Abstract
Monoamine oxidase (MAO) A and MAO B are a crucial pair of isoenzymes, which oxidatively deaminate monoamine neurotransmitters and dietary amines with a production of hydrogen peroxide. These two isoenzymes have different but overlapping substrate and inhibitor specificities. MAO A and MAO B share 70% amino acid sequence identity and show different temporal and spatial expressions in both humans and mice. Abnormal MAO A or MAO B activity has been implicated in numerous neurological and psychiatric disorders. A better understanding of the transcriptional regulation of MAO A and MAO B genes may help explain the differential tissue-specific expression of these two isoenzymes and provide insights into the molecular basis of the disorders associated with MAO dysfunction. This review discusses the recent progress in the transcriptional regulation and multiple functions of MAO A and MAO B genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
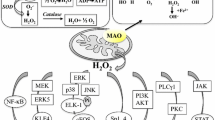
Monoamine oxidase (MAO) [amine: oxygen oxidoreductase (deaminating) (flavin-containing); MAO; E.C. 1.4.3.4.] oxidatively deaminates a number of biogenic and dietary amines in the brain and peripheral tissues and generates the byproduct hydrogen peroxide. MAO exists in two isoenzymes, MAO A and MAO B, with different but overlapping substrate and inhibitor specificities. MAO A preferentially oxidizes serotonin (5-hydroxytryptamine, 5-HT), norepinephrine (NE) and epinephrine and is irreversibly inhibited by low doses of clorgyline. MAO B preferentially oxidizes phenylethylamine (PEA) and is irreversibly inhibited by low doses of deprenyl (selegiline). Dopamine (DA) and tyramine are common substrates for both MAO A and MAO B (Shih et al. 1999). The success in cDNA cloning of MAO A and MAO B has demonstrated unequivocally that MAO A and MAO B are made of two different proteins, coded by two genes (Bach et al. 1988). They have identical exon–intron organization, suggesting that they are derived from the same ancestral gene by duplication (Bach et al. 1988; Grimsby et al. 1991). They are both located on the X chromosome (Xp11.23) (Lan et al. 1989). MAO A and MAO B proteins share 70% amino acid sequence identity and are located in the outer membrane of mitochondria. Although MAO A and MAO B are widely co-distributed in the central and peripheral nervous systems, MAO A is predominantly found in catecholaminergic neurons, whereas MAO B is more abundant in serotonergic and histaminergic neurons and glial cells (Shih et al. 1999).
Abnormal MAO activity has been implicated in a variety of neurological and psychiatric disorders, such as depression and social anxiety (Bortolato et al. 2008). MAO A deficiency caused by a spontaneous mutation in the MAO A gene led to impulsive aggressive behaviors and mild mental retardation in affected males in a Dutch family (Brunner et al. 1993). Consistent with humans, MAO A knock-out (KO) mice also show aggressive behaviors (Cases et al. 1995; Scott et al. 2008). Low platelet MAO B activity with the increased levels of PEA is associated with alcoholism and stress-related disorders (Devor et al. 1993; Faraj et al. 1994; Grimsby et al. 1997). Moreover, MAO B KO mice show behavioral disinhibition and reduced anxiety-like behaviors (Bortolato et al. 2009). In addition, MAO B activity is significantly increased in the brain with age in rats (Arai and Kinemuchi 1988) and humans (Fowler et al. 1997), suggesting that MAO B may play a role in aging process.
MAO inhibitors have long been developed and widely used in clinics for the treatment of many neuropsychiatric and neurodegenerative disorders. MAO A inhibitors have been shown to be effective antidepressant drugs (Bortolato et al. 2008). Recent evidence shows that MAO A inhibitors, such as moclobemide, also have antiparkinsonian effects by improving motor function (Sieradzan et al. 1995; Youdim and Riederer 2004). MAO B inhibitors, such as selegiline (deprenyl) and rasagiline (Azilect®, N-propargyl-1-(R)-aminoindan), have been effectively used for the treatment of Parkinson’s disease (PD), and their neuroprotective mechanisms have been substantially studied over the past decades (Foley et al. 2000; Mandel et al. 2003). These inhibitors protect neurons by preventing cell damage from neurotoxins (Naoi et al. 2000; Wu et al. 1995), free radical formation (Carrillo et al. 2000; Chiueh et al. 1992) and apoptosis (Maruyama et al. 2000; Weinreb et al. 2004) as well as stimulating the expression of neurotrophic factors (e. g. NGF, BDNF and GDNF) (Mizuta et al. 2000; Weinreb et al. 2004). Recently, structure–activity studies provide evidence indicating the important association of neuroprotection with the intrinsic pharmacological action of the propargylamine moiety in MAO B inhibitors (e.g. rasagiline), which leads to the development of multifunctional chimeric propargylamine-derivatives, such as M30 (Bar-Am et al. 2005; Zheng et al. 2005; Youdim 2006). M30, an iron-chelator, possesses the same neuroprotective propargylamine moiety as rasagiline but is a brain selective MAO A and MAO B inhibitor, which shows no cheese effect in response to tyramine, an unwanted side effect associated with earlier inhibitors (Gal et al. 2005, 2010a, b). In addition, these propargylamine-derivative compounds regulate the processing of amyloid-β protein precursor by the non-amyloidogenic α-secretase pathway (Avramovich-Tirosh et al. 2007a, b; Bar-Am et al. 2010), suggesting the potential use in Alzheimer’s disease. Undoubtedly, the studies on the development and the molecular mechanisms of MAO inhibitors not only provided the basis for the clinical treatment of neuropsychiatric and neurodegenerative disorders, but also demonstrated the important role of MAO in the brain.
The transcriptional regulation of MAO A and MAO B genes has been extensively studied in recent years after both MAO A and MAO B genes were cloned (Bach et al. 1988). Using a series of 5′-flanking sequences linked to a human growth hormone receptor gene, we identified the maximum promoter activities for MAO A and MAO B in a 0.14-kb PvuII/DraII and 0.15-kb PstI/NaeI fragment, respectively. Both fragments are GC-rich, contain potential Sp1-binding sites and share approximately 60% sequence identity (Zhu et al. 1992). Sp1 and Sp4 trans-activate MAO A and MAO B promoter activities by directly interacting with Sp1 sites, whereas this activation can be repressed by Sp3 and a related family member BTEB2 via the competition for binding to Sp1 sites (Wong et al. 2001; Zhu et al. 1994). However, the organization of transcription factor binding elements is different between the two promoters. The MAO A 0.14-kb promoter lacks a TATA box, consists of four Sp1-binding sites and exhibits bi-directional promoter activity (Zhu et al. 1994). The MAO B 0.15-kb promoter consists of two clusters of overlapping Sp1-bindng sites separated by a CACCC element (Ou et al. 2004). Moreover, a 30-bp variable number tandem repeat (VNTR) polymorphism is found 1.2 kb upstream of the MAO A coding sequences in human MAO A promoter (Zhu and Shih 1997). This 30-bp VNTR is present in 3, 3.5, 4 or 5 copies in different individuals across ethnic groups. The polymorphism has been associated with MAO A promoter/enzymatic activity, and alleles with 3.5 or 4 copies of the repeat are more efficiently transcribed than those with 3 or 5 copies of the repeat (Sabol et al. 1998). The different promoter organization of MAO A and MAO B genes may underline their different tissue- and cell-specific expressions.
Regulation of MAO B gene by TIEG2
The CACCC element flanked by two clusters of overlapping Sp1 sites in MAO B promoter was demonstrated to be a repressor element. The basal MAO B promoter activity increases by eightfold when this element is mutated. Moreover, transforming growth factor-β-inducible early gene (TIEG) 2 exhibits dual functions at the MAO B promoter. TIEG2 acts as a repressor at the CACCC element but an activator at the distal Sp1 sites of MAO B promoter. TIEG2 is capable of directly interacting with the CACCC element and distal Sp1 sites both in vitro and in vivo. Since TIEG2 has a higher affinity to the Sp1 sites than CACCC element, TIEG2 exerts an overall activating effect on MAO B promoter activity and mRNA level (Ou et al. 2004).
Recently, we and our collaborators have demonstrated that glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and MAO B play a contributory role in alcoholism associated with ethanol-induced brain cell damage, which is mediated by the up-regulation of MAO B by TIEG2. Ethanol significantly increases the levels of nuclear GAPDH and MAO B in neuronal cells as well as in human and rat brains. Moreover, nuclear GAPDH interacts with TIEG2 and further augments TIEG2-mediated MAO B trans-activation, which results in cell damage, correlating with increased levels of hydrogen peroxide, when exposed to ethanol (Ou et al. 2009, 2010). These studies suggest that the interference with TIEG2-MAO B interaction could be an alternative approach in addition to the MAO B inhibitors for treating neuropsychiatric symptoms associated with alcoholism.
Identification of a novel transcriptional repressor R1 (RAM2/CDCA7L/JPO2) for MAO A gene
To search for additional novel transcription factors, which may interact with Sp1 sites and regulate MAO promoters, we used three copies of Sp1-binding motifs derived from MAO B core promoter as bait to screen a human cDNA library in the yeast one-hybrid system. Two novel transcription factors have been identified, one of which was named R1 (RAM2/CDCA7L/JPO2) by us. The other one is currently under investigation.
The R1 cDNA encodes a protein with 454 amino acids, and the C-terminal of R1 protein encompassing 77 amino acid residues (349–425) shows 87% identity with c-Myc targeting protein JPO1, which ultimately leads to tumorigenesis (Chen et al. 2005; Huang et al. 2005). Further analysis of this region reveals 12 conserved cysteine residues and 4 CXXC zinc finger putative DNA-binding domains, which provides bases for proper protein confirmation for DNA binding. This region is highly conserved between human and mouse R1 with 94% sequence identity. Moreover, a nuclear targeting sequence (amino acids 301–318) and multiple potential phosphorylation sites have been identified, suggesting the cellular location of R1 may be dependent on its phosphorylation level. Similar to MAO genes, R1 is widely expressed in the human brain and peripheral tissues. At the cellular level, R1 is found in both the nucleus and cytosol (Chen et al. 2005). Our studies and that of other groups have further shown that R1 is a c-Myc interacting protein that potentiates and complements c-Myc transforming activity (Huang et al. 2005; Ou et al. 2006b).
Subsequent experiments showed that R1 acts as a transcriptional repressor of MAO A gene and represses MAO A promoter activity by competing with Sp1 for binding to Sp1 sites. R1 is capable of directly interacting with Sp1 sites under both in vitro and in vivo conditions. Moreover, over-expression of R1 in stable neuronal cell lines down-regulates MAO A enzymatic activity, suggesting its physiological effect (Chen et al. 2005). Given the multiple functions of R1 in certain cellular events, such as cell proliferation, the possible indirect mechanisms, i. e. other than direct transcriptional regulation, by which R1 regulates MAO A gene, remain to be studied. The regulatory effect/mechanism of R1 on MAO B gene is currently under investigation as well.
Regulation of X-located MAO A gene by sex-determining region gene on the Y chromosome (SRY)
The fact that MAO is located on the X chromosome has driven us to hypothesize whether there is a sex-specific transcription factor regulating MAO gene expression, which provides us bases to speculate the sex dimorphism as observed in several neuropsychiatric disorders associated with abnormal MAO activity, such as depression (Dulcan 1997; Williams et al. 2008; Wright et al. 2009) and autism (Williams et al. 2008). Recently, we and our collaborators have identified MAO A as a novel neural target for the SRY gene in a genome-wide ChIP and promoter tiling microarray analysis. The SRY gene, encoding a putative transcription factor, is the master switch regulator responsible for initiating the testis determination and differentiation during embryogenesis (Wilhelm et al. 2007).
SRY activates both MAO A promoter and enzymatic activities via a functional SRY-binding site (−117/−111, AT-rich) in the MAO A core promoter. Sp1 synergistically enhances the SRY activation of MAO A promoter in a dose-dependent manner. Moreover, Sp1 interacts and forms a transcriptional regulatory complex with SRY at the natural MAO A core promoter, which potentiates SRY binding to MAO A promoter (Wu et al. 2009a). This is the first study showing that the Y-encoded transcription factor SRY is capable of regulating an X-located gene, suggesting a novel molecular mechanism for sexual dimorphism in neural development, brain functions and initiation/progression of neural disorders associated with MAO A dysfunction.
Previous studies have demonstrated that various cofactors interact with SRY to form transcriptional complex that regulate SRY target genes (Li et al. 2006; Oh et al. 2005). Since other factors, such as Sp3, Sp4 and R1, also utilize the same Sp1 sites, neighboring the SRY site, in their transcriptional regulation of MAO A, it would be interesting to determine whether these MAO A regulators could also interact with SRY, thereby exerting potentially complex transcriptional interplay and sexually dimorphic physiological effects (Fig. 1). Moreover, recent studies have suggested that some neuropsychiatric disorders, such as autism, may originate in early embryonic development (Ploeger et al. 2010). Hence, understanding how the transcriptional network coordinated by SRY or other sex-determining factors regulates MAO A gene during brain development may provide insights into addressing the role of MAO genes in these mental disorders which show sexual dimorphism and MAO dysfunction. These studies are currently under investigation.
Regulation of MAO genes by methylation
Several lines of direct and indirect evidence suggest DNA methylation can be a factor/mechanism contributing to the regulation of MAO genes. For example, variations of MAO activity have been associated with their methylation status of promoters in smoking groups (Launay et al. 2009). The most direct evidence is the identification of functional CpG islands (CGIs) in the MAO promoters. There is a putative CGI containing 22 potential CpG methylation sites in the MAO B promoter (−261/−58). In vitro demethylation of MAO B promoter with 5-aza-2′-deoxycytidine, a DNA methyltransferase inhibitor, up-regulates MAO B gene expression in both HeLa and Caco-2 cells (Wong et al. 2003). Since the CGI in the MAO B promoter encompasses several Sp1 sites (GC-rich), the lower MAO B promoter activity under methylation status could also be likely due to its altered chromatin structure, which impedes the recruitment of transcription factors, such as Sp1, to this region.
Recently, other groups report that the extended MAO A regulatory region contains two CGIs, one of which overlaps with the canonical MAO A promoter and the other is located further upstream. Both CGIs exhibit sensitivity to differential methylation (Shumay and Fowler 2010). Moreover, the effect of VNTR on the MAO A transcription may have epigenetic nature, as this polymorphic region resides within the CGI of MAO A promoter and itself contains CpG sites. Thus, the number of repeats, correlating with the number of methylatable cytosines in the MAO A promoter, could exert a possible effect on the transcriptional activity of MAO A.
Hormonal regulation of MAO A and MAO B genes
Emerging evidence has shown that steroid and non-steroid hormones are involved in the regulation of many neuropsychiatric processes in which MAO also plays a critical role, such as responses to stress, behavioral adaption and mood (de Kloet et al. 1990). For instance, significant hypersecretion of glucocorticoids has been shown to be associated with depression (Duval et al. 2006), and anti-glucocorticoid agents have been used in the treatment of depression (Wolkowitz and Reus 1999). We have been dedicated to investigating the hormonal regulation of MAO genes and the corresponding mechanisms at the molecular level in recent years. To date, we have demonstrated that androgen, glucocorticoid (Ou et al. 2006a) and retinoic acid (RA) (Wu et al. 2009b) activate both MAO A and MAO B gene expression albeit to different extents in human neuronal cell models, whereas estrogen shows tissue-specific effect on MAO A and MAO B genes (Holschneider et al. 1998). Since these hormones play a pivotal role(s) in many physiological and pathological states, our findings may thus also provide insights into the potential new functions of MAO genes in these hormone-controlled steps/processes.
Androgen and glucocorticoid
Androgen and glucocorticoid show their capabilities to up-regulate MAO A gene in a canonical way by directing their receptors to bind to a functional androgen/glucocorticoid response element (−289/−275) in the MAO A promoter. On the other hand, both androgen and glucocorticoid receptors interact with Sp1 sites indirectly via Sp1, and such interaction is enhanced in response to ligands. Moreover, glucocortcoid but not androgen induces R1 translocation. R1 is translated into the nucleus upon 12-h dexamethasone (a synthetic glucocorticoid) and re-located into the cytosol after 24- or 48-h treatment (Ou et al. 2006a).
Retinoic acid
Similarly, RA activates the MAO B transcription by both ligand-receptor interaction and crosstalk between the receptor and transcription factor Sp1 as well. Retinoic acid receptor α (RAR α), but not retinod X receptor α, binds to a functional RA response element (−303/−287) in the MAO B promoter. Mutation of Sp1 sites or interference of Sp1-binding ability down-regulates the RA activation of MAO B promoter, suggesting a mediating role of Sp1 in this activation. Further analysis shows that RAR α interacts with Sp1 via the zinc finger domains in Sp1. Furthermore, RAR α is able to be recruited by Sp1 and forms a transcriptional regulation complex with Sp1 at the Sp1 sites in the MAO B promoter in vivo, which enhances Sp1-binding ability (Wu et al. 2009b).
Estrogen
The interplay of estrogen receptors (ERs) and estrogen-related receptors (ERRs) in the regulation of MAO B promoter activity has been studied in breast cancer cells. ERR α recognizes a variety of estrogen response elements (EREs) and shares many target genes and cofactors with ER α. ERR α and ERRΥ up-regulate MAO B gene expression, whereas this up-regulation is repressed by ER α and ER β in both ligand-dependent and -independent manners. In contrast to ER-negative HeLa cells, the ability of ERRs to stimulate MAO B promoter activity is reduced in ER-positive MCF-7 and T47D cells. Several EREs responsible for the up-regulation by ERRs are located in the MAO B promoter, and ERs compete with ERRs for binding to the MAO B promoter at selective ERR motifs (−1,762, −1,468, and −289/−286), thereby changing the chromatin status and cofactor recruitment to a repressed state (Zhang et al. 2006).
MAO genes in apoptosis and stem cell proliferation
MAO A has long been suggested as a pro-apoptotic gene. MAO A expression increases during the apoptosis induced by the withdrawal of neurotrophic factors in PC12 cells, which is mediated via p38 kinase pathway (De Zutter and Davis 2001). Moreover, clogyline, a MAO A inhibitor, shows protective effects from serum starvation-induced apoptosis in human melanoma M14 cells (Malorni et al. 1998). Recently, the roles of MAO A and its novel transcriptional repressor R1 in apoptosis and cell proliferation are further studied in a human neuronal cell model and MAO A KO mice. In response to serum starvation, the expression of p38, MAO A and caspase-3 increases, whereas Bcl-2 and R1 levels decrease. MAO A and R1 are demonstrated downstream of p38 kinase and Bcl-2 but upstream of caspase-3. Consistently, the serum starvation-induced apoptosis is reduced in cortical brain cells from MAO A-deficient mice in comparison with the wild-type control. Furthermore, cyclin D1 and E2F show negative correlation with MAO A in an R1-modulated manner and act as downstream targets of MAO A- and R1-mediated cellular proliferative pathway (Ou et al. 2006b).
In contrast to MAO A, MAO B is induced by phorbol-12-myristate 13-acetate (PMA), a tumor-promoting agent, which is mediated via the activation of PKC and MAPK signaling pathways. Further studies show that transcription factor Egr-1 and c-Jun are ultimately responsible for the induction of MAO B gene by directly interacting with the overlapping Sp1/Egr-1/Sp1-binding sites (−246/−225) in MAO B core promoter (Wong et al. 2002). Since PMA has shown to stimulate cell proliferation in a number of cell models (Amos et al. 2005; Isakov et al. 1993), this regulation may suggest a potential role of MAO B in regulating cell proliferation.
One possible mechanism by which MAO genes influence cell proliferation is via the control of specific neurotransmitter levels, such as serotonin. Numerous studies have well documented the role of serotonin along with its receptors in stimulating cell proliferation and tumor growth in an array of cancer cell models, such as prostate and hepatocellular carcinomas (Siddiqui et al. 2006; Soll et al. 2010). Moreover, our recent study shows that mice lacking both MAO A and MAO B (MAO AB double KO) exhibit diminished proliferation of neural stem cells in late embryonic and early postnatal development, which is mediated by serotonin (Cheng et al. 2010). It would be interesting to study the transcriptional regulatory patterns of MAO genes during embryonic and early postnatal developmental stages, which may provide new clues for understanding the profound changes in cell proliferation capacity caused by altered neurotransmitter levels.
Summary and future prospective
The current knowledge on the promoter organization and transcriptional regulation of MAO A and MAO B genes has been reviewed here. The Sp1-binding sites and Sp-family/Sp-family-like transcription factors were identified to be the major players for regulating both MAO A and MAO B promoters. On the other hand, MAO A and MAO B also show differential regulatory mechanisms in terms of their responses to different transcription factors, cellular signal transductions and hormones. For example, MAO A is up-regulated by SRY, whereas MAO B shows the induced responses to TIEG2. In contrast to MAO A, which is involved in a c-Myc- and R1-mediated apoptotic signaling pathway, MAO B is activated by PKC and MAPK cascades. Moreover, MAO A and MAO B are distinctly regulated by diverse hormones in terms of their extent of responses in different cell models and tissue regions. These differential regulations may contribute to the differences in the temporal/spatial expression and physiological functions between these two isoenzymes. In addition, the identification of novel/unique transcription factors of MAO genes (e.g. R1, a c-Myc interacting protein which enhances c-Myc transforming activity) may provide insights into the new functions of MAO genes (e.g. a potential role in tumor progression) as well as the molecular basis of neuropsychiatric disorders associated with MAO dysfunction. Modulation of endogenous levels of such factors could be considered as an alternative approach to maintain normal MAO activity in addition to using MAO inhibitors. The translational values of these findings will be further investigated.
References
Amos S, Martin PM, Polar GA, Parsons SJ, Hussaini IM (2005) Phorbol 12-myristate 13-acetate induces epidermal growth factor receptor transactivation via protein kinase Cdelta/c-Src pathways in glioblastoma cells. J Biol Chem 280(9):7729–7738
Arai Y, Kinemuchi H (1988) Differences between monoamine oxidase concentrations in striatum and forebrain of aged and young rats. J Neural Transm 72(2):99–105
Avramovich-Tirosh Y, Amit T, Bar-Am O, Zheng H, Fridkin M, Youdim MB (2007a) Therapeutic targets and potential of the novel brain-permeable multifunctional iron chelator-monoamine oxidase inhibitor drug, M-30, for the treatment of Alzheimer’s disease. J Neurochem 100(2):490–502
Avramovich-Tirosh Y, Reznichenko L, Mit T, Zheng H, Fridkin M, Weinreb O, Mandel S, Youdim MB (2007b) Neurorescue activity, APP regulation and amyloid-beta peptide reduction by novel multi-functional brain permeable iron-chelating-antioxidants, M-30 and green tea polyphenol, EGCG. Curr Alzheimer Res 4(4):403–411
Bach AW, Lan NC, Johnson DL, Abell CW, Bembenek ME, Kwan SW, Seeburg PH, Shih JC (1988) cDNA cloning of human liver monoamine oxidase A and B: molecular basis of differences in enzymatic properties. Proc Natl Acad Sci USA 85(13):4934–4938
Bar-Am O, Weinreb O, Amit T, Youdim MB (2005) Regulation of Bcl-2 family proteins, neurotrophic factors, and APP processing in the neurorescue activity of propargylamine. FASEB J 19(13):1899–1901
Bar-Am O, Amit T, Weinreb O, Youdim MB, Mandel S (2010) Propargylamine containing compounds as modulators of proteolytic cleavage of amyloid-beta protein precursor: involvement of MAPK and PKC activation. J Alzheimers Dis 21(2):361–371
Bortolato M, Chen K, Shih JC (2008) Monoamine oxidase inactivation: from pathophysiology to therapeutics. Adv Drug Deliv Rev 60(13–14):1527–1533
Bortolato M, Godar SC, Davarian S, Chen K, Shih JC (2009) Behavioral disinhibition and reduced anxiety-like behaviors in monoamine oxidase B-deficient mice. Neuropsychopharmacology 34(13):2746–2757
Brunner HG, Nelen M, Breakefield XO, Ropers HH, van Oost BA (1993) Abnormal behavior associated with a point mutation in the structural gene for monoamine oxidase A. Science 262(5133):578–580
Carrillo MC, Minami C, Kitani K, Maruyama W, Ohashi K, Yamamoto T, Naoi M, Kanai S, Youdim MB (2000) Enhancing effect of rasagiline on superoxide dismutase and catalase activities in the dopaminergic system in the rat. Life Sci 67(5):577–585
Cases O, Seif I, Grimsby J, Gaspar P, Chen K, Pournin S, Muller U, Aguet M, Babinet C, Shih JC et al (1995) Aggressive behavior and altered amounts of brain serotonin and norepinephrine in mice lacking MAO A. Science 268(5218):1763–1766
Chen K, Ou XM, Chen G, Choi SH, Shih JC (2005) R1, a novel repressor of the human monoamine oxidase A. J Biol Chem 280(12):11552–11559
Cheng A, Scott AL, Ladenheim B, Chen K, Ouyang X, Lathia JD, Mughal M, Cadet JL, Mattson MP, Shih JC (2010) Monoamine oxidases regulate telencephalic neural progenitors in late embryonic and early postnatal development. J Neurosci 30(32):10752–10762
Chiueh CC, Huang SJ, Murphy DL (1992) Enhanced hydroxyl radical generation by 2′-methyl analog of MPTP: suppression by clorgyline and deprenyl. Synapse 11(4):346–348
de Kloet ER, Reul JM, Sutanto W (1990) Corticosteroids and the brain. J Steroid Biochem Mol Biol 37(3):387–394
De Zutter GS, Davis RJ (2001) Pro-apoptotic gene expression mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Proc Natl Acad Sci USA 98(11):6168–6173
Devor EJ, Cloninger CR, Hoffman PL, Tabakoff B (1993) Association of monoamine oxidase (MAO) activity with alcoholism and alcoholic subtypes. Am J Med Genet 48(4):209–213
Dulcan M (1997) Practice parameters for the assessment and treatment of children, adolescents, and adults with attention-deficit/hyperactivity disorder. American Academy of Child and Adolescent Psychiatry. J Am Acad Child Adolesc Psychiatry 36(10 Suppl):85S–121S
Duval F, Mokrani MC, Monreal-Ortiz JA, Fattah S, Champeval C, Schulz P, Macher JP (2006) Cortisol hypersecretion in unipolar major depression with melancholic and psychotic features: dopaminergic, noradrenergic and thyroid correlates. Psychoneuroendocrinology 31(7):876–888
Faraj BA, Davis DC, Camp VM, Mooney AJ 3rd, Holloway T, Barika G (1994) Platelet monoamine oxidase activity in alcoholics, alcoholics with drug dependence, and cocaine addicts. Alcohol Clin Exp Res 18(5):1114–1120
Foley P, Gerlach M, Youdim MB, Riederer P (2000) MAO-B inhibitors: multiple roles in the therapy of neurodegenerative disorders? Parkinsonism Relat Disord 6(1):25–47
Fowler JS, Volkow ND, Wang GJ, Logan J, Pappas N, Shea C, MacGregor R (1997) Age-related increases in brain monoamine oxidase B in living healthy human subjects. Neurobiol Aging 18(4):431–435
Gal S, Zheng H, Fridkin M, Youdim MB (2005) Novel multifunctional neuroprotective iron chelator-monoamine oxidase inhibitor drugs for neurodegenerative diseases. In vivo selective brain monoamine oxidase inhibition and prevention of MPTP-induced striatal dopamine depletion. J Neurochem 95(1):79–88
Gal S, Abassi ZA, Youdim MB (2010a) Limited potentiation of blood pressure in response to oral tyramine by the anti-Parkinson brain selective multifunctional monoamine oxidase-AB inhibitor, M30. Neurotox Res 18(2):143–150
Gal S, Zheng H, Fridkin M, Youdim MB (2010b) Restoration of nigrostriatal dopamine neurons in post-MPTP treatment by the novel multifunctional brain-permeable iron chelator-monoamine oxidase inhibitor drug, M30. Neurotox Res 17(1):15–27
Grimsby J, Chen K, Wang LJ, Lan NC, Shih JC (1991) Human monoamine oxidase A and B genes exhibit identical exon–intron organization. Proc Natl Acad Sci USA 88(9):3637–3641
Grimsby J, Toth M, Chen K, Kumazawa T, Klaidman L, Adams JD, Karoum F, Gal J, Shih JC (1997) Increased stress response and beta-phenylethylamine in MAOB-deficient mice. Nat Genet 17(2):206–210
Holschneider DP, Kumazawa T, Chen K, Shih JC (1998) Tissue-specific effects of estrogen on monoamine oxidase A and B in the rat. Life Sci 63(3):155–160
Huang A, Ho CS, Ponzielli R, Barsyte-Lovejoy D, Bouffet E, Picard D, Hawkins CE, Penn LZ (2005) Identification of a novel c-Myc protein interactor, JPO2, with transforming activity in medulloblastoma cells. Cancer Res 65(13):5607–5619
Isakov N, Galron D, Mustelin T, Pettit GR, Altman A (1993) Inhibition of phorbol ester-induced T cell proliferation by bryostatin is associated with rapid degradation of protein kinase C. J Immunol 150(4):1195–1204
Lan NC, Heinzmann C, Gal A, Klisak I, Orth U, Lai E, Grimsby J, Sparkes RS, Mohandas T, Shih JC (1989) Human monoamine oxidase A and B genes map to Xp 11.23 and are deleted in a patient with Norrie disease. Genomics 4(4):552–559
Launay JM, Del Pino M, Chironi G, Callebert J, Peoc’h K, Megnien JL, Mallet J, Simon A, Rendu F (2009) Smoking induces long-lasting effects through a monoamine-oxidase epigenetic regulation. PLoS One 4(11):e7959
Li Y, Oh HJ, Lau YF (2006) The poly(ADP-ribose) polymerase 1 interacts with Sry and modulates its biological functions. Mol Cell Endocrinol 257–258:35–46
Malorni W, Giammarioli AM, Matarrese P, Pietrangeli P, Agostinelli E, Ciaccio A, Grassilli E, Mondovi B (1998) Protection against apoptosis by monoamine oxidase A inhibitors. FEBS Lett 426(1):155–159
Mandel S, Grunblatt E, Riederer P, Gerlach M, Levites Y, Youdim MB (2003) Neuroprotective strategies in Parkinson’s disease: an update on progress. CNS Drugs 17(10):729–762
Maruyama W, Yamamoto T, Kitani K, Carrillo MC, Youdim M, Naoi M (2000) Mechanism underlying anti-apoptotic activity of a (−)deprenyl-related propargylamine, rasagiline. Mech Ageing Dev 116(2–3):181–191
Mizuta I, Ohta M, Ohta K, Nishimura M, Mizuta E, Hayashi K, Kuno S (2000) Selegiline and desmethylselegiline stimulate NGF, BDNF, and GDNF synthesis in cultured mouse astrocytes. Biochem Biophys Res Commun 279(3):751–755
Naoi M, Maruyama W, Akao Y, Zhang J, Parvez H (2000) Apoptosis induced by an endogenous neurotoxin, N-methyl(R)salsolinol, in dopamine neurons. Toxicology 153(1–3):123–141
Oh HJ, Li Y, Lau YF (2005) Sry associates with the heterochromatin protein 1 complex by interacting with a KRAB domain protein. Biol Reprod 72(2):407–415
Ou XM, Chen K, Shih JC (2004) Dual functions of transcription factors, transforming growth factor-beta-inducible early gene (TIEG)2 and Sp3, are mediated by CACCC element and Sp1 sites of human monoamine oxidase (MAO) B gene. J Biol Chem 279(20):21021–21028
Ou XM, Chen K, Shih JC (2006a) Glucocorticoid and androgen activation of monoamine oxidase A is regulated differently by R1 and Sp1. J Biol Chem 281(30):21512–21525
Ou XM, Chen K, Shih JC (2006b) Monoamine oxidase A and repressor R1 are involved in apoptotic signaling pathway. Proc Natl Acad Sci USA 103(29):10923–10928
Ou XM, Lu D, Johnson C, Chen K, Youdim MB, Rajkowska G, Shih JC (2009) Glyceraldehyde-3-phosphate dehydrogenase-monoamine oxidase B-mediated cell death-induced by ethanol is prevented by rasagiline and 1-R-aminoindan. Neurotox Res 16(2):148–159
Ou XM, Stockmeier CA, Meltzer HY, Overholser JC, Jurjus GJ, Dieter L, Chen K, Lu D, Johnson C, Youdim MB, Austin MC, Luo J, Sawa A, May W, Shih JC (2010) A novel role for glyceraldehyde-3-phosphate dehydrogenase and monoamine oxidase B cascade in ethanol-induced cellular damage. Biol Psychiatry 67(9):855–863
Ploeger A, Raijmakers ME, van der Maas HL, Galis F (2010) The association between autism and errors in early embryogenesis: what is the causal mechanism? Biol Psychiatry 67(7):602–607
Sabol SZ, Hu S, Hamer D (1998) A functional polymorphism in the monoamine oxidase A gene promoter. Hum Genet 103(3):273–279
Scott AL, Bortolato M, Chen K, Shih JC (2008) Novel monoamine oxidase A knock out mice with human-like spontaneous mutation. Neuroreport 19(7):739–743
Shih JC, Chen K, Ridd MJ (1999) Monoamine oxidase: from genes to behavior. Annu Rev Neurosci 22:197–217
Shumay E, Fowler JS (2010) Identification and characterization of putative methylation targets in the MAOA locus using bioinformatic approaches. Epigenetics 5(4):325–342
Siddiqui EJ, Shabbir M, Mikhailidis DP, Thompson CS, Mumtaz FH (2006) The role of serotonin (5-hydroxytryptamine 1A and 1B) receptors in prostate cancer cell proliferation. J Urol 176(4 Pt 1):1648–1653
Sieradzan K, Channon S, Ramponi C, Stern GM, Lees AJ, Youdim MB (1995) The therapeutic potential of moclobemide, a reversible selective monoamine oxidase A inhibitor in Parkinson’s disease. J Clin Psychopharmacol 15(4 Supp 2):51S–59S
Soll C, Jang JH, Riener MO, Moritz W, Wild PJ, Graf R, Clavien PA (2010) Serotonin promotes tumor growth in human hepatocellular cancer. Hepatology 51(4):1244–1254
Weinreb O, Bar-Am O, Amit T, Chillag-Talmor O, Youdim MB (2004) Neuroprotection via pro-survival protein kinase C isoforms associated with Bcl-2 family members. FASEB J 18(12):1471–1473
Wilhelm D, Palmer S, Koopman P (2007) Sex determination and gonadal development in mammals. Physiol Rev 87(1):1–28
Williams JG, Allison C, Scott FJ, Bolton PF, Baron-Cohen S, Matthews FE, Brayne C (2008) The Childhood Autism Spectrum Test (CAST): sex differences. J Autism Dev Disord 38(9):1731–1739
Wolkowitz OM, Reus VI (1999) Treatment of depression with antiglucocorticoid drugs. Psychosom Med 61(5):698–711
Wong WK, Chen K, Shih JC (2001) Regulation of human monoamine oxidase B gene by Sp1 and Sp3. Mol Pharmacol 59(4):852–859
Wong WK, Ou XM, Chen K, Shih JC (2002) Activation of human monoamine oxidase B gene expression by a protein kinase C MAPK signal transduction pathway involves c-Jun and Egr-1. J Biol Chem 277(25):22222–22230
Wong WK, Chen K, Shih JC (2003) Decreased methylation and transcription repressor Sp3 up-regulated human monoamine oxidase (MAO) B expression during Caco-2 differentiation. J Biol Chem 278(38):36227–36235
Wright SL, Langenecker SA, Deldin PJ, Rapport LJ, Nielson KA, Kade AM, Own LS, Akil H, Young EA, Zubieta JK (2009) Gender-specific disruptions in emotion processing in younger adults with depression. Depress Anxiety 26(2):182–189
Wu RM, Murphy DL, Chiueh CC (1995) Neuronal protective and rescue effects of deprenyl against MPP+ dopaminergic toxicity. J Neural Transm Gen Sect 100(1):53–61
Wu JB, Chen K, Li Y, Lau YF, Shih JC (2009a) Regulation of monoamine oxidase A by the SRY gene on the Y chromosome. FASEB J 23(11):4029–4038
Wu JB, Chen K, Ou XM, Shih JC (2009b) Retinoic acid activates monoamine oxidase B promoter in human neuronal cells. J Biol Chem 284(25):16723–16735
Youdim MB (2006) The path from anti Parkinson drug selegiline and rasagiline to multifunctional neuroprotective anti Alzheimer drugs ladostigil and m30. Curr Alzheimer Res 3(5):541–550
Youdim MB, Riederer PF (2004) A review of the mechanisms and role of monoamine oxidase inhibitors in Parkinson’s disease. Neurology 63(7 Suppl 2):S32–S35
Zhang Z, Chen K, Shih JC, Teng CT (2006) Estrogen-related receptors-stimulated monoamine oxidase B promoter activity is down-regulated by estrogen receptors. Mol Endocrinol 20(7):1547–1561
Zheng H, Weiner LM, Bar-Am O, Epsztejn S, Cabantchik ZI, Warshawsky A, Youdim MB, Fridkin M (2005) Design, synthesis, and evaluation of novel bifunctional iron-chelators as potential agents for neuroprotection in Alzheimer’s, Parkinson’s, and other neurodegenerative diseases. Bioorg Med Chem 13(3):773–783
Zhu Q, Shih JC (1997) An extensive repeat structure down-regulates human monoamine oxidase A promoter activity independent of an initiator-like sequence. J Neurochem 69(4):1368–1373
Zhu QS, Grimsby J, Chen K, Shih JC (1992) Promoter organization and activity of human monoamine oxidase (MAO) A and B genes. J Neurosci 12(11):4437–4446
Zhu QS, Chen K, Shih JC (1994) Bidirectional promoter of human monoamine oxidase A (MAO A) controlled by transcription factor Sp1. J Neurosci 14(12):7393–7403
Acknowledgments
This work was supported by National Institute of Mental Health Grant R37MH39085 (MERIT Award), R01MH67968, and the Boyd and Elsie Welin Professorship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shih, J.C., Wu, J.B. & Chen, K. Transcriptional regulation and multiple functions of MAO genes. J Neural Transm 118, 979–986 (2011). https://doi.org/10.1007/s00702-010-0562-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00702-010-0562-9