Abstract
Purpose
Patients with hepatocellular carcinoma (HCC) can only be treated curatively at early stages and then have a favorable prognosis of this often fatal disease. For this reason, an early detection and diagnostic confirmation are crucial. Raman imaging spectroscopy is a promising technology for high-resolution visualization of the spatial distribution of molecular composition in tissue sections. The aim of this study was to investigate molecular information of liver tissue by Raman imaging for classification and diagnostic prediction.
Methods
Unstained cryosections of human hepatic tissues (23 patients) were measured by Raman spectroscope in the regions of HCC (n = 12) and fibrosis (n = 17). The acquired data set was used to generate a random forest classification model with 101 iterations of sevenfold cross-validation. The models obtained during cross-validation were also used to predict regions of tumor margin (n = 8) aside from independent testing.
Results
Raman spectra differed between malignant and non-malignant tissue regions. Based on these spectral data, a random forest classification model calculated a prediction accuracy of 86 % (76 % sensitivity and 93 % specificity). The ten most important variables were identified at 2895, 2856, 1439, 1298, 1080, 1063, 1023, 937, 920, and 719 cm−1.
Conclusions
In this study, Raman imaging spectroscopy was applied successfully for liver tissue to differentiate, classify, and predict with high accuracy malignant and non-malignant tissue regions. Furthermore, the most important differences were identified as the Raman signature of fatty acids. The demonstrated results highlight the enormous potential which vibrational spectroscopy techniques have for the future diagnostics and prognosis estimation of HCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The dramatic increase in the incidence of hepatocellular carcinoma (HCC) causes worldwide 692,000 deaths per year (Iavarone and Colombo 2013). Surveillance strategies and the early diagnosis of the tumor are essential for curative treatment option. Histopathological diagnosis of HCC is based essentially on the cytohistologic criteria according to the International Consensus Group for Hepatocellular Neoplasia. However, morphological criteria alone still pose problems for the differential diagnosis and depend upon location and size of liver biopsy as well as doctors’ expertise. Therefore, gene expression profiles and more cost-efficient immunohistochemical stainings for glypican 3 (GPC3), heat shock protein 70 (HSP-70), and glutamine synthetase (GS) are recommended by current international clinical practice guidelines to establishing a diagnosis (European Association for the Study of the Liver and European Organisation for Research and Treatment of Cancer 2012). Nevertheless, always a panel of several markers is necessary to achieve a required level of accuracy and a more standardized diagnosis of HCC, but the rate of false negative diagnosis is still high (Di Tommaso et al. 2009; Bruix and Sherman 2011).
Against this background, completely new approaches should be explored that are able to detect various markers in the tissue at once and to use this “fingerprint” for an objective tumor differentiation. Moreover, these innovative approaches could also be used for a classification of the severity of the disease and perhaps even for a risk stratification of the patients. Vibrational spectroscopic techniques such as infrared or Raman spectroscopy are efficient tools for investigating rapidly and non-destructively a wide variety of biochemical compounds in parallel directly in cells or tissue and without any need of labeling. Raman spectroscopy uses the inelastic scattering of visible or near-infrared light to analyze vibrational modes of molecules. One advantage of Raman imaging spectroscopy in comparison with the conventional Raman spectroscopy is high resolution (<1 µm, i.e., subcellular resolution) which allows to determine the spatial distribution of biochemical compounds in tissue sections and to correlate the obtained molecular information with the morphological structures at the very same sample location. Using pattern recognition and classification algorithms, tissue-specific patterns can be discovered and used to develop predictive models. To date, Raman imaging has already been successfully applied in several studies to elucidate the molecular information in colon, cervical, brain, and stomach malignant tissues (Beljebbar et al. 2009; Kamemoto et al. 2010; Bergner et al. 2012; Bergholt et al. 2010). As Raman spectroscopic data will become available for various tumor tissue types, this approach will provide a common disease-wide methodology that can be applied to a variety of clinical questions.
Materials and methods
Tissue collection and sample preparation
The research project was approved by the local medical ethics committee, and written informed consent was obtained from all patients. In the Department of General, Visceral and Vascular Surgery (Jena University Hospital, Germany) surgically resected liver tissue samples were obtained from 23 patients diagnosed with HCC. Immediately after liver resection, tissue samples of the tumor margins were shock frozen in liquid nitrogen and stored at −80 °C. For diagnosis, a histological examination of the resected tissue was performed in the Institute of Pathology (Jena University Hospital, Germany). Table 1 shows the pathological characteristics of these samples.
On the day of Raman measurements, a microtome (Cryostat Leica 3050 S, Leica Biosystems, Germany) was used to prepare two parallel sections of each tissue sample. The first section was mounted onto microscope glass slide and processed with standard hematoxylin and eosin (HE) staining to define the areas of interest for Raman analysis. The second tissue section was deposited onto a calcium fluoride slide (CaF2; Vacuum-UV quality, Crystal GmbH, Berlin) and transferred to the Raman spectrometer in a vacuum desiccator. According to the pre-defined region of the HE-stained tissue section, Raman images were acquired from the correlating region of the parallel section. After the measurements, the tissue sections on CaF2 slides remained intact and were also stained with HE, microscopic images were taken and correlated with the obtained Raman maps.
Raman image acquisition
Raman images were collected using a confocal Raman microscope (WITec, Ulm, Germany, Model CRM 2000) with 300 lines/mm grating (blaze wavelength 750 nm) and a 785 nm diode laser as excitation. The laser power was 50 mW. The laser light was focused on the sample with a 50× NA 0.95 objective (EC Epiplan-Apochromat, Zeiss, Germany) coupled to the microscope by a single mode optical fiber. The scattered Raman signal was detected by a back-illuminated deep-depletion CCD camera operating at −65 °C. The Raman system was calibrated to the 520.7 cm−1 spectral line of silicon. Raman spectral images of the selected tissue regions were acquired with a measurement area of 75 × 75 µm2 for tumor center and fibrotic regions and a measurement area of 75 × 100 µm2 for the tumor margin. As lateral resolution of the Raman image a 1 µm step size in the spectral region of 3200–200 cm−1 was set in the mapping mode of WITec Control. Dark tissues such as kidney and liver have often caused problems for Raman spectroscopy because they are highly absorbing and fluorescent at most visible and NIR wavelengths (Matousek and Stone 2009). Therefore, pre-bleaching of the autofluorescence of 2 s followed by an integration time for each spectrum of 5 s was necessary. The mounting material CaF2 was chosen to avoid interfering fluorescence background from usually used glass slides.
Data analysis
The chemometric analysis of the Raman data set of all maps was performed with CytoSpec (CytoSpec Inc., USA), OPUS (Bruker Optik GmbH, Germany) and R (R Core Team 2014).
As an important first step of our data analysis, a data pre-processing was performed. Therefore, interfering signals such as fluorescence, burning artifacts, and present contaminants were determined and removed according to defined upper and lower thresholds for intensity of the CH-stretching region using a quality test included in the CytoSpec software. Artifacts from the recording due to cosmic radiation were removed from the data set by suitable algorithms. Furthermore, baseline correction and vector normalization were applied to the spectral region of 3100–600 cm−1. These spectral pre-processing steps are appropriate for vibrational spectroscopy and were already successfully applied for Raman imaging (Neugebauer et al. 2010; Mazur et al. 2013; Bielecki et al. 2012). A detailed description is published by Lasch (2012).
After that, average spectra from Raman maps of the tumor center and fibrotic tissue region were calculated with the CytoSpec software. In addition, by use of the OPUS software, a difference spectrum of these average spectra was generated by subtraction.
All further data analysis was performed in R using packages hyperSpec (Beleites and Sergo 2014) for import and handling of the data as well as lattice (Sarkar 2008) and ggplot2 (Wickham 2009) for graphical display. For the analyses in R, further preprocessing steps were used: firstly, the uninformative spectral range between 1800 and 2800 cm−1 was deleted, and secondly, the spectra were laterally binned 2 × 2 in order to gain signal to noise ratio. Last but not least, the spectra were normalized to the mean intensity of the C–H stretching region between 2800 and 3050 cm−1.
In order to receive a first overview of the inter-individual tissue variability and to detect different chemical components in the tissue samples, all spectral image data were initially subjected to N-FINDR analysis (Winter 1999). N-FINDR is an unsupervised so-called endmember extraction or spectral unmixing algorithm. It is based on the assumptions that the spectra are stemming from a mixture of a known (pre-specified) number of pure component spectra, and that somewhere in the data these pure component spectra (“endmembers”) are available. Internally, N-FINDR uses a representation of the data which corresponds to a mixture diagram and the endmembers are determined as corners of that mixture diagram. Since this algorithm does not perform an exhaustive search, the results may be sensitive to the selection of initial endmembers (Plaza and Chang 2005). However, this is typically a symptom that the assumptions are not sufficiently well met, in particular that the specified number of endmembers may have not been appropriate. When replicating the N-FINDR analysis 100 times with randomly chosen initial conditions, we always obtained the same set of endmembers, indicating that the obtained solution is stable. In a second step, for each spectrum the so-called abundances which correspond to the concentrations in the mixture are calculated. Abundances were subjected to a non-negativity constraint as concentrations can take positive values only.
N-FINDR has been used to visualize the variability of Raman spectra within malignant and non-malignant brain tissue regions (Bergner et al. 2012) and within single cells (Hedegaard et al. 2011; Stiebing et al. 2014). Our calculations were carried out using package unmixR (McManus et al. 2013) specifying that 6 endmembers should be extracted. The endmember spectra were compared with Raman spectra of reference substances (proteins, collagen, triglycerides, and cholesterol ester) (De Gelder et al. 2007; Bonifacio et al. 2010; Bonifacio and Sergo 2010; Krafft et al. 2005; Köhler et al. 2009).
As the N-FINDR results indicate a high variability of the biochemical composition within each tissue class (see “Results” section), we chose a nonlinear algorithm for classification. High variability between a comparably small number of patients implies that any kind of spectroscopic classification can become unstable, that is, the model and its predictions change substantially if, e.g., new patients become available in the training data. We meet this difficulty in two ways: Firstly, our choice of classification algorithm is an ensemble model. Ensemble models account for this instability by internally varying the training data base. Briefly, the process is like obtaining the average of multiple noisy measurements—just that here it is the model that is considered “noisy” (Beleites and Salzer 2008). Ensemble models of vibrational spectroscopic data have been successfully used for detection of a variety of diseases (Beleites and Salzer 2008; Menze et al. 2007; Teh et al. 2009; Kallenbach-Thieltges et al. 2013; Ollesch et al. 2013). The random forest algorithm applied here was first described by Breiman (1996). Briefly, decision trees are grown that partition the data according to thresholds for individual wavenumber positions. The “forest” consists of a number of such decision trees which are grown on a randomly chosen (“bootstrapped”) subset of spectra and wavenumbers. We used package random Forest (Liaw and Wiener 2002) to grow an ensemble of 500 trees, limiting the leaf node size to not less than 100 spectra. It should be noted that the algorithm is not aware of the hierarchical nature of the data, namely that many spectra are available of each patient. Therefore, the usual out-of-bag classification error estimate (Breiman 1996) cannot be used.
Instead, the performance of the classifier was measured with 101× iterated patient-wise sevenfold cross-validation, (Kohavi 1995; Beleites et al. 2005) which was parallelized using package snow (Tierney et al. 2013). The iterated setup here allows to measure not only the average performance but also the stability of the predictions (Beleites and Salzer 2008).
Results
Comparative analysis of Raman maps
Three regions of interest, including tumor center, fibrotic tissue and tumor margin, of in total 23 tissue samples from 23 patients were measured with Raman imaging spectroscopy, pre-processed, analyzed and correlated with HE-stained microscopy images of the same tissue regions. After the Raman measurements all cryosections were examined a second time by a pathologist and classified into malignant regions of HCC (n = 12, 243713 spectra), cancer-free regions of fibrosis (n = 17, 330876 spectra) and regions of tumor margin (n = 8, 44709 spectra). Raman spectroscopy allows detecting all substances in the sample at the same time. Despite the strong similarities between the spectra of many biomolecules, there are significant differences in the band positions for different cellular compounds such as lipids and proteins. The chemometric data analysis tools can be used to evaluate precisely the spectral data and find spectral differences for the classification of malignant tumors.
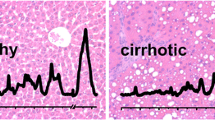
Figure 1 shows the Raman spectra of the main biological components that were found in the tissue sections. The protein spectrum reflects the typical Raman features of the protein backbone and its residues. The CH-stretching vibrations are located between 2800 and 3100 cm−1, the stretching of the C=O of the amide groups (usually referred to as Amide I band) is centered around 1655 cm1, CH2 deformations are observed at 1450 cm−1. The spectral region between 1200 and 1350 cm−1 is due to bending vibrations of the amide bonds (usually referred to as Amide II band) and other CH deformations. The small but reproducible band at 1003 cm−1 is associated with phenylalanine. The spectrum of collagen is very similar to those other proteins, but exhibits some additional features due to its unique triple helical structure as for instance the bands at 940 and 920 cm−1. The bands of the Raman spectra of triglycerides and cholesterol esters can all be assigned to deformations of the alkyl chains and the steran skeleton.
Abundance plots of HCC regions (n = 12) and fibrosis regions (n = 17) using the N-FINDR algorithm: proteins (row No 1, yellow), collagen (row No 2, green), triglycerides (row No 3, blue) and cholesterol ester (row No 4, red). The Raman spectra of the reference substances were acquired with the same Raman instrument and can be seen in the upper-left corner
For each Raman map, the average spectra were calculated and grouped according to the tissue type. Higher band intensities appeared at 2932, 2898, 1654, 1441, 1299, 1261, 1123, 1076, 1001, 953, 923, 851, 748, 721, and 645 cm−1 in regions of HCC (Fig. 2a) and 2944, 1661, 1448, 1341, 1257, 1001, 937, 856, 747, and 668 cm−1 in regions of fibrosis (Fig. 2b).
Average Raman spectra of (a) HCC (n = 12) and (b) fibrosis (n = 17) regions in the spectral range of 3100–600 cm−1. c Difference spectrum (×5) of HCC versus fibrosis. Identified Raman bands are indicated by the corresponding wavenumber (in cm−1). The numbers colored in blue can be assigned to lipids and colored in green to collagen. Shaded areas of the spectra a and b represents the standard deviations
The spectral changes of the two tissue types can be analyzed more precisely by difference spectra, which are plotted in Fig. 2c. The positive and negative bands of the difference spectra correspond to the main differences in cancer and fibrotic tissue, respectively. The identified band positions of the intracellular constituents were correlated with reference databases of Raman spectra (De Gelder et al. 2007; Movasaghi et al. 2007). The positive bands relating to regions of HCC showed characteristic bands of lipids or rather saturated and unsaturated fatty acids at the CH-stretching region 2800–3200 cm−1 and at the wavelengths of 1655, 1439, 1298, 1087, 1063, and 890 cm−1. The negative bands of the difference spectra relating to regions of fibrosis showed the characteristic features of collagen near 1387, 1343, 1242, 937, 857 cm−1. In addition, in both tissue types (HCC and fibrosis), different protein bands were identified (1677, 1017, 851, 754, 668, 645, 637 cm−1).
Unsupervised unmixing of Raman microspectroscopic images using N-FINDR
Using the N-FINDR algorithm, four endmembers were obtained in all tissue regions that reveal spectral signatures of proteins, triglycerides, collagen, and cholesterol esters. The color-coded Raman maps of the obtained endmembers in regions of HCC and fibrotic tissue can be seen in Fig. 1. HCC tissue mostly included proteins or triglycerides as main components and only small amount of collagen or cholesterol esters were found. The fibrotic tissue consists of collagen, proteins, and small amount of cholesterol ester. No triglyceride signatures were found in the Raman maps of fibrotic regions. These results correlate well with the typical chemical composition of HCC and fibrotic tissue (George and Chandrakasan 2000). Although none of these identified endmembers showed a significant difference in the visualized Raman maps, the results of the analysis demonstrates the heterogeneity and complexity of our data set. Because the N-FINDR analysis resulted in a high inter-individual variability of the tissue samples and a heterogeneous distribution of typical chemical components of HCC and fibrotic tissue was identified, the data analysis was supplemented by a nonlinear classification algorithm.
Random forest classification model and variables importance plot
In order to identify relevant signatures and to classify malignant and non-malignant liver tissue, a random forest classification model was applied to the complete data set of regions of HCC and fibrosis. A 101× iterated sevenfold cross-validation estimating classifier performance characteristics resulted in an accuracy of 86 % (sensitivity of 76 % and specificity of 93 %) to predict HCC. The spectral bands used by the random forest classifier were ranked by importance. In Fig. 3, the variables importance plot of the random forests is demonstrated. Based on the importance intensities of variables, the ten most discriminating bands were determined at 2895, 2856, 1439, 1298, 1080, 1063, 1023, 937, 920, and 719 cm−1.
Prediction maps
In addition, the calculated cross-validated classification model was used to generate color-coded prediction maps of unknown regions of tumor margins. Consequently, each Raman map of the region in-between cancer and fibrosis was predicted and assembled as a color-coded image (Fig. 4). The color-intensity scale illustrates the positive decisions of the tree predictors in percentage. The dark blue color is consistent with 0 % positive decisions for HCC and the color red is consistent with 100 % positive decisions for HCC. In total, 8 unknown regions of tumor margin were predicted into HCC and fibrosis. By correlating the color intensity of the prediction maps with the corresponding tissue architecture of the same sample location in the HE-stained tissue section, the classifier discriminates between regions of HCC and fibrosis with high accuracy using a cut-off value of 50 %.
Discussion
To develop innovative and effective diagnostic methods and therapeutic strategies, it is crucial to first identify the target molecules of the underlying disease. By the combination of highly specific Raman spectroscopic information and the spatial resolution of microscopic imaging, the novel Raman imaging technology is a very promising and powerful tool, especially in terms of sensitivity and specificity, to detect and localize a variety of unknown molecules at the cellular level in a label-free tissue manner. Thus, this technique provides spectral information at molecular level of the underlying tissue structure.
Following on a previous study, Tolstik et al. (2013) in which we investigated liver cancer cell lines of HCC (HepG2) and adenocarcinoma (SK-Hep1) by Raman imaging, we analyzed in this study a number of different regions of human HCC and fibrosis in sections of cirrhotic liver tissue with this spectroscopic technique. For this approach, acquired Raman imaging data of predefined malignant and non-malignant tissue regions were subjected to a comparative analysis and were used to establish a classification model based on a random forest algorithm. A cross-validation technique was performed to estimate the prediction accuracy of this model. Furthermore, we applied the developed algorithm to Raman imaging data of tumor margins and the adjacent fibrotic tissue to verify the predictive performance of the resulted classifier.
For diagnostic use, the obtained Raman pattern was applied in the above-described random forests model. Estimating classifier performance characteristics, a cross-validation was performed that resulted in a sensitivity of 76 % [95 % c.i. using an effective sample size of 12 patients (Dorai-Raj 2014): 49–93 %] and specificity of 93 % (73–99 %). The prediction maps of the tumor margin regions (Fig. 4) visualize the prediction accuracy of the classification algorithm in form of a color-coded map. Besides the tumor area, even minimal amounts of tumor cells dispersed over the tumor margin might be detectable in the tissue section using this high-resolution detection method. For some other tumor entities, the assessment of tumor margin by Raman spectroscopy has already been successfully demonstrated for in vivo (using an optical fiber Raman probe) and ex vivo experimental approaches (Hughes et al. 2010; Haka et al. 2006). The results of these studies and our findings indicate that vibrational spectroscopy techniques, such as Raman spectroscopy, have an enormous potential for the future to assess intraoperatively the tumor margin of HCC in real time during liver resection and with potential application in other malignancies.
In addition to these results, the random forest model provides the so-called variable importance measure, which is shown in Fig. 3. Here, the measure depends on the increase in classification errors if the wavenumber channel in question is not available (Liaw and Wiener 2002). It is important to realize that the vibrational spectrum of one substance consists of a number of different bands due to the different vibrational modes that are possible within the molecule. In principle, all these bands are equivalent to each other. Thus, while high importance implies that the band in question and therefore also the biochemical substance giving raise to this band is important for the distinction of HCC from fibrosis, we cannot conclude anything from low importance values: Not allowing the use of one specific band (say, a symmetric stretching band), the random forest may just switch over to use one of the other vibrations (e.g., the antisymmetric stretching or a deformation) of the molecule in question.
The ten most important spectral channels were identified at the wavenumbers 2856, 1298, 937, 920, 1439, 1080, 2895, 1023, 1063, and 719 cm−1 (ranked in order of importance). By comparison of these most important bands with the difference spectrum of malignant and non-malignant tissue regions, four bands at the wavelength 2856, 1298, 1439, and 1063 cm−1 were assigned to regions of HCC, whereas only one band (937 cm−1) among the ten most important bands corresponds to regions of fibrosis (Table 2, column 1, 2). In addition to the ten most important variables, two other Raman bands 1655 and 889 cm−1 of the variables importance plot were positive (HCC) and five other important variables at 1677, 1387, 1242, 856, and 668 cm−1 were negative (fibrosis) in the difference spectrum. The Raman bands at 2895, 1080, 1023, 920, and 719 cm−1 of the importance plot could not be attributed to a specific tissue region. However, the bands at 1080 and 719 cm−1 were assigned to the phospholipid phosphatidylcholine. The occurrence of the choline band at 719 cm−1 was not expected because a difference band was not evident (Fig. 2). The role of phosphatidylcholine in detection of fibrosis and HCC will be studied in more detail, after more specimens are available.
Using independent reference databases of Raman spectra for biological molecules (De Gelder et al. 2007b; Bonifacio et al. 2010; Bonifacio and Sergo 2010; Krafft et al. 2005; Köhler et al. 2009) (Table 2, column 3, 4), the most discriminating bands detected in HCC tissue regions could be correlated with Raman signature of the palmitic acid at 1438, 1296, 1063, and 893 cm−1. In contrast to this, the most discriminating band at 937 cm−1 detected in fibrotic tissue regions was assigned to collagen. Furthermore, additional bands of the variables importance plot that simultaneously appeared in the difference spectrum were attributed to unsaturated or saturated fatty acids (vaccenic acid near 889 cm−1 and oleic acid near 1080, 1023 cm−1, or stearic acid near 890 cm−1) in regions of HCC and other Raman bands of collagen (1246, 940, 820, and 857 cm−1) in regions of fibrosis (Table 2) (De Gelder et al. 2007).
The results of this study are in good agreement with our previously obtained spectral data and classification algorithm of liver cancer cell lines using the Raman imaging technique Tolstik et al. (2013) (Table 2, column 5) and are in accordance with the current state of research relating to the “de novo lipogenesis” in cancer (Menendez and Lupu 2007; Currie et al. 2013). In a very recently published study, Tolstik et al. (2013) we were able to discriminate and classify liver cancer cells, including cell lines of HCC (HepG2) and adenocarcinoma of the liver (SK-Hep1), and their proliferative stages with Raman imaging. The main detected differences in Raman spectra were attributed to a higher expression of fatty acids in the HCC cell line and in the exponential phase of tumor cell growth.
Currently, it is already known that aberrant lipid biosynthesis is an important mechanism in pathogenesis of liver diseases and HCC. A small-scale study of 10 HCC tissue samples published in 2005 described already an elevated expression of mRNA for fatty acid synthase (FASN), acetyl-CoA carboxylase (ACAC) and ATP citrate lyase (ACLY) in HCC tissue (Yahagi et al. 2005; Calvisi et al. 2011; Gao et al. 2006). In 2011, a large-scale study demonstrated a progressive upregulation from non-malignant liver tissue toward the HCC of all relevant lipogenic enzymes, which are involved in the fatty acid synthesis (FASN, ACAC, ACLY, ME, and SCD1), the cholesterol biosynthesis (SREBP2, HMGCR, MVK, and SQS) and their upstream inductors (chREBP, SREBP1, and LXR-β). This latter study showed as well that a higher expression of these enzymes correlate with an activation of a lipogenic pathway (AKT-mTORC1-RPS6) and the clinical aggressiveness, characterized by shorter and longer patient survival (Calvisi et al. 2011).
These results have recently been substantiated by another published study that identified lipid biomarkers associated with progression of HCC and patient outcomes using a combination of gene expression and metabolic profile analysis. In this study, the authors identified 28 metabolites and 169 genes associated with aggressive HCC. Lipid metabolites of stearoyl-CoA-desaturase (SCD) activity were associated with aberrant palmitate signaling in aggressive HCC samples. Expression of gene products associated with these metabolites, including SCD, were associated independently with survival times and tumor recurrence in the test and validation sets. Combined expression of SCD and α-fetoprotein were associated with outcomes of patients with early-stage HCC. Levels of palmitic acid, the product of SCD activity, were increased in aggressive HCCs; palmitic acid increased migration and invasion of cultured HCC cells and colony formation by HCC cells. HCC cells that expressed small interfering RNA against SCD had decreased cell migration and colony formation in culture and reduced tumorigenicity in mice (Budhu et al. 2013).
Despite of the promising data demonstrated by this study, some limitations should be mentioned. First, the size of the sample collective of the study is limited. This is attributed to the fact that in this study frozen tissue samples predefined for HCC were used to avoid potential artifacts caused by sample modification by some kind of fixation. Clearly, before considering the use of a Raman imaging algorithm for molecular classification of HCC patients suited for confirmation of diagnosis or risk stratification, future large-scale studies will be required to confirm our findings and to correlate the results of the spectral data with testing results of approved methods. Second, Raman measurements were taken from ex vivo tissue.
Conclusion
In summary, we have demonstrated that the use of Raman imaging can provide a classification and prediction model for HCC in cirrhotic liver tissue. Furthermore, the most discriminating bands detected in HCC tissue were identified as palmitic acid. Increased levels of saturated palmitic acid, as well as its activating enzyme (SCD), were associated with aggressive HCC, characterized by survival times, tumor recurrence and outcomes of patients with early-stage HCC plus increased migration and invasion of cultured HCC cells (Budhu et al. 2013). Due to the fact that aberrant lipid biosynthesis, upregulation of relevant lipogenic enzymes and activation of lipogenic pathways with a resulting imbalance of lipogenic components are important factors in pathogenesis and aggressiveness of HCC, effective diagnostic methods, including Raman spectroscopy, could be helpful to identify these lipid biomarkers. Even a future use of Raman spectroscopy for intraoperative risk stratification would be conceivable that may cause a change in the surgical procedure. Although subject to validation of our findings in larger HCC cohorts, the demonstrated results affirm that a classifier based on Raman imaging data could provide additional molecular information for diagnostics and risk stratification in HCC.
References
Beleites C, Salzer R (2008) Anal Bioanal Chem 390:1261–1271
Beleites C, Sergo V (2014) HyperSpec: a package to handle hyperspectral data sets in R, version 0.98-20140612. http://hyperspec.r-forge.r-project.org
Beleites C, Baumgartner R, Bowman C, Somorjai R, Steiner G, Salzer R, Sowa MG (2005) Chemom Intell Lab Syst 79:91–100
Beljebbar A, Bouché O, Diébold MD, Guillou PJ, Palot JP, Eudes D, Manfait M (2009) Crit Rev Oncol Hematol 72:255–264
Bergholt MS, Zheng W, Lin K, Ho KY, Teh M, Yeoh KG, So JBY, Huang Z (2010) Analyst 135:3162–3168
Bergner N, Krafft C, Geiger KD, Kirsch M, Schackert G, Popp J (2012) Anal Bioanal Chem 403:719–725
Bielecki C, Bocklitz TW, Schmitt M, Krafft C, Marquardt C, Gharbi A, Knösel T, Stallmach A, Popp J (2012) J Biomed Opt 17:076030
Bonifacio A, Sergo V (2010) Vib Spectrosc 53:314–317
Bonifacio A, Beleites C, Vittur F, Marsich E, Semeraro S, Paoletti S, Sergo V (2010) Analyst 135:3193–3204
Breiman L (1996) Mach Learn 24:123–140
Bruix J, Sherman M (2011) Hepatology 53:1020–1022
Budhu A, Roessler S, Zhao X, Yu Z, Forgues M, Ji J, Karoly E, Qin L-X, Ye Q-H, Jia H-L, Fan J, Sun H-C, Tang Z-Y, Wang XW (2013) Gastroenterology 144:1066–1075 e1
Calvisi DF, Wang C, Ho C, Ladu S, Lee SA, Mattu S, Destefanis G, Delogu S, Zimmermann A, Ericsson J, Brozzetti S, Staniscia T, Chen X, Dombrowski F, Evert M (2011) Gastroenterology 140:1071–1083
Currie E, Schulze A, Zechner R, Walther TC, Farese RV Jr (2013) Cell Metab 18:153–161
De Gelder J, De Gussem K, Vandenabeele P, Moens L (2007a) J Raman Spectrosc 38:1133–1147
De Gelder J, De Gussem K, Vandenabeele P, Moens L (2007b) J Raman Spectrosc 38:1133–1147
Di Tommaso L, Destro A, Seok JY, Balladore E, Terracciano L, Sangiovanni A, Iavarone M, Colombo M, Jang JJ, Yu E, Jin SY, Morenghi E, Park YN, Roncalli M (2009) J Hepatol 50:746–754
Dorai-Raj S (2014) Binom: binomial confidence intervals for several parameterizations, version 1.1-1. http://CRAN.R-project.org/package=binom
European Association for the Study of the Liver and European Organisation for Research and Treatment of Cancer (2012) J Hepatol 56:908–943
Gao Y, Lin L-P, Zhu C-H, Chen Y, Hou Y-T, Ding J (2006) Cancer Biol Ther 5:978–985
George J, Chandrakasan G (2000) Clin Biochem 33:563–570
Haka AS, Volynskaya Z, Gardecki JA, Nazemi J, Lyons J, Hicks D, Fitzmaurice M, Dasari RR, Crowe JP, Feld MS (2006) Cancer Res 66:3317–3322
Hedegaard M, Matthäus C, Hassing S, Krafft C, Diem M, Popp J (2011) Theor Chem Acc 130:1249–1260
Hughes OR, Stone N, Kraft M, Arens C, Birchall MA (2010) Head Neck 32:1544–1553
Iavarone M, Colombo M (2013) Clin Liver Dis 17:375–397
Kallenbach-Thieltges A, Großerüschkamp F, Mosig A, Diem M, Tannapfel A, Gerwert K (2013) J Biophotonics 6:88–100
Kamemoto LE, Misra AK, Sharma SK, Goodman MT, Luk H, Dykes AC, Acosta T (2010) Appl Spectrosc 64:255–261
Kohavi R (1995) A study of cross-validation and bootstrap for accuracy estimation and model selection. In: Proceedings of the 14th international joint conference on artificial intelligence. Morgan Kaufmann, USA, pp 1137–1145
Köhler M, Machill S, Salzer R, Krafft C (2009) Anal Bioanal Chem 393:1513–1520
Krafft C, Neudert L, Simat T, Salzer R (2005) Spectrochim Acta A Mol Biomol Spectrosc 61:1529–1535
Lasch P (2012) Chemometr Intell Lab Syst 117:100–114
Liaw A, Wiener M (2002) R News 2:18–22
Matousek P, Stone N (2009) Analyst 134:1058–1066
Mazur AI, Monahan JL, Miljković M, Laver N, Diem M, Bird B (2013) J Biophotonics 6:101–109
McManus C, Fuller S, Hanson B, Beleites C (2013) UnmixR: spectral unmixing methods, version 0.1-6. http://github.com/Chathurga/unmixR
Menendez JA, Lupu R (2007) Nat Rev Cancer 7:763–777
Menze BH, Petrich W, Hamprecht FA (2007) Anal Bioanal Chem 387:1801–1807
Movasaghi Z, Rehman S, Rehman IU (2007) Appl Spectrosc Rev 42:493–541
Neugebauer U, Clement JH, Bocklitz T, Krafft C, Popp J (2010) J Biophotonics 3:579–587
Ollesch J, Drees SL, Heise HM, Behrens T, Brüning T, Gerwert K (2013) Analyst 138:4092–4102
Plaza A, Chang C-I (2005) An improved N-FINDR algorithm in Implementation, algorithms and technologies for multispectral, hyperspectral, and ultraspectral imagery XI. Proc SPIE 5806:298–306. doi:10.1117/12.602373
R Core Team (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Sarkar D (2008) Lattice: multivariate data visualization with R. Springer, New York
Stiebing C, Matthäus C, Krafft C, Keller A-A, Weber K, Lorkowski S, Popp J (2014) Complexity of fatty acid distribution inside human macrophages on single cell level using Raman micro-spectroscopy. Anal Bioanal Chem. doi:10.1007/s00216-014-7927-0
Teh SK, Zheng W, Lau DP, Huang Z (2009) Analyst 134:1232–1239
Tierney L, Rossini AJ, Li N, Sevcikova H (2013) Snow: simple network of workstations, version 0.3-13. http://CRAN.R-project.org/package=snow
Wickham H (2009) ggplot2: elegant graphics for data analysis. Springer, New York. ISBN 978-0387981406. http://had.co.nz/ggplot2/book
Winter ME (1999) N-FINDR: an algorithm for fast autonomous spectral end-member determination in hyperspectral data. Proc SPIE 3753:266–275
Tolstik T, Marquardt C, Matthäus C, Bergner N, Bielecki C, Krafft C, Stallmach A, Popp J (2013) Discrimination and classification of liver cancer cells and proliferation states by raman spectroscopic imaging. Anal (submitted)
Yahagi N, Shimano H, Hasegawa K, Ohashi K, Matsuzaka T, Najima Y, Sekiya M, Tomita S, Okazaki H, Tamura Y, Iizuka Y, Ohashi K, Nagai R, Ishibashi S, Kadowaki T, Makuuchi M, Ohnishi S, Osuga J, Yamada N (2005) Eur J Cancer 41:1316–1322
Acknowledgments
We are grateful to all co-workers from the Gastroenterology Laboratory of the Jena University Hospital and the Spectroscopy/Imaging group in IPHT for their support and help. Financial support from the “Deutsche Forschungsgemeinschaft (DFG)”, the “Europäischer Fonds für Regionale Entwicklung (EFRE)” and the “Thüringer Ministerium für Bildung, Wissenschaft und Kultur (TMBWK)” are gratefully acknowledged. CB Acknowledges Funding by the BMBF via project RamanCTC (AZ 13N12685).
Conflict of interest
We declare that we have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Tolstik, T., Marquardt, C., Beleites, C. et al. Classification and prediction of HCC tissues by Raman imaging with identification of fatty acids as potential lipid biomarkers. J Cancer Res Clin Oncol 141, 407–418 (2015). https://doi.org/10.1007/s00432-014-1818-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-014-1818-9