involved in neuronal loss (Mallajosyula et al. 2008). Neurotoxicity of MAO-B against dopamine neurons is due not only to oxidative stress, but also to dopaminochrome produced from dopamine with hydrogen peroxide, a reaction product by MAO, resulting in impaired mitochondrial function (Siddiqui et al. 2010). On the other hand, selegiline, desmethylselegiline and dopamine agonists induce pro-survival BDNF and GDNF in cultured mouse astrocytes (Mizuta et al. 2000; Ohta et al. 2010). These results present bilateral function of glial MAO-B in neurodegeneration.
MAO-B and MAO-A contradictorily regulate gene induction by selegiline and rasagiline in U118MG and SH-SY5Y cells. Selegiline and rasagiline increased Mao-B, Mao-A and Bcl-2 expression. siMao-A suppressed mRNA induction of MAO-B by selegiline, and siMao-B enhanced that of MAO-A. siMao-A treatment inhibited rasagiline-dependent Mao-A induction, but not Mao-B, whereas siMao-b did not affect Mao-A and Mao-B induction. siMao-B treatment enhanced and siMao-A suppressed the mRNA levels of Bcl-2 and neurotrophic factors. Selegiline synergistically enhanced BDNF and GDNF in Mao-B-knockdown cells, and siMao-A treatment inhibited BDNF and GDNF induction. Rasagiline also increased BDNF and GDNF in U118MG cells, which was inhibited by siMao-A treatment. In SH-SY5Y cells, rasagiline induced Bcl-2 and BDNF, which was suppressed by siMao-A treatment. Selegiline increased BDNF, which siMao-A treatment synergistically increased
MAO-A regulates the homeostasis of serotonin, norepinephrine and dopamine in the brain, and its activity is closely associated with emotional, cognitive, perceptive and behavioral functions (Shih et al. 1999; Bortolato et al. 2008; Naoi et al. 2016, 2017). MAO-A mRNA and activity were increased in apoptosis induced in SH-SY5Y and PC12 cells, suggesting the involvement of MAO-A in neuronal death (De Zutter and Davis 2001). An endogenous dopaminergic neurotoxin, N-methyl(R)salsolinol, binds to MAO-A and induces apoptosis in SH-SY5Y cells (Yi et al. 2006). MAO-A knockdown with microRNA protected SH-SY5Y cells from cell death by mitochondrial toxins (Fitzgerald et al. 2014). However, protection by clorgyline has been reported only in non-neuronal cells: human melanoma M14 cells (Malorni et al. 1998) and mouse N2a neuroblastoma cells (De Girolamo et al. 2001). Clorgyline prevented tyramine-induced mitochondrial permeability transition and calcium release in rat liver mitochondria (Marcocci et al. 2002). Clorgyline increased Bcl-2 in irradiated HaCaT cells, a human keratinocyte cell line, but the protective effects were limited (Seymour et al. 2003). In this paper, clorgyline induced BDNF and GDNF in U118MG cells, suggesting that MAO-A inhibitors may show neuroprotective activity through induction of neuroprotective genes in glial cells.
In in vivo and in vitro experiments, selegiline and rasagiline increase Bcl-2 and neurotrophic factors. In non-human primate, rasagiline increased GDNF levels in the cerebrospinal fluid (CSF) more markedly than selegiline, and vice versa selegiline increased BDNF levels in the CSF from parkinsonian patients (Maruyama and Naoi 2010). These results are comparable to our results on the preferential induction of GDNF and BDNF by rasagiline and selegiline in SH-SY5Y cells. In clinical studies, selegiline shows the beneficial effects in depression, and rasagiline in Parkinson’s disease and other neurodegenerative disorders. The difference is considered to ascribe to their metabolites, namely cytotoxic methamphetamine from selegiline, and protective aminoindan and N-propargylamine from rasagiline (Bar Am et al. 2004). Our results in U118MG cells present that MAO-B functioned as a repressor for the constitutive expression of neuroprotective genes, whereas MAO-A as an enhancer, and that selegiline and rasagiline induced BDNF or GDNF by different signal pathways, either depending on MAO-A or MAO-B.
Molecular mechanism underlying how MAOs are associated with gene induction by MAO inhibitors remains elusive. Increase of monoamines by MAO inhibitors induced BDNF and GDNF in glial cells (Juric et al. 2006). Dopamine upregulated BDNF protein levels in cultured rat astrocytes (Miklic et al. 2004), serotonin induced GDNF mRNA in rat C6 glioma cells via fibroblast growth factor receptor 2 (FGFR2) (Tsuchioka et al. 2008) and norepinephrine stimulated BDNF synthesis in cultured rat cortical astrocytes through α1- and β1/β2-adrenergic receptors (Juric et al. 2008). In U118MG cells, these monoamines were not detected in the cell lysate and culture medium, as observed in the increased neurotrophic factors of rat astrocytes by antidepressants (Kajitani et al. 2012). These results suggest that selegiline and rasagiline might increase GDNF and BDNF expression through signal pathway not dependent on monoamines.
Rasagiline increased the mRNA, protein and enzymatic activity of MAO-A in SH-SY5Y cells though suppression of R1 (RAM2/CDCA7L/JPO2) (Inaba-Hasegawa et al. 2013). R1 represses MAO-A promoter activity by competing the zinc finger transcription factor specificity protein 1 (Sp1) for the binding to Sp1 sites (Chen et al. 2005; Shih et al. 2011). Expression of MAO-A and MAO-B is activated by a transcription factor, Kruppel-like factor 11 [KLF11, also called as transforming growth factor-β-inducible early gene (TIEG2)], which forms a transcriptional complex with Sp1 and enhances MAO transcription (Ou et al. 2004). The KLF11–Sp1 pathway was involved in MAO-B induction by ethanol in U118MG and SH-SY5Y cells, and the induction was inhibited by selegiline and rasagiline (Ou et al. 2010). These results suggest that this KLF11–Sp1 pathway might be associated with regulation of gene expression by MAO-B and MAO-A, and also the gene induction by MAO-B inhibitors. This issue should be further investigated.
Future study on the more detailed molecular mechanism of gene induction by MAO-B and MAO-A inhibitors may promote the development of novel anti-apoptotic and neuroprotective drugs applicable for the treatment of neurodegenerative disorders and depression.
Abbreviations
- MAO-A and MAO-B:
-
Type A and B monoamine oxidase
- MAOB-SH cells:
-
MAO-B overexpressed SH-SY5Y cells
- siRNA:
-
Short interfering RNA
- siMao-A and siMao-B :
-
siRNA against Mao-A and Mao-B
- siNS :
-
Non-specific siRNA
References
Bar Am O, Amit T, Youdim MBH (2004) Contrasting neuroprotective and neurotoxic actions of respective metabolites of anti-Parkinson drugs rasagiline and selegiline. Neurosci Lett 355(3):169–172
Bortolato M, Chen K, Shih JC (2008) Monoamine inactivation: from pathophysiology to therapeutics. Adv Drug Deliv Rev 60(13–14):1527–1533
Chen Z, Trapp B (2016) Microglia and neuroprotection. J Neurochem 136(Suppl 1):10–17
Chen K, Ou XM, Chen G, Choi SH, Shih JC (2005) R1, a novel receptor of the human monoamine oxidase A. J Biol Chem 280(12):11552–11559
Damier P, Kastner A, Agid Y, Hirsch EC (1996) Does monoamine oxidase type B play a role in dopaminergic nerve cell death in Parkinson’s disease? Neurology 46(5):1262–1269
De Girolamo LA, Hargreaves AJ, Billett EE (2001) Protection from MPTP-induced neurotoxicity in differentiating mouse N2a neuroblastoma cells. J Neurochem 78(3):650–660
De Zutter GS, Davis RJ (2001) Pro-apoptotic gene expression mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Proc Natl Acad Sci USA 98(11):6168–6173
Ekblom J, Oreland L, Chen K, Shih JC (1998) Is there a “non-MAO” macromolecular target for l-deprenyl?: studies on MAOB mutant mice. Life Sci 63(2):181–186
Fitzgerald JC, Ugun-Klusek A, Allen G, De Cirolamo LA, Hargreaves I, Ufer C, Abramov AY, Billett EE (2014) Monoamine oxidase-A knockdown in human neuroblastoma cells reveals protection against mitochondrial toxins. FASEB J 28(1):218–229
Holschneider DP, Scremin OU, Huynh L, Chen K, Shih JC (1999) Lack of protection from ischemic injury of monoamine oxidase B-deficient mice following middle cerebral artery occlusion. Neurosci Lett 259(3):161–164
Inaba-Hasegawa K, Akao Y, Maruyama W, Naoi M (2012) Type A monoamine oxidase is associated with induction of neuroprotective Bcl-2 by rasagiline, an inhibitor of type B monoamine oxidase. J Neural Transm 119(4):405–414
Inaba-Hasegawa K, Akao Y, Maruyama W, Naoi M (2013) Rasagiline and selegiline, inhibitors of type B monoamine oxidase, induce type A monoamine oxidase in human SH-SY5Y cells. J Neural Transm 120(3):435–444
Jenner P, Olanow CW (1996) Oxidative stress and the pathogenesis of Parkinson’s disease. Neurology 47(Suppl 3):S161–S170
Juric DM, Miklic S, Carman-Krzan M (2006) Monoaminergic neuronal activity up-regulates BDNF synthesis in cultured neonatal rat astrocytes. Brain Res 1108:54–62
Juric DM, Loncar D, Carman-Lrzan M (2008) Noradrenergic stimulation of BDNF synthesis in astrocytes: mediation via α1- and β1/β2-adrenergic receptors. Neurochem Int 52(1–2):297–306
Kajitani N, Hisaoka-Nakanishi K, Morioka N, Okada-Tsuchiuoka M, Kaneko M, Kasai M, Shibasaki C, Nakata Y, Takebayashi M (2012) Antidepressant acts on astrocytes leading to an increase in the expression of neurotrophic/growth factors: different regulation of FGF-2 by noradrenalin. PLoS One 7(12):e51197
Kraml M (1965) A rapid microfluorometric determination of monoamine oxidase. Biochem Pharmacol 14:1684–1686
Magyar K (2011) Pharmacology of selegiline. Int Rev Neurobiol 100:65–84
Mallajosyula JK, Kauer AD, Chinta SJ, Rajagopalan S, Rane A, Nicholis DG, Di Monte DA, Macarthour H, Andersen JA (2008) MAO-B elevation in mouse brain astrocytes results in Parkinson’s pathology. PLoS One 3:e1616
Malorni W, Giammarioli AM, Matarrese P, Pieryrangeli P, Agostinelli E, Cianccio A, Grassilli E, Mondovi B (1998) Protection against apoptosis by monoamine oxidase A inhibitors. FEBS Lett 426(1):155–159
Marcocci L, de Marchi U, Salvi M, Milella ZG, Nocera S, Agostinelli E, Mondovi B, Toninello A (2002) Tyramine and monoamine oxidase inhibitors as modulators of the mitochondrial membrane permeability transition. J Membr Biol 188(1):12–31
Maruyama W, Naoi M (2010) “70th birthday Professor Riederer” induction of glial cell-line-derived and brain-derived neurotrophic factors by rasagiline and (−)deprenyl: a way to a disease-modifying therapy? J Neural Transm 120(1):83–89
Maruyama W, Nitta A, Shamoto-Nagai M, Hirata Y, Akao Y, Youdim M, Furukawa S, Nabeshima T, Naoi M (2004) N-Propargyl-1-(R)-aminoindan, rasagiline, increases glial cell line-derived neurotrophic factor (GDNF) in neuroblastoma SH-SY5Y cells through activation of NF-κB transcription factor. Neurochem Int 44(6):293–400
Miklic S, Juric DM, Caman-Krzan M (2004) Differences in the regulation of BDNF and NGF synthesis in cultured neonatal rat astrocytes. Int J Dev Neurosci 22(3):119–130
Mizuta I, Ohta M, Ohta K, Nishimura M, Mizuta E, Hayashi K, Kuno S (2000) Selegiline and desmethylselegiline stimulate NGF, BDNF and GDNF synthesis in cultured mouse astrocytes. Biochem Biophys Res Commun 279(3):751–755
Mosley RL, Benner EJ, Kadiu I, Thomas M, Boska MD, Hasan K, Laurie C, Gendelman HE (2006) Neuroinflammation, oxidative stress and the pathogenesis of Parkinson’s disease. Clin Neurosci Res 6(5):261–281
Naoi M, Maruyama W, Inaba-Hasegawa K (2012) Type A and B monoamine oxidase in age-related neurodegenerative disorders. Their distinct roles in neuronal death and survival. Curr Top Med Chem 12(20):2177–2188
Naoi M, Maruyama W, Inaba-Hasegawa K (2013a) Revelation in neuroprotective functions of rasagiline and selegiline: the induction of distinct genes by different mechanisms. Expert Rev Neurother 13(6):671–684
Naoi M, Maruyama W, Yi H (2013b) Rasagiline prevents apoptosis induced by PK11195, a ligand of the outer membrane translocator protein (18 kDa), in SH-SH5Y cells through suppression of cytochrome c release from mitochondria. J Neural Transm 120(11):1539–1551
Naoi M, Riederer P, Maruyama W (2016) Modulation of monoamine oxidase (MAO) expression in neuropsychiatric disorders: genetic and environmental factors involved in type A MAO expression. J Neural Transm 123(2):91–106
Naoi M, Maruyama W, Shamoto-Nagai M (2017) Type A monoamine oxidase and serotonin are coordinately involved in depressive disorders: from neurotransmitter imbalance to impaired neurogenesis. J Neural Transm. doi:10.1007/s00702-017-1709-8
Ohta K, Kuno S, Inoue S, Ikeda E, Fujinami A, Ohta M (2010) The effect of dopamine agonists; the expression of GDNF, NGF, and BDNF in cultured mouse astrocytes. J Neurol Sci 291(1–2):12–26
Ou X-M, Chen K, Shih JC (2004) Dual functions of transcription factors, transforming growth factor-beta-inducible early gene (TIEG)2 and Sp3, are mediated by CACCC element and Sp1 sites of human monoamine oxidase (MAO) B gene. J Biol Chem 279(20):21021–21028
Ou X-M, Stockmeier CA, Meltzer HY et al (2010) A novel role for GADPDH-MAO B cascade in ethanol-induced cellular damage. Biol Psychiatry 67(9):855–863
Rappold PM, Tieu K (2010) Astrocytes and therapeutics for Parkinson’s disease. Neurotherapeutics 7(4):413–423
Saura J, Andres N, Andrade C, Ojuel J, Eriksson K, Mahy N (1997) Biphasic and region-specific MAO-B response to aging in normal human brain. Neurobiol Aging 18(5):497–507
Seymour CB, Mothersill C, Mooney R, Moriarty M, Tipton KF (2003) Monoamine oxidase inhibitors l-deprenyl and clorgyline protect nonmalignant human cells from ionising radiation and chemotherapy toxicity. Br J Cancer 89(10):20
Shih JC, Chen K, Ridd MJ (1999) Monoamine oxidase: from genes to behavior. Ann Rev Neurosci 22:197–217
Shih JC, Wu JB, Chen K (2011) Transcriptional regulation and multiple functions of MAO genes. J Neural Transm 118(7):979–986
Siddiqui A, Mallajosyula JK, Rane A, Anderson JK (2010) Ability to delay neuropathological events associated with astrocytic MAO-B increase in a Parkinsonian mouse model: implications for early intervention on disease progression. Neurobiol Dis 40(2):444–448
Tsuchioka M, Takebayashi M, Hisaoka K, Maeda N, Nakata Y (2008) Serotonin (5-HT) induces glial cell line-derived neurotrophic factor (GDNF) mRNA expression via the transactivation of fibroblast growth factor 2 (FGR2) in rat C6 glioma cells. J Neurochem 106(1):244–257
Weinreb O, Amit T, Bar-Am O, Youdim MB (2007) Induction of neurotrophic factors GDNF and BDNF associated with the mechanism of neurorescue action of rasagiline and ladostigil: new insights and implications for therapy. Ann N Y Acad Sci 1122:155–168
Wu Y, Kazumura K, Maruyama W, Osawa T, Naoi M (2015) Rasagiline and selegiline suppress calcium efflux from mitochondria by PK11195-induced opening of mitochondrial permeability transition pore: a novel anti-apoptotic function for neuroprotection. J Neural Transm 122(10):1399–1407
Wu Y, Shamoto-Nagai M, Maruyama W, Osawa T, Naoi M (2016) Rasagiline prevents cyclosporin A-sensitive superoxide flashes induced by PK11195, the initial signal of mitochondrial membrane permeabilization and apoptosis. J Neural Transm 123(5):491–494
Yi H, Akao Y, Maruyama W, Chen K, Shih J, Naoi M (2006) Type A monoamine oxidase is the target of an endogenous dopaminergic neurotoxin, N-methyl(R)salsolinol, leading to apoptosis in SH-SY5Y cells. J Neurochem 96(2):541–549
Youdim MBH, Bakhle YS (2006) Monoamine oxidase: isoforms and inhibitors in Parkinson’s disease and depressive illness. Br J Pharmacol 147(Suppl 1):S287–S296
Youdim MBH, Edmondson D, Tipton KF (2006) The therapeutic potential of monoamine oxidase inhibitors. Nat Rev Neurosci 7(4):295–309
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no competing financial interests in relation to the work described.
Rights and permissions
About this article
Cite this article
Inaba-Hasegawa, K., Shamoto-Nagai, M., Maruyama, W. et al. Type B and A monoamine oxidase and their inhibitors regulate the gene expression of Bcl-2 and neurotrophic factors in human glioblastoma U118MG cells: different signal pathways for neuroprotection by selegiline and rasagiline. J Neural Transm 124, 1055–1066 (2017). https://doi.org/10.1007/s00702-017-1740-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00702-017-1740-9