Abstract
Higher alcohols significantly influence the quality and flavor profiles of Chinese Baijiu. ILV1-encoded threonine deaminase, LEU1-encoded α-isopropylmalate dehydrogenase, and LEU2-encoded β-isopropylmalate dehydrogenase are involved in the production of higher alcohols. In this work, ILV1, LEU1, and LEU2 deletions in α-type haploid, a-type haploid, and diploid Saccharomyces cerevisiae strains and ILV1, LEU1, and LEU2 single-allele deletions in diploid strains were constructed to examine the effects of these alterations on the metabolism of higher alcohols. Results showed that different genetic engineering strategies influence carbon flux and higher alcohol metabolism in different manners. Compared with the parental diploid strain, the ILV1 double-allele-deletion diploid mutant produced lower concentrations of n-propanol, active amyl alcohol, and 2-phenylethanol by 30.33, 35.58, and 11.71%, respectively. Moreover, the production of isobutanol and isoamyl alcohol increased by 326.39 and 57.6%, respectively. The LEU1 double-allele-deletion diploid mutant exhibited 14.09% increased n-propanol, 33.74% decreased isoamyl alcohol, and 13.21% decreased 2-phenylethanol production, which were similar to those of the LEU2 mutant. Furthermore, the LEU1 and LEU2 double-allele-deletion diploid mutants exhibited 41.72 and 52.18% increased isobutanol production, respectively. The effects of ILV1, LEU1, and LEU2 deletions on the production of higher alcohols by α-type and a-type haploid strains were similar to those of double-allele deletion in diploid strains. Moreover, the isobutanol production of the ILV1 single-allele-deletion diploid strain increased by 27.76%. Variations in higher alcohol production by the mutants are due to the carbon flux changes in yeast metabolism. This study could provide a valuable reference for further research on higher alcohol metabolism and future optimization of yeast strains for alcoholic beverages.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chinese Baijiu, also known as Chinese liquor, is one of the six most well-known distillates in the world. The alcoholic beverage is a traditional drink in China for more than a thousand years. This beverage presents a unique smell and taste that attract numerous customers. During the fermentation of Chinese Baijiu, flavor compounds result from the metabolic activity of the microbial community (Wang et al. 2008; Wu et al. 2015; Zheng et al. 2011). These compounds exhibit interesting organoleptic properties that determine the taste and quality of Chinese Baijiu. Higher alcohol (fusel alcohol) is one of the most abundant and most important groups of these aroma compounds (Fan and Qian 2006; Xiao et al. 2014; Zhu et al. 2007).
Higher alcohols significantly influence the organoleptic property of alcoholic beverages for their pungent and strong flavor (Eden et al. 2001; Pires et al. 2014; Stribny et al. 2016; Swiegers and Pretorius 2005). These compounds exhibit different flavor characteristics. Different proportions and contents of these compounds in coexistence with other flavor compounds are required to obtain different flavor types of Chinese Baijiu (Fan and Qian 2005, 2006). With the appropriate content and proportion of higher alcohols, Chinese Baijiu is mellow and savory. In contrast, excessive amounts of higher alcohols result in a strong fusel oil flavor and can potentially harm human health, causing cerebral paralysis and other symptoms (Valero et al. 2002; Yang et al. 2014). Higher alcohols are the substrate for synthesis of acetate esters (Yuan et al. 2016), another important group of volatile flavor substances in Chinese Baijiu.
The production of higher alcohols is mainly the function of Saccharomyces cerevisiae during the traditional fermentation of Chinese Baijiu (Wu et al. 2015). These compounds are synthesized from α-keto acids via decarboxylation and dehydrogenation reactions. As shown in Fig. 1, n-propanol, isobutanol, isoamyl alcohol, and active amyl alcohol are synthesized from α-ketobutyrate, α-ketoisovalerate, α-ketoisocaproate, and α-keto-β-methylvalerate through steps comprising decarboxylation and dehydrogenation reactions (Chen et al. 2011; Kobayashi et al. 2008; Park et al. 2014). α-Keto acids originate from the biosynthetic pathway (Dickinson and Norte 1993) or from the degradation of amino acids via the Ehrlich pathway (Derrick and Large 1993; Gietz and Woods 2002). Threonine deaminase, α-isopropylmalate dehydrogenase, and β-isopropylmalate dehydrogenase are three key enzymes involved in α-keto acid synthesis.
The ILV1 gene encodes threonine deaminase, which converts threonine to α-ketobutyrate. This deaminase is the first enzyme in the anabolic pathway of isoleucine (Petersen et al. 1983). α-Keto-β-methylvalerate, the isoleucine precursor, is synthesized in the anabolic pathway. Previous studies showed that S. cerevisiae mutants with ILV1 deletion are isoleucine auxotrophic (Holmberg and Petersen 1988; Ida et al. 2015; Petersen et al. 1983). S. cerevisiae mutants with ILV1 deletion exhibited 3.5-fold increased isobutanol production (Ida et al. 2015). Moreover, α-ketobutyrate can be synthesized by β-methyl malate, which is synthesized from citraconate by LEU1-encoded α-isopropylmalate dehydrogenase (Vollbrecht 1974). α-Isopropylmalate dehydrogenase and LEU2-encoded β-isopropylmalate dehydrogenase are involved in the conversion from α-ketoisovalerate to α-ketoisocaproate in the leucine anabolic pathway (Baichwal et al. 1983; Kobayashi et al. 2008; Park et al. 2014). S. cerevisiae mutants with LEU1 or LEU2 deletion are leucine auxotrophic (Hsu and Schimmel 1984; Nigavekar and Cannon 2002; Sakai and Tani 1992). The additional overexpression of LEU1 exhibited a negative influence on the production of isoamyl alcohol. The introduction of the LEU2 gene into a leucine auxotrophic strain with a LEU2 gene mutation markedly increased the concentration of isoamyl alcohol (Park et al. 2014). The reconstruction of a chromosome-based leucine biosynthetic pathway under the control of galactose-inducible promoters and the overexpression of the mitochondrial α-isopropylmalate transporter could result in the overexpression of LEU1 and LEU2 (Yuan et al. 2017a). Rewiring of the cytosolic α-isopropylmalate synthesis pathway could result in the overexpression of all leucine biosynthetic pathway genes including ILV2, ILV5, ILV3, LEU9, LEU1, and LEU2 (Yuan et al. 2017b). Furthermore, ILV1, LEU1, and LEU2 are all involved in n-butanol biosynthesis (Shi et al. 2016). Research on the three genes has concentrated mainly on the production of one type of higher alcohol as a biorefinery target alcohol in S. cerevisiae. However, the effects of ILV1, LEU1, and LEU2 deletion and single-allele deletion on the metabolism of several higher alcohols and the change in carbon flux remain unclear.
In this study, α-type and a-type haploid S. cerevisiae mutants with ILV1, LEU1, and LEU2 deletions were constructed, respectively. Gene double-allele-deletion diploid mutants were constructed by hybridizing α-type and a-type haploid mutants. Moreover, engineered diploid strains with ILV1, LEU1, and LEU2 single-allele deletions were successfully constructed. The effects of genetic engineering of α-type haploid, a-type haploid, and diploid strains on flavor compounds were investigated in a simulated alcohol fermentation process. Our results reveal that different genetic engineering strategies influence metabolism related to higher alcohol in different manners. Moreover, this study attempts to explain the effects of genetic engineering strategies on synthetic and metabolic pathways. This study could provide a valuable reference for metabolism research and the future optimization of yeast strains for Chinese Baijiu and other alcoholic beverages.
Materials and methods
Strains and plasmids
All strains and plasmids used in this study are summarized in Table 1.
Media and cultivation
Escherichia coli strains were grown in Luria-Bertani (LB) medium (1% NaCl, 0.5% yeast extract, and 1% tryptone) at 37 °C. LB media containing 100 mg/L ampicillin were used to select positive transformants.
S. cerevisiae strains were grown in yeast extract peptone dextrose (YEPD) medium (1% yeast extract, 2% glucose, and 2% peptone) at 30 °C. YEPD media added with G418 (Promega, Madison, WI, USA) were used to select positive transformants harboring the kanMX gene. YEPD media added with 500 mg/L Zeocin (Promega, Madison, WI, USA) were used to select Zeocin-resistant strains. Then, yeast extract peptone D-galactose (YEPG) medium (2% peptone, 2% galactose, and 1% yeast extract) was used to express Cre in the transformants. Mutants and parental strains were grown at 30 °C on synthetic dropout (SD) medium agar plates (0.67% yeast nitrogen base without amino acids and ammonium sulfate, 2% glucose, 2% agar powder, and 0.083% amino acid dropout mix) and SD medium agar plates containing the appropriate essential amino acid (isoleucine or leucine) to verify auxotrophy.
All solid media used in this study contained 2% agar powder (Solarbio, Beijing, China).
Construction of plasmids
The plasmid pUG6 was employed as amplification template to obtain the kanMX gene for G418 resistance. The plasmid pUC19 was employed as the backbone to construct recombinant plasmids. Plasmid and genomic yeast DNAs were extracted from E. coli strain DH5α by the Plasmid Mini Kit II (D6945, Omega, Norcross, GA, USA) and from industrial S. cerevisiae strain α5 by the yeast DNA kit (D3370-01, Omega, Norcross, GA, USA). Table S1 in the Supplementary Material summarizes all primers used in this study.
Plasmid pUC-VABK was structured as follows: the VA (389 bp) and VB (472 bp) fragments were homologous to the upstream and downstream areas of the ILV1 gene, respectively. Fragments were amplified from genomic yeast DNA with primers VA-U/VA-D and VB-U/VB-D via polymerase chain reaction (PCR). The VA and VB fragments were digested with the appropriate endonucleases and inserted into the HindIII/BamHI and BamHI/EcoRI sites of pUC19 to construct the plasmid pUC-VAB. Then, the loxP-kanMX-loxP fragment (1613 bp), which was amplified from the plasmid pUG6 with primers K-U/K-D via PCR, was inserted into the BamHI site of the plasmid pUC-VAB to create the recombinant plasmid pUC-VABK. Plasmids pUC-LABK and pUC-L2ABK were constructed similarly. The PCR-generated LA (383 bp) and LB (402 bp) fragments were successively inserted into the HindIII/BamHI and BamHI/EcoRI sites of pUC19 to construct the plasmid pUC-LAB. The loxP-kanMX-loxP fragment was inserted into the BamHI site of the plasmid pUC-LAB to construct the recombinant plasmid pUC-LABK. The PCR-generated L2A (287 bp) and L2B (510 bp) fragments were successively inserted into the BamHI/HindIII and EcoRI/BamHI sites of pUC19 to construct the plasmid pUC-L2AB. The loxP-kanMX-loxP fragment was inserted into the BamHI site of plasmid pUC-LAB to construct the recombinant plasmid pUC-L2ABK.
Yeast transformation and selection
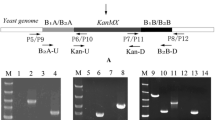
The constructed fragments VA-loxP-kanMX-loxP-VB, LA-loxP-kanMX-loxP-LB, and L2A-loxP-kanMX-loxP-L2B were amplified via PCR from the recombinant plasmids pUC-VABK, pUC-LABK, and pUC-L2ABK by using the VA-U/VB-D, L1A-U/L1B-D, and L2A-U/L2B-D primers, respectively. Then, the constructed cassettes were transferred into yeast cells by means of the lithium acetate/polyethylene glycol (PEG) strategy (Gietz and Woods 2002). Recombinants were screened using YEPD media containing G418 (diploid, 1200 mg/L; a-type haploid, 1400 mg/L; and α-type haploid, 1000 mg/L) and PCR with the primers summarized in Table S1 in the Supplementary Material. The resistance gene kanMX of the mutants was eliminated by utilizing the Cre/loxP procedure (Güldener et al. 1996).
Construction of the diploid recombinants
Diploid recombinants were constructed by hybridizing a-type and α-type haploid recombinant strains. Haploid a-type and α-type cells were grown in YEPD media (5 mL) for 24 h at 30 °C. Then, the cells (0.5 mL each) were transferred into fresh YEPD medium (5 mL) and grown and hybridized for 24 h at 30 °C. The diploid recombinants were screened by utilizing MacConkey medium (0.82% sodium acetate, 0.25% yeast extract, 0.18% NaCl, 0.1% glucose, and 2% agar) and PCR with the primers MAT-F/MAT-a/MAT-α. Spore formation of the obtained diploid recombinant strains was observed using a microscope (Olympus, Tokyo, Japan).
Fermentation experiments
S. cerevisiae cells were fermented in a medium (16% glucose, 1% yeast extract, and 2% peptone) in the simulated alcohol fermentation process at 30 °C. Yeast strains were precultured in 8 °Bx YEPD medium (4 mL) at 30 °C for 24 h. Yeast cells and the medium were subsequently transferred into 12 °Bx YEPD medium (36 mL) in a 100-mL conical flask and cultured at 30 °C for 16 h. The second precultured yeast (15 mL) was transferred into 16 °Bx YEPD medium (135 mL) in a 250-mL conical flask. Fermentation was carried out at 30 °C until a CO2 weight loss less than 1 g was achieved after an interval of 12 h. Fermentation experiments were all executed in triplicate.
Fermentation performance in terms of CO2 weight loss, ethanol production, and residual sugar was measured using an analytical balance, an oenometer, and a Brix hydrometer, respectively. The produced higher alcohols were determined using gas chromatography (GC) analysis.
GC analysis
GC has been widely used in the analysis of volatile compounds in Chinese Baijiu (Fan and Qian 2005). Samples from simulated alcohol fermentation process were distilled after fermentation. Then, the distilled samples were used for GC analysis.
Volatile substances were analyzed on an Agilent (Palo Alto, CA, USA) 7890C gas chromatograph with HP-INNOWax polyethylene glycol column (0.5-μm coating thickness and 30 m × 320 μm internal diameter; Lab Alliance, Fayetteville, NY, USA). The machine was provided with a flame ionization detector (FID), an injector, and an Agilent (Palo Alto, CA, USA) G4513A autosampler. Nitrogen at a constant flow rate of 2 mL/min was used as the carrier gas. The injection parameters were 200 °C injector temperature, 10:1 split ratio, and 1 μL injection volume. The FID operating temperature was 200 °C. The oven temperature procedure was as follows: 50 °C for 8 min, followed by an increase to 120 °C at 5 °C/min, and the final temperature was sustained for 5 min. n-Butyl acetate was applied as the internal standard. For each measured compound, an internal calibration curve was constructed using a specific number of authentic standards. The chemicals were purchased from Merck (Darmstadt, Germany).
Real-time quantitative PCR
Total yeast RNA was obtained using the Yeast RNAiso Kit (Takara Biotechnology, Dalian, China). Afterward, the RNA was reverse-transcribed using the PrimeScript™ RT Reagent Kit with gDNA Eraser (Perfect Real Time) (Takara Biotechnology, Dalian, China). Changes in the expression levels of key genes were investigated by RT-qPCR using the SYBR Premix Ex Taq II (Tli RNaseH Plus) (Takara Biotechnology, Dalian, China). The primers are summarized in Table S1 in the Supplementary Material. The PCR procedure was composed of predenaturation at 95 °C for 30 s, amplification using 40 cycles of denaturation at 95 °C for 5 s, annealing and polymerization at 60 °C for 30 s, and a melt curve stage at 95 °C for 15 s and at 60 °C for 1 min. The 2−ΔΔCt method was used in quantitative analysis. The ACT1 gene was used as a housekeeping gene.
Determination of the growth curve
Yeast strains were cultured at 30 °C in YEPD media (5 mL) for 12 h. Then, appropriate amounts of cells were transferred into fresh YEPD medium and grown at 30 °C for 15 h. Optical density (OD) was determined at 600 nm by using the Bioscreen Automated Growth Curves analysis system (OY Growth Curves Ab Ltd., Helsinki, Finland).
Statistical analysis
Data were presented as the mean ± standard deviation (SD). Student’s t test was used to analyze the differences of recombinant strains compared with the original strains. Differences were considered to be statistically significant at P < 0.05.
Results
Constructions of haploid and diploid mutants
Strains α5 and a8 are high ethanol-producing α mating type and a mating type haploid strains, respectively, derived from the commercial liquor yeast strain AY15 (Li et al. 2017). The ILV1, LEU1, and LEU2 genes of the α-type haploid strain α5 were replaced with the constructed cassettes ILV1A-loxP-kanMX-loxP-ILV1B, LEU1A-loxP-kanMX-loxP-LEU1B, and LEU2A-loxP-kanMX-loxP-LEU2B, respectively. Recombinant haploid yeast strains α5-ILV1, α5-LEU1, and α5-LEU2 were obtained. Similarly, the ILV1, LEU1, and LEU2 genes of the a-type haploid strain a8 were replaced with the constructed cassettes, and recombinant haploid strains a8-ILV1, a8-LEU1, and a8-LEU2 were obtained. α5-ILV1 and a8-ILV1 were verified to be isoleucine auxotrophic, and α5-LEU1, α5-LEU2, a8-LEU1, and a8-LEU2 were leucine auxotrophic.
α-Type and a-type haploid mutants were hybridized to construct gene double-allele-deletion diploid strains. The ILV1 double-allele-deletion diploid recombinant AY15-ILV1, LEU1 double-allele-deletion diploid recombinant AY15-LEU1, and LEU2 double-allele-deletion diploid recombinant AY15-LEU2 were obtained successfully. AY15-ILV1 was verified to be isoleucine auxotrophic, and AY15-LEU1 and AY15-LEU2 were leucine auxotrophic.
A single allele of ILV1 in the diploid strain AY15 was replaced with the constructed cassette ILV1A-loxP-kanMX-loxP-ILV1B. The ILV1 single-allele-deletion diploid recombinant AY15-ILV1SA was constructed successfully. In the same manner, we obtained the LEU1 single-allele-deletion diploid recombinant AY15-LEU1SA and LEU2 single-allele-deletion diploid recombinant AY15-LEU2SA. AY15-ILV1SA, AY15-LEU1SA, and AY15-LEU2SA were verified to be non-auxotrophic.
Production of higher alcohols by ILV1, LEU1, and LEU2 deletion haploid strains
The effects of ILV1, LEU1, and LEU2 deletion on production of higher alcohols by haploid strains were investigated in the simulated alcohol fermentation. Data on fermentation samples analyzed by GC are presented in Fig. 2. As shown in Fig. 2a, the production of n-propanol by recombinant α5-ILV1 declined to 6.82 mg/L, a 35.3% decrease compared with that of the parental strain α5. Moreover, the concentrations of isobutanol and isoamyl alcohol produced by α5-ILV1 were 240.79 and 77.05 mg/L, respectively, which were approximately 4.5-fold and 1.52-fold higher than those produced by α5. The production of active amyl alcohol by α5-ILV1 diminished to 7.94 mg/L, representing a 34.49% decrease compared with that of α5.
Higher alcohol productions by gene deletion α-type and a-type haploid recombinants and the parental strains. a The production of higher alcohols by ILV1, LEU1, and LEU2 deletion α-type haploid strains (α5-ILV1, α5-LEU1, and α5-LEU2) and α5 in the simulation of ethanol fermentation. b The production of higher alcohols by ILV1, LEU1, and LEU2 deletion a-type haploid strains (a8-ILV1, a8-LEU1, and a8-LEU2) and a8 in the simulation of ethanol fermentation. Error bars indicate standard deviations (SD) of three independent replicate fermentations. Significant difference of the recombinants from their parental strains was confirmed by Student’s t test (★★P < 0.01, ★P < 0.05)
Different from α5-ILV1, recombinant α5-LEU1 and α5-LEU2 showed increased n-propanol production by 18.03 and 15.28% with that of α5, respectively. The isobutanol concentration produced by α5-LEU1 and α5-LEU2 increased by 47.02 and 62.18%, respectively. The isoamyl alcohol concentration produced by α5-LEU1 and α5-LEU2 decreased by 35.77 and 36.82%, respectively. The active amyl alcohol concentration produced by α5-LEU1 and α5-LEU2 were approximately the same as that produced by α5. Moreover, the production of 2-phenylethanol by the recombinant α5-ILV1, α5-LEU1, and α5-LEU2 decreased by 11.16, 11.82, and 13.26%, respectively, compared with that of α5.
Similar results were observed in a-type haploid strains (Fig. 2b). Compared with the parental strain, a8-ILV1 exhibited 29.03% decreased n-propanol, 340.44% increased isobutanol, 53.88% increased isoamyl alcohol, 39.68% decreased active amyl alcohol, and 13.06% decreased 2-phenylethanol production. Similar to a8-LEU2, a8-LEU1 exhibited 15.28% increased n-propanol productivity and 27.34% decreased isoamyl alcohol production. Furthermore, a8-LEU1 and a8-LEU2 exhibited 39 and 62.38% increased isobutanol production, respectively.
These findings indicate that ILV1, LEU1, and LEU2 deletions resulted in different influences on the production of higher alcohols by the haploid yeast strain. Significant differences in the production of isobutanol by LEU1 deletion haploid strains α5-LEU1 and a8-LEU1 and LEU2 deletion haploid strains α5-LEU2 and a8-LEU2.
Production of higher alcohols by ILV1, LEU1, and LEU2 double-allele-deletion diploid strains
The effects of ILV1, LEU1, and LEU2 double-allele deletion on the production of higher alcohols by diploid strains were investigated in the simulated alcohol fermentation. Data on fermentation samples analyzed by GC are presented in Fig. 3.
Higher alcohol productions by the gene deletion diploid recombinants and the parental strain. The production of higher alcohols by ILV1, LEU1, and LEU2 deletion diploid strains (AY15-ILV1, AY15-LEU1, and AY15-LEU2) and AY15 in the simulation of ethanol fermentation. Error bars indicate standard deviations (SD) of three independent replicate fermentations. Significant difference of the recombinants from the parental strains was confirmed by Student’s t test (★★P < 0.01, ★P < 0.05)
The effects of ILV1, LEU1, and LEU2 double-allele deletion on higher alcohol production by diploid strains were similar to that in α-type and a-type haploid mutants with gene deletion (Fig. 3). Compared with the parental strain, the ILV1 double-allele-deletion diploid strain AY15-ILV1 yielded 30.33% decreased n-propanol, 326.39% increased isobutanol, 57.6% increased isoamyl alcohol, 35.58% decreased active amyl alcohol, and 11.71% decreased 2-phenylethanol production. Similar to the LEU2 double-allele-deletion diploid strain AY15-LEU2, the LEU1 double-allele-deletion diploid strain AY15-LEU1 exhibited 14.09% increased n-propanol, 33.74% decreased isoamyl alcohol, and 13.21% decreased 2-phenylethanol production. Furthermore, AY15-LEU1 and AY15-LEU2 exhibited 41.72 and 52.18% increased isobutanol production, respectively.
Production of higher alcohols by ILV1, LEU1, and LEU2 single-allele-deletion diploid strains
The effects of ILV1, LEU1, and LEU2 single-allele deletion on higher alcohol production by diploid strains were investigated in the simulated alcohol fermentation. Data on fermentation samples analyzed by GC are presented in Fig. 4. Compared with the parental strain AY15, ILV1 single-allele-deletion recombinant AY15-ILV1SA showed 31.43% increased production of isobutanol. Negligible differences of other higher alcohol contents were observed in the fermentation samples of AY15-ILV1SA, AY15-LEU1SA, AY15-LEU2SA, and AY15.
Higher alcohol productions by single-allele-deletion diploid recombinants and the parental strain. The production of higher alcohols by ILV1, LEU1, and LEU2 single-allele-deletion diploid strains (AY15-ILV1SA, AY15-LEU1SA, AY15-LEU2SA, and AY15-LEU1SA-LEU2SA) and AY15 in the simulation of ethanol fermentation. Error bars indicate standard deviations (SD) of three independent replicate fermentations. Significant difference of the recombinants from the parental strain was confirmed by Student’s t test (★★P < 0.01)
The kanr marker gene was removed from the engineered strain AY15-LEU1SA using the Cre/loxP recombination system to produce the mutant strain AY15-LEU1SA-1. A single allele of LEU2 in AY15-LEU1SA-1 was replaced with the constructed cassette LEU2A-loxP-kanMX-loxP-LEU2B. The engineered strain AY15-LEU1SA-LEU2SA was constructed. As shown in Fig. 4, the double deletion of LEU1 and LEU2 single alleles did not markedly affect the production of higher alcohols.
These results indicate that the single-allele deletion of the ILV1 gene can increase the production of isobutanol.
mRNA levels of genes related to higher alcohol metabolism
Significant differences were observed in the production of higher alcohols among α5-ILV1, α5-LEU1, α5-LEU2, and AY15-ILV1SA and their parental strains. We quantified the mRNA expression levels of key genes in metabolic pathways to clarify the differences (Figs. 5 and 6). The biosynthesis processes of isoleucine and valine are parallel pathways catalyzed by the same enzymes, namely, acetolactate synthase (encoded by ILV2), ketol-acid reductoisomerase (encoded by ILV5), and dihydroxyacid dehydratase (encoded by ILV3) (Holmberg and Petersen 1988; Kondo et al. 2012; Ryan and Kohlhaw 1974).
Determination of gene expression levels in gene deletion α-type haploid recombinants and the parental strain α5. a The expression levels of ILV1, ILV2, ILV5, and ILV3 in α5-ILV1 and α5. b The expression levels of ILV1 and LEU1 in α5-LEU1 and α5. c The expression levels of ILV1 and LEU2 in α5-LEU2 and α5. Error bars indicate standard deviations (SD) of three independent replicate fermentations. Significant difference of the recombinants from the parental strain was confirmed by Student’s t test (★★P < 0.01, ★P < 0.05)
Determination of gene expression levels in single-allele-deletion diploid recombinants and the parental strain. a The expression levels of ILV1 in AY15-ILV1SA and AY15. b The expression levels of LEU1 in AY15-LEU1SA and AY15. c The expression levels of LEU2 in AY15-LEU2SA and AY15. d The expression levels of ILV2, ILV5, and ILV3 in AY15-ILV1SA and AY15. Error bars indicate standard deviations (SD) of three independent replicate fermentations. Significant difference of the recombinants from the parental strain was confirmed by Student’s t test (★★P < 0.01, ★P < 0.05)
The results of RT-qPCR (Fig. 5) exhibited that ILV1 gene deletion resulted in 2.26-fold, 1.95-fold, and 1.75-fold increases in the expression levels of ILV2, ILV5, and ILV3, respectively. Moreover, the expression levels of ILV1 in α5-LEU1 and α5-LEU2 were 3.31-fold and 3.49-fold higher than that in α5, respectively. As shown in Fig. 6, the expression levels of ILV1, LEU1, and LEU2 in AY15-ILV1SA, AY15-LEU1SA, and AY15-LEU2SA were approximately half of those in AY15, respectively. The expression levels of ILV2, ILV5, and ILV3 in AY15-ILV1SA were slightly higher than those in AY15.
Growth and fermentation characteristics
In the fermentation of Chinese Baijiu, the growth and fermentation performance of yeast strain directly determine liquor yield and fermentation period. Compared with parental strains, the gene deletion strains showed slightly weaker growth characteristics (Fig. 7a–c). The gene single-allele-deletion diploid strains showed similar growth characteristics to their parental strain AY15 (Fig. 7d). Fermentation properties, including residual sugar and weight loss of CO2, were further monitored (Table 2). The engineered strains showed similar fermentative capabilities to their parental strains.
Growth curves of recombinant strains and their parental strains. a The growth curves of the gene deletion α-type haploid recombinants and α5. b The growth curves of the gene deletion a-type haploid recombinants and a8. c The growth curves of the gene double-allele-deletion diploid recombinants and AY15. d The growth curves of the single-allele-deletion diploid recombinants and AY15. Growth curves (in triplicate) were monitored at 30 °C by measuring the optical density (OD) 600 of the cultures every 1.5 h
Discussion
Higher alcohols produced by yeast cells exert a significant influence on the flavor and taste of alcoholic beverages (Swiegers and Pretorius 2005). In different flavor types of Chinese Baijiu, different proportions and contents of higher alcohols are required. This study showed for the first time that deletion of ILV1, LEU1, and LEU2 could alter carbon flux for various productions of higher alcohols by S. cerevisiae. ILV1 deletion resulted in significant changes in the production of n-propanol, isobutanol, isoamyl alcohol, active amyl alcohol, and 2-phenylethanol. LEU1 or LEU2 deletions led to significant changes in the production of n-propanol, isobutanol, isoamyl alcohol, and 2-phenylethanol. Moreover, this study showed for the first time that the single-allele deletion of ILV1 could increase the production of isobutanol, and single-allele deletion of LEU1 or LEU2 did not significantly influence the production of higher alcohols.
The decrease in n-propanol caused by ILV1 deletion was due to the prevented generation of α-ketobutyrate from threonine. Meanwhile, the prevention of α-ketobutyrate generated from threonine resulted in the decrease of α-keto-β-methylvalerate and its corresponding higher alcohol (active amyl alcohol). Besides, ILV1 deletion significantly affected the isobutanol, isoamyl alcohol, and 2-phenylethanol biosynthesis pathways, which were not directly related to the ILV1 gene. A higher concentration of isobutanol relative to that of parental strains was obtained with ILV1 deletion strains (Figs. 2 and 3). These results are in accordance with those of Ida et al. (2015), who reported that S. cerevisiae mutants with ILV1 deletion showed 3.5-fold increased production of isobutanol. However, the group did not investigate the effects of ILV1 deletion on other higher alcohols. Furthermore, our work showed that the expression levels of ILV2, ILV5, and ILV3 in α5-ILV1 increased significantly compared with those of strain α5 (Fig. 5a). It has been reported that overexpression of the ILV2, ILV3, and ILV5 genes led to a significant increase in the production of isobutanol (Chen et al. 2011; Yuan and Ching 2015). We can infer that ILV1 deletion resulted in the prevention of the competitive carbon outflow from the pyruvate pathway to the isoleucine pathway. ILV1 deletion also reinforced the biosynthesis of α-ketoisovalerate, which is a precursor in the isobutanol and isoamyl alcohol biosynthesis; this outcome may explain the increase in isobutanol and isoamyl alcohol production. Increases in ILV2, ILV5, and ILV3 expression levels in the ILV1 deletion strain are consistent with the conjecture. Furthermore, the decrease in 2-phenylethanol in ILV1 deletion strains, as well as in LEU1 and LEU2 deletion strains, may be attributed to the reinforcement of competitive carbon outflow or weak cell growth capability.
The biosynthesis of α-ketoisocaproate is terminated by LEU1 or LEU2 deletion in yeast, resulting in the decrease in isoamyl alcohol production. This finding is in accordance with that of Park et al. (2014), who reported that the decrease in isoamyl alcohol production was affected by LEU2 deletion. Furthermore, the elimination of the α-ketoisovalerate degradation pathway leads to the increase in produced isobutanol. LEU1 and LEU2 deletions exert different influences on isobutanol production. The evident difference may be due to different feedback regulations of α-isopropylmalate and β-isopropylmalate, which should be further investigated. Furthermore, we speculate that the increase in n-propanol production was due to the reinforcement of threonine deamination, considering that the expression levels of ILV1 in α5-LEU1 and α5-LEU2 increased significantly compared with those in α5 (Fig. 5b, c).
The deletion of three genes all resulted in considerable changes in metabolism involved in higher alcohol synthesis; by contrast, single-allele deletion exerted less influence on higher alcohol metabolism. The increase in isobutanol by ILV1 single-allele deletion may also be attributed to the reinforcement of α-ketoisovalerate biosynthesis in the valine pathway. No significant differences were observed in the production of other higher alcohols among the single-allele deletion mutants and the parental strain.
This study attempts to explore the effects of genetic engineering strategies in synthetic and metabolic pathways. Variations in the production of higher alcohols by the engineered mutants are due to carbon flux changes affected by the deletion of ILV1, LEU1, or LEU2 and single-allele deletion of ILV1 in yeast metabolism. This study could provide a valuable reference for further research on higher alcohol metabolism and future optimization of yeast strains for Chinese Baijiu and other alcoholic beverages. Furthermore, the engineered strains showed similar fermentative capabilities to their parental strains. Given the various requirements of flavor and taste in different aroma types of Chinese Baijiu, ILV1, LEU1, and LEU2 double-allele-deletion yeast strains can be used to obtain different concentrations and proportions of higher alcohols. The ILV1 single-allele-deletion strain can be applied to fine-tune the isobutanol proportion without influencing on other flavor substances in Chinese Baijiu.
References
Baichwal VR, Cunningham TS, Gatzek PR, Kohlhaw GB (1983) Leucine biosynthesis in yeast: identification of two genes (LEU4, LEU5) that affect α-isopropylmalate synthase activity and evidence that LEU1 and LEU2 gene expression is controlled by α-isopropylmalate and the product of a regulatory gene. Curr Genet 7(5):369–377. https://doi.org/10.1007/BF00445877
Chen X, Nielsen KF, Borodina I, Kielland-Brandt MC, Karhumaa K (2011) Increased isobutanol production in Saccharomyces cerevisiae by overexpression of genes in valine metabolism. Biotechnol Biofuels 4(1):21. https://doi.org/10.1186/1754-6834-4-21
Derrick S, Large PJ (1993) Activities of the enzymes of the Ehrlich pathway and formation of branched-chain alcohols in Saccharomyces cerevisiae and Candida utilis grown in continuous culture on valine or ammonium as sole nitrogen source. J Gen Microbiol 139(11):2783–2792. https://doi.org/10.1099/00221287-139-11-2783
Dickinson JR, Norte V (1993) A study of branched-chain amino acid aminotransferase and isolation of mutations affecting the catabolism of branched-chain amino acids in Saccharomyces cerevisiae. FEBS Lett 326(1-3):29–32. https://doi.org/10.1016/0014-5793(93)81754-N
Eden A, Nedervelde LV, Drukker M, Benvenisty N, Debourg A (2001) Involvement of branched-chain amino acid aminotransferases in the production of fusel alcohols during fermentation in yeast. Appl Microbiol Biotechnol 55(3):296–300. https://doi.org/10.1007/s002530000506
Fan W, Qian MC (2005) Headspace solid phase microextraction and gas chromatography-olfactometry dilution analysis of young and aged Chinese “Yanghe Daqu” liquors. J Agric Food Chem 53(20):7931–7938. https://doi.org/10.1021/jf051011k
Fan W, Qian MC (2006) Characterization of aroma compounds of Chinese “Wuliangye” and “Jiannanchun” liquors by aroma extract dilution analysis. J Agric Food Chem 54(7):2695–2704. https://doi.org/10.1021/jf052635t
Gietz RD, Woods RA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350:87–96. https://doi.org/10.1016/S0076-6879(02)50957-5
Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24(13):2519–2524. https://doi.org/10.1093/nar/24.13.2519
Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH (2002) A second set of loxP marker cassettes for Cre-mediated multiple gene deletions in budding yeast. Nucleic Acids Res 30(6):88–94. https://doi.org/10.1093/nar/30.6.e23
Holmberg S, Petersen JG (1988) Regulation of isoleucine-valine biosynthesis in Saccharomyces cerevisiae. Curr Genet 13(3):207–217. https://doi.org/10.1007/BF00387766
Hsu YP, Schimmel P (1984) Yeast LEU1. Repression of mRNA levels by leucine and relationship of 5′-noncoding region to that of LEU2. J Biol Chem 259(6):3714–3719
Ida K, Ishii J, Matsuda F, Kondo T, Kondo A (2015) Eliminating the isoleucine biosynthetic pathway to reduce competitive carbon outflow during isobutanol production by Saccharomyces cerevisiae. Microb Cell Factories 14(1):1–9. https://doi.org/10.1186/s12934-015-0240-6
Kobayashi M, Shimizu H, Shioya S (2008) Beer volatile compounds and their application to low-malt beer fermentation. J Biosci Bioeng 106(4):317–323. https://doi.org/10.1263/jbb.106.317
Kondo T, Tezuka H, Ishii J, Matsuda F, Ogino C, Kondo A (2012) Genetic engineering to enhance the Ehrlich pathway and alter carbon flux for increased isobutanol production from glucose by Saccharomyces cerevisiae. J Biotechnol 159(1-2):32–37. https://doi.org/10.1016/j.jbiotec.2012.01.022
Li W, Wang JH, Zhang CY, Ma HX, Xiao DG (2017) Regulation of Saccharomyces cerevisiae genetic engineering on the production of acetate esters and higher alcohols during Chinese baijiu fermentation. J Ind Microbiol Biotechnol 44(6):949–960. https://doi.org/10.1007/s10295-017-1907-2
Lu J, Dong J, Wu D, Chen Y, Guo X, Shi Y, Sun X, Xiao D (2012) Construction of recombinant industrial brewer’s yeast with lower diacetyl production and proteinase A activity. Eur Food Res Technol 235(5):951–961. https://doi.org/10.1007/s00217-012-1821-9
Nigavekar SS, Cannon JF (2002) Characterization of genes that are synthetically lethal with ade3 or leu2 in Saccharomyces cerevisiae. Yeast 19(2):115–122. https://doi.org/10.1002/yea.807
Park SH, Kim S, Hahn JS (2014) Metabolic engineering of Saccharomyces cerevisiae for the production of isobutanol and 3-methyl-1-butanol. Appl Microbiol Biotechnol 98(21):9139–9147. https://doi.org/10.1007/s00253-014-6081-0
Petersen JGL, Holmberg S, Nilsson-Tillgren T, Kielland-Brandt MC (1983) Molecular cloning and characterization of the threonine deaminase (ILV1) gene of Saccharomyces cerevisiae. Carlsb Res Commun 48(3):149–159. https://doi.org/10.1007/BF02907764
Pires EJ, Teixeira JA, Brányik T, Vicente AA (2014) Yeast: the soul of beer's aroma-a review of flavour-active esters and higher alcohols produced by the brewing yeast. Appl Microbiol Biotechnol 98(5):1937–1949. https://doi.org/10.1007/s00253-013-5470-0
Ryan ED, Kohlhaw GB (1974) Subcellular localization of isoleucine-valine biosynthetic enzymes in yeast. J Bacteriol 120(2):631–637
Sakai Y, Tani Y (1992) Directed mutagenesis in an asporogenous methylotrophic yeast: cloning, sequencing, and one-step gene disruption of the 3-isopropylmalate dehydrogenase gene (LEU2) of Candida boidinii to derive doubly auxotrophic marker strains. J Bacteriol 174(18):5988–5993. https://doi.org/10.1128/jb.174.18.5988-5993.1992
Shi S, Tong S, Liu Z, Zhang H, Ang EL, Zhao H (2016) Metabolic engineering of a synergistic pathway for n-butanol production in Saccharomyces cerevisiae. Sci Rep 6(1):25675. https://doi.org/10.1038/srep25675
Stribny J, Romagnoli G, Pérez-Torrado R, Daran JM, Querol A (2016) Characterisation of the broad substrate specificity 2-keto acid decarboxylase Aro10p of Saccharomyces kudriavzevii and its implication in aroma development. Microb Cell Factories 15(1):1–12. https://doi.org/10.1186/s12934-016-0449-z
Swiegers JH, Pretorius IS (2005) Yeast modulation of wine flavor. Adv Appl Microbiol 57:131–175. https://doi.org/10.1016/S0065-2164(05)57005-9
Valero E, Moyano L, Millan MC, Medinab M, Ortega JM (2002) Higher alcohols and esters production by Saccharomyces cerevisiae. Influence of the initial oxygenation of the grape must. Food Chem 78(1):57–61. https://doi.org/10.1016/S0308-8146(01)00361-2
Vollbrecht D (1974) Three pathways of isoleucine biosynthesis in mutant strains of Saccharomyces cerevisiae. Biochim Biophys Acta 362(2):382–389. https://doi.org/10.1016/0304-4165(74)90231-1
Wang CL, Shi DJ, Gong GL (2008) Microorganisms in Daqu: a starter culture of Chinese Maotai-flavor liquor. World J Microbiol Biotechnol 24(10):2183–2190. https://doi.org/10.1007/s11274-008-9728-0
Wu Q, Kong Y, Xu Y (2015) Flavor profile of Chinese liquor is altered by interactions of intrinsic and extrinsic microbes. Appl Environ Microbiol 82(2):422–430. https://doi.org/10.1128/AEM.02518-15
Xiao ZB, Yu D, Niu YW, Chen F, Song SQ, Zhu JC, Zhu GY (2014) Characterization of aroma compounds of Chinese famous liquors by gas chromatography-mass spectrometry and flash GC electronic-nose. J Chromatogr B 945-946:92–100. https://doi.org/10.1016/j.jchromb.2013.11.032
Yang D, Luo X, Wang X (2014) Characteristics of traditional Chinese shanlan wine fermentation. J Biosci Bioeng 117(2):203–207. https://doi.org/10.1016/j.jbiosc.2013.07.010
Yuan J, Ching CB (2015) Combinatorial assembly of large biochemical pathways into yeast chromosomes for improved production of value-added compounds. ACS Synth Biol 4(1):23–31. https://doi.org/10.1021/sb500079f
Yuan J, Mishra P, Ching CB (2016) Metabolically engineered Saccharomyces cerevisiae for branched-chain ester productions. J Biotechnol 239:90–97. https://doi.org/10.1016/j.jbiotec.2016.10.013
Yuan J, Chen X, Mishra P, Ching CB (2017a) Metabolically engineered Saccharomyces cerevisiae for enhanced isoamyl alcohol production. Appl Microbiol Biotechnol 101(1):465–474. https://doi.org/10.1007/s00253-016-7970-1
Yuan J, Mishra P, Ching CB (2017b) Engineering the leucine biosynthetic pathway for isoamyl alcohol overproduction in Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 44(1):107–117. https://doi.org/10.1007/s10295-016-1855-2
Zheng XW, Tabrizi MR, Nout MJR, Han BZ (2011) Daqu—a traditional Chinese liquor fermentation starter. J I Brewing 117(1):82–90. https://doi.org/10.1002/j.2050-0416.2011.tb00447.x
Zhu S, Lu X, Ji K, Guo K, Li Y, Wu C, Xu G (2007) Characterization of flavor compounds in Chinese liquor Moutai by comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry. Anal Chim Acta 597(2):340–348. https://doi.org/10.1016/j.aca.2007.07.007
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2016YFD0400505), the National Natural Science Foundation of China (31471724), the Major Project of Research Program on Applied Fundamentals and Advanced Technologies of Tianjin (14JCZDJC32900), and the National High Technology Research and Development Program of China (2013AA102108).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethical statement
This manuscript is in compliance with ethical standards. This manuscript does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
ESM 1
(PDF 410 kb)
Rights and permissions
About this article
Cite this article
Li, W., Chen, SJ., Wang, JH. et al. Genetic engineering to alter carbon flux for various higher alcohol productions by Saccharomyces cerevisiae for Chinese Baijiu fermentation. Appl Microbiol Biotechnol 102, 1783–1795 (2018). https://doi.org/10.1007/s00253-017-8715-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8715-5