Abstract
The characteristic buttery taste of diacetyl has long been a major problem in the brewing industry, and the foam stability of unpasteurized beer is often influenced by proteinase A (PrA), which is encoded by PEP4 and released from yeast cells into beer during brewing. A recombinant industrial brewer’s yeast strain that reduces the diacetyl content of beer and improves foam stability was constructed. We constructed a PGK1p-ILV5-PGK1t expression cassette, which was introduced into one of the PEP4 alleles via PCR-mediated homologous recombination. Then, the second PEP4 allele was disrupted using the Cre-loxP recombination system, and the recombinant strain was designated as S-CSIK12. The results show that the diacetyl production of S-CSIK12 is always lower than that of the host strain at all stages of beer fermentation. In addition, brewing with S-CSIK12 reduced the PrA activity of the final beer by 44 % compared with that using the wild-type strain. The head retention of the beer brewed with S-CSIK12 (260 ± 2 s) was better than that of the host strain S-6 (212 ± 3 s). Considering that more PrA is released from yeast cells during the final stage of main fermentation and that the timing of yeast cropping is determined by diacetyl reduction, brewing with strains that have low diacetyl production also reduced the PrA activity of the beer and improved its head retention. The present study provides reference for the brewing industry as well as research on the diacetyl reduction and foam stability of beer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
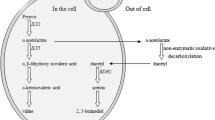
Diacetyl (2,3-butanedione) has an unpleasant butter-like flavor, and its content is crucial to beer maturation. This compound is particularly undesirable in lagers, and the threshold for diacetyl is 0.15 ppm or even lower [1, 2]. Diacetyl is a vicinal diketone formed by the non-enzymatic oxidative decarboxylation of α-acetolactate that diffuses from the isoleucine–valine (ILV) biosynthetic pathway in yeast during the main fermentation [3]. Diacetyl is reabsorbed into yeast cells and reduced to acceptable levels during maturation. Diacetyl removal is the major reason for the 1–3-week maturation period of beer. Therefore, preventing diacetyl formation would help shorten the maturation process. Numerous strategies have been proposed to eliminate this costly process, including improvement of the fermentation craft, genetic approaches to decrease the diacetyl content in yeast during beer production [4], and the elimination of the diacetyl production from its precursor α-acetolactate. Two approaches have been applied. First, α-acetolactate decarboxylase (ALDC) can be added to green beer, which will convert α-acetolactate into acetoin. Second, the genes that encode ALDC from different bacterial species can be expressed in brewer’s yeast using genetic methods. The genetically modified brewer’s yeast would then decrease the diacetyl content during beer production [5–8].
Diacetyl is a side product of the ILV pathway. Blocking the formation of α-acetolactate and increasing the flux toward valine formation would reduce the diacetyl concentration in green beer [9]. Several genes are closely related to diacetyl content in the ILV biosynthetic pathway. Previous studies on brewers’ yeast strains with ILV2 and ILV6 gene deletions, as well as overexpression of the ILV5 gene or both of ILV3 and ILV5 result in decreased diacetyl concentrations (40–70 %) during beer production [9–11].
Foam stability is a critical characteristic of beer quality, and a solid head of foam is essential to the presentation of a well-crafted beer [12, 13]. During beer production, raw materials and the brewing process play major roles in the development and maintenance of foam. The most important compounds involved in beer foam formation are proteins and polypeptides. In terms of beer foam stability, several beer proteins have been identified as foam positive, such as protein Z, lipid transfer protein 1 (LTP1), and various hordein-derived polypeptides [14–16]. Researchers have also shown that beers with different heads of foam influence the taste in sensory evaluation tests [17].
Yeast proteinase A (PrA, EC3.4.23.25) is detrimental to beer foam, especially in unpasteurized beer [18–21]. PrA is a vacuolar aspartic proteinase encoded by the PEP4 gene [22, 23] that can be excreted into beer under stress conditions or released through yeast autolysis or mechanical damage [19, 24, 25]. During beer production, yeast cells release PrA that actively degrades LTP1, which decreases beer head retention [26]. Previous studies have shown that deleting the PEP4 gene in industrial brewer’s yeast helps maintain beer foam stability [20, 21, 26, 27].
ILV5 gene overexpression decreases diacetyl production by 50–60 % compared with the control strain and significantly shortens the maturation period of beer production [28, 29]. In addition, PrA is primarily released form yeast cells during the final stage of fermentation and maturation because of nutrition-deficient conditions, especially under nitrogen starvation; pressure and temperature are also thought to affect yeast cell vitality and subsequent PrA excretion [30]. Yeast strains should be removed once the diacetyl concentration in the worts decreases below the taste threshold during beer fermentation. Otherwise, the low-vitality yeast and enzymes released will adversely affect beer flavor and foam stability. Hence, shortening the maturation time would decrease the PrA activity of beer. In the current study, we constructed a recombinant industrial brewer’s yeast strain with partial PEP4 allelic genes replaced with a constructed ILV5 expression cassette to develop an improved strain with lower diacetyl production and PrA activity. The diacetyl content, PrA activity, and fermentation performance of the recombinant industrial brewer’s yeast strains were examined.
Materials and methods
Strains, vectors, and cultivation conditions
The genetic properties of all strains and plasmids used in the present study are summarized in Table 1. The industrial brewer’s yeast S-6 was obtained from the Yeast Collection Center of the Tianjin Key Laboratory of Industrial Microbiology, Tianjin University of Science and Technology, China.
Escherichia coli DH5α strain was grown at 37 °C in Luria–Bertani broth (1 % NaCl, 1 % tryptone, and 0.5 % yeast extract) supplemented with ampicillin (100 mg l−1). The yeast strain was grown at 28 °C in YEPD medium (1 % yeast extract, 2 % peptone, and 2 % glucose). To select the Geneticin (G418)-resistant yeast strains after transformation, the YEPD plate was supplemented with 800 mg l−1 G418. To select Zeocin-resistant yeast strains, 500 mg l−1 Zeocin (Promega, Madison, United States) was added to the YEPD plates for yeast culture. Then, the YEPG medium (1 % yeast extract, 2 % peptone, and 2 % galactose) was used for Cre expression in the yeast transformants.
Plasmid construction
Plasmid DNA was prepared from E. coli DH5α as described by Sambrook and Russell [31]. Genomic yeast DNA was prepared from industrial brewer’s yeast S-6 as described by Sambrook and Russell [31]. The primers used in this study are listed in Table 2.
An upstream homologous fragment of the PEP4 gene was amplified by PCR using S-6 genomic DNA as templates with the A-UP and A-DOWN primers. A downstream homologous fragment was similarly amplified using the PR-U and PR-D primers. Then, the PCR products were digested with the appropriate endonucleases and cloned into the pUC19 cloning vector (Invitrogen, China) at Hind III and Kpn I sites to construct plasmid pUCAR. An XhoI fragment of the ILV5 gene was amplified from S-6 genomic DNA by PCR using the primers ILV-U and ILV-D to construct the recombinant plasmid. The ILV5 fragment was cloned into vector pPGK1 [32] after both were digested with XhoI, resulting in pPILV. The fragment containing the PGK1 promoter, the ILV5 gene, and the PGK1 terminator was amplified from pPILV using the primers PGK1p and PGK1t. The PCR product was digested with PstI and inserted into pUCAR to construct pIAR, which contains the ILV5 expression cassette. Finally, the KanMX cassette amplified by PCR using pUG6 [33] as the template with the primers K-U and K-D was cloned into pUCAR and pPILV after digestion with the appropriate endonucleases to produce the final plasmids, which were designated as pKAR and pIKAR (Table 1). Based on the aforementioned strategy, the pKSAR plasmid (Table 1) was constructed by inserting the upstream homologous fragment PEP4 SA, the KanMX cassette, and the downstream homologous fragment PEP4 SR into the pUC19 cloning vector. The homologous fragments of this plasmid were amplified using the designed retractive primers, as previously described [34].
Disruption and transformation strategy
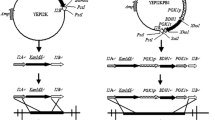
Industrial brewer’s yeast strains are diploid or polyploid. Thus, a retractive primer disruption strategy was used to repeat the deletion of the PEP4 gene efficiently as previously described [34]. The PEP4 A-loxP-KanMX-loxP-PEP4 R and PEP4 A-PGK1p-ILV5-PGK1t-loxP-KanMX-loxP-PEP4 R disruption cassettes were amplified from the plasmids pKAR and pIKAR, respectively. The disruption strategy is shown in Figs. 1a, b. A Cre-loxP recombination system with gene disruption strategy was used to rescue the KanMX cassette. Then, PEP4 SA-loxP-KanMX-loxP-PEP4 SR disruption cassette was used to disrupt the second copy of the PEP4 gene. The retractive primer disruption strategy is shown in Fig. 1c.
Strategy for homologous recombination in a brewers’ yeast strain and the primer binding region for PCR confirmation of successful homologous recombination. The horizontal arrows indicate the primer sites. a The homologous recombination of PEP4 A-PGK1p-ILV5-PGK1t-KanMX-PEP4 R with the brewers’ yeast S-6. b The homologous recombination of PEP4 A-KanMX-PEP4 R with the brewers’ yeast S-6. c Retractive primer design strategy for deleting the second copies of the PEP4 gene in the brewers’ yeast. Homologous sequences located on the outboard side of the dotted line were used for deleting the first allele, whereas those located on the inboard side of the dotted line were used for deleting the second allele. Homologous regions were segregated by the dotted line
Brewer’s yeast transformation was performed using the lithium acetate/PEG method [35]. Transformants were screened on YEPD plate containing 800 mg l−1 G418. PCR was applied to verify the recombinant strains with accurate site integration. Several different pairs of primers were designed, and the primer binding regions are shown in Fig. 1. The genetic stability of the yeast strain was determined using the method of Cha et al. [36].
Construction of recombinant industrial brewer’s yeast
The DNA fragment of PEP4 A-PGK1p-ILV5-PGK1t-loxP-KanMX-loxP-PEP4 R was amplified and transformed into the industrial brewer’s yeast S-6. The fragment was integrated into the chromosome at the PEP4 locus of S-6 by homologous recombination to construct single PEP4 allele disruption and ILV5 overexpression strains (Fig. 1a). The transformants were screened on G418 selective plates after transformation and verified by PCR using primer pairs YZ-SUP/YZ-SDOWN and NYZ-AUP/NYZ-ADOWN. Amplified 1,625- and 1,235-bp DNA fragments were obtained, respectively (Fig. 2). These results proved that the constructed DNA was inserted into the PEP4 locus correctly. After the PCR verification, 10 verified transformants were selected randomly for initial fermentation tests. One transformant with the lowest PrA activity and diacetyl content was selected and was designated as S-IK12. We also constructed single PEP4 allele disruption strains, which resulted from transforming PEP4 A-loxP-KanMX-loxP-PEP4 R disruption cassette into S-6 using the same method described above (Fig. 1b). The strains were verified using the primers YZ-SUP/YZ-2A and YZ-AUP/YZ-ADOWN obtained from the amplification of the 1,626- and 1,751-bp DNA fragments (Fig. 2). A recombinant strain with the lowest PrA activity and diacetyl content was selected and designated as S-K10.
PCR analysis of the yeast recombinants. DNA templates: genome of the single PEP4 allele disruption and ILV5 overexpression transformants (lanes 1 and 2), genome of the single PEP4 allele disruption transformants (lanes 3 and 4), genome of the two PEP4 allele disruption and ILV5 overexpression transformants (lanes 5, 6, and 8), genome of the two PEP4 allele disruption transformants (lane 7). Primers: YZ-SUP/YZ-SDOWN (lane 1), NYZ-AUP/NYZ-ADOWN (lane 2), YZ-SUP/YZ-2A (lanes 3 and 5), YZ-AUP/YZ-ADOWN (lanes 4 and 6), and A-UP/PR-D (lanes 7 and 8)
Cre recombinase was expressed and KanMX was excised after introducing the plasmid pSH-Zeocin into S-IK12 and S-K10. Loss of pSH-Zeocin changed the genotype of the mutants into PEP4 (n-1)/pep4 (73, 750)::PGK1p-ILV5-PGK1t-loxP and PEP4 (n-1)/pep4 (73, 750)::loxP, which were designated as S-CIK12 and S-CK10, respectively. The DNA fragment of PEP4 SA-loxP-KanMX-loxP-PEP4 SR disruption cassette was used to transform S-CIK12 and S-CK10 to delete the second PEP4 allele. To verify whether KanMX was correctly integrated into the second PEP4, the primer pairs YZ-SUP/YZ-2A and YZ-AUP/YZ-ADOWN were used, and the 1,932- and 2,092-bp amplification fragments indicated correct recombination (Fig. 2). To verify further the disruption of the second PEP4 allele, A-UP/PR-DOWN was used as primers with the genome of two PEP4 allele disruption transformants and those of two PEP4 allele disruption and ILV5 overexpression transformants as templates. The PCR products were 3,042 + 887 + 1,407 bp and 3,042 + 3,851 + 1,407 bp, respectively (Fig. 2). A 3,042-bp fragment indicated amplification of the sequence that includes PEP4 SA-loxP-KanMX-loxP-PEP4 SR for both yeasts. By contrast, a 3,851-bp fragment indicated PEP4 A-PGK1p-ILV5-PGK1t-loxP-PEP4 R for S-SIK12 and 887-bp fragment indicated PEP4 A-loxP-PEP4 R for S-SK10. A 1,407-bp fragment indicated the presence of intact PEP4 because the same fragment was obtained when the primer pair A-UP/PR-DOWN was used to amplify the S-6 DNA template. S-SIK12 and S-SK10 were selected from 20 verified transformants through an initial fermentation test.
All of the 50 single-grown colonies of the 10th generation verified that the recombinant strain could grow on YEPD with 800 mg l−1 G418. These results indicate that the KanMX gene might have been integrated into the PEP4 locus, which increased the kanamycin resistance of the recombinant strains, with their genetic stability reaching 100 %.
Finally, the KanMX gene was excised using the Cre-loxP recombination system to obtain the mutants S-CSIK12 and S-CSK10.
Analytical methods
Headspace gas chromatography coupled with electron capture detection was used to measure diacetyl content [37]. Before analysis, the samples were heated to 60 °C for 1 h to convert completely α-acetolactate into diacetyl. Assay for the ILV5 gene product acetohydroxyacid reductoisomerase (RI) activity has been described previously [28]. PrA activity was assayed using a fluorescent method [30]. Head retention was determined with a NIBEM-T foam stability tester (Haffmans, VENLO Holland) according to the manufacturer’s instructions. The esters and higher alcohols were measured using headspace gas chromatography coupled with flame ionization detection [37]. Headspace instrument conditions were as follows: Samples were heated for 20 min at 60 °C in the headspace autosampler before injection; sample temperature, 60 °C; needle temperature, 95 °C; transfer line temperature, 105 °C; GC-cycle time, 26 min; equilibration time, 20 min; pressurize time, 1 min; injection time, 1 min; withdrawal time, 0.5 min; GC conditions were as follows: The oven temperature was held at 35 °C (3 min), followed by an increase to 80 °C (0.5 min) at a rate of 10 °C per min, then increasing to 220 °C (0.5 min) at a rate of 10 °C per min. The FID temperatures were kept constant at 240 °C. The analyses were performed in triplicate. Statistical analysis was done by using the Student’s t test.
Real-time PCR
Yeast strains were cultured in a YEPD medium for 18 h and collected for RNA extraction. The mRNA was extracted using a yeast RNA kit (Omega, Madison, United States), and the changes in ILV5 gene expression were assessed via real-time quantitative PCR (qRT-PCR) using an Ultra SYBR Two-Step qRT-PCR kit with ROX (reference dye for real-time PCR) (CWBIO, China). The primers used to amplify the small parts of ILV5 and the reference gene GAPDH (GPD1) are listed in Table 1. The ΔΔC (t) method was applied for the relative quantification of transcription of ILV5 in the overexpressing strains compared with the reference strain.
Fermentation test
The yeast strains were first grown in 5 ml of 11°P wort at 28 °C for 24 h, and the suspension was inoculated into 50 ml of wort (11°P). Then, the preculture was incubated without shaking at 16 °C for 48 h to adapt the yeast to a lower temperature. After this cultivation process, it was inoculated into 500 ml of 11°P wort in conical flask with fermentation bung. The density of the inoculum was approximately 2 × 107 cells ml−1. Fermentation was conducted at 10 °C for approximately 15 days. After the diacetyl level in the wort decreased below the taste threshold, the yeast was cropped immediately, and an additional 10 days of cold conditioning process was started. Apparent wort extracts were recorded daily. The diacetyl concentration and the PrA activity of the fermented worts during brewing were assayed at different stages of fermentation such as the vigorous stage of main fermentation (day 4), after yeast cropping (days 13–15), after cold conditioning (days 23–25), and other stages. The esters and higher alcohols were assayed after cold conditioning. Microscale fermentation (5 l) and pilot scale brewing (100 l) trials were conducted to evaluate the flavor and taste of the finished beer products brewed with the host strain and the recombinant strains [46]. The finished beer samples were evaluated by an expert panel consisting of six tasting experts from the Tianjin University of Science and Technology Beer Fermentation Pilot Base.
Results and discussion
Diacetyl content assay during beer fermentation
The diacetyl production of the host strain (S-6), the two PEP4 allele disruption and ILV5 overexpression strain (S-CSIK12), and the two PEP4 allele disruption strain (S-CSK10) were measured at different stages of fermentation. Samples were taken for analysis on days 2, 4, 6, 8, 10, 11, 12, 13, 14, and 15. The fermentation was performed in triplicate for each strain. The results show that the peak diacetyl production of each strain was observed on day 6 of fermentation (Fig. 3). The peak diacetyl content of the beer brewed with S-6, S-CSIK12, and S-CSK10 was 0.855, 0.581, and 0.946 mg l−1, respectively. Then, the diacetyl concentration decreased moderately and dropped to the 0.1–0.15 mg l−1 threshold on days 13 (brewed with S-CSIK12) and 15 (brewed with S-6 or S-CSK10). As shown in Fig. 3, the diacetyl production of S-CSIK12 was always lower than that of S-6 and S-CSK10 at all sampling stages. This finding indicates that ILV5 gene overexpression effectively decreases the diacetyl production in beer. The results support the conclusion by Villanueba et al. [10], who stated that the presence of extra ILV5 copies in yeast strains decreases diacetyl production during wort fermentation. In this study, an extra copy of the ILV5 gene was integrated into the brewer’s yeast chromosome to produce a stable, decreased diacetyl production strain. The two copies of the PEP4 disruption strain S-CSK10 produced slightly more diacetyl than the S-6 host strain during fermentation, as shown in Fig. 3. However, the explanation for this phenomenon is beyond the scope of the current knowledge. We speculate that altering a single gene could have some unexpected effects on a cell. Although the effect is not evident, the disruption of two PEP4 alleles may have influence on diacetyl production by the yeast cells. Further studies are needed to elucidate this condition.
RI activity and the ILV5 expression levels
The host strain (S-6), the two PEP4 allele disruption, and ILV5 overexpression strain (S-CSIK12) were grown in wort medium for 18 h. The RI activities of S-6 and S-CSIK12 were 0.38 and 0.62 μM NADPH oxidized per hour per milligram of protein. These results are the means of three independent experiments. Furthermore, the qRT-PCR results show that the cDNA of the recombinant ILV5 overexpression strain (S-CSIK12) increased to 1.88-fold that of the host strain (S-6), as shown in Fig. 4. The real-time PCR results demonstrate higher ILV5 expression levels and larger transcript sizes in S-CSIK12 than those of the control. The RI activity assay results indicate that ILV5 gene overexpression decreased diacetyl production.
Determination of ILV5 gene expression levels in the recombinant strain S-CSIK12 using real-time PCR and the acetohydroxyacid reductoisomerase activity of the recombinant strain S-CSIK12 and the host strain S-6. The GAPDH (GPD1) gene was used as the internal control. RT-qPCRs and measurement of acetohydroxyacid reductoisomerase activity were performed with three biological replicates and technical duplicates of each sample. Error bars represent the standard deviation of the mean
PrA activity and beer head retention
The wild-type brewer’s yeast S-6 and the two recombinant strains S-CSIK12 and S-CSK10 were inoculated for investigation. The PrA activity of the worts was assayed on days 4 (vigorous stage of main fermentation), 13 (S-CSIK12 was cropped), and 15 (S-CSK10 and S-6 were cropped), and then after cold conditioning. The PrA activity gradually increased from the vigorous stage to the final stage of main fermentation (Fig. 5). However, the PrA activity slightly decreased after cold conditioning. The PrA activities of the three yeast strains were also compared during the different stages of brewing (Fig. 5). The PrA activities of the recombinant strains S-CSIK12 and S-CSK10 were always lower than those of the wild-type strain at all sampling stages. The PrA activities of the recombinant strains S-CSIK12 and S-CSK10 after cold conditioning were only 56 and 65 % compared to the wild-type strain.
Proteinase A activity of the recombinant strains and their host during fermentation. Sampling stage I: vigorous stage of main fermentation (day 4 of brewing); sampling stage II: after S-CSIK12 was cropped (day 13 of brewing); sampling stage III: after S-CSK10 and S-6 were cropped (day 15 of brewing); sampling stage IV: after cold conditioning (10 days after yeast cropping). The values are the means of triplicate experiments with standard deviations lower than 5 %
The construction of a PEP4 disruption strain was a logical approach to improving foam stability [26, 34]. First, a single PEP4 allele disruption strain was constructed. Then, the PrA activity was determined by disrupting one copy of the PEP4 gene of S-6; however, no significant difference was observed between transformants and the host. All transformants exhibited at least 90 % PrA activity compared with the host (data not shown). Two PEP4 allele disruption strains were then constructed by applying the Cre-loxP recombination system, and their PrA activities were significantly lower than that of the host strain. This decrease may have been caused by the tetraploidy of S-6. Three PEP4 allele disruption strains and four PEP4 allele disruption strains were also constructed, and their PrA activities were even lower than that of the two PEP4 allele disruption strains; however, a long growth lag phase and significantly reduced wort sugar consumption were observed when the third and fourth allelic PEP4 genes were disrupted (data not shown). Thus, the three PEP4 allele disruption strain and four PEP4 allele disruption strains were not adopted because of their negative effects on beer fermentation. The qPCR results indicated that S-6 contained four copies of the PEP4 gene (data not shown). The PrA activity test showed that a lower copy number of the PEP4 gene in the mutants led to lower PrA activity production. In addition, we could not detect PrA activity when all copies of the PEP4 gene were disrupted. Thus, we conclude that the PrA activity is related to PEP4 gene copy number and polyploidy of a yeast strain. Some authors have reported that the disruption of single PEP4 alleles in brewer’s yeast significantly decreases PrA activity [38, 39]. This discrepancy with the present results may have been caused by the different PEP4 copy numbers and the evolutionary origins of the host strains. S-6 contained four copies of the PEP4 gene, which might be more than its host strains. Previous studies on brewer’s yeast have shown that their genomes are hybrid and polyploid, such that the copy numbers of certain genes in various brewer’s yeast strains may range from haploid to tetraploid [40]. Brewer’s yeast strains with different PEP4 gene copy numbers might produce different amounts of PrA.
The stage following the end of diacetyl removal is an important stage of brewing. The worts during this stage of beer fermentation present a hostile environment for yeast cells because of nitrogen starvation, high ethanol concentrations, high osmotic pressure, and high carbon dioxide concentrations [19, 24]. These conditions may decrease cell viability or even cause the autolysis of yeast cells, which will lead to a significant increase in the release of PrA into the beer [19, 25]. Yeast strains should be removed once the diacetyl concentration in the worts decreases below the taste threshold during beer fermentation for the reasons described above. Thus, shortening the time of diacetyl removal means the yeast can be cropped ahead of time. Once the yeast is removed, there is no way to release PrA into the beer. Hence, the timing of yeast cropping can be optimized to minimize PrA release into the beer if a brewer’s yeast strain with lower diacetyl production is used. In this study, the yeast cropping time was performed 2 days earlier using the ILV5 overexpression strain (S-CSIK12) instead of the ILV5 wild-type strains (S-CSK10 and S-6). The assay results show that the PrA activity of the wort brewed with S-CSIK12 was lower than that of S-CSK10 on day 15 and after cold conditioning (Fig. 5). The S-CSIK12 yeast was cropped on day 13 such that it could not release PrA into the wort during subsequent brewing. Hence, the yeast should be cropped immediately once the diacetyl level in the wort is below the taste threshold. Optimized yeast handling decreases the PrA content of beer. Longer yeast bleed-off times do not contribute to diacetyl removal, the higher the probabilities of high PrA release into the finished beer.
The beer head retention of S-6 and the recombinant strains S-CSIK12 and S-CSK10 was determined after cold conditioning. As shown in Table 3, the head retention of beer brewed with S-CSIK12 (260 ± 2 s) and S-CSK10 (244 ± 3 s) was better than that with the host strain S-6 (212 ± 3 s). Based on the PrA activity assay, the PrA activity of beer is closely related to beer head retention. The head retention of the beers brewed with S-CSIK12 and S-CSK10 indicates that brewing using strains with low diacetyl production positively affects beer foam stability. This result shows that disruption of two PEP4 alleles reduces the PrA activity of the finished beer by 35 % and increases its head longevity by 32 s. By contrast, ILV5 gene overexpression reduces the PrA activity of the finished beer by 44 % and increases its head longevity by 48 s.
The main beer performance indices
The flavor component profiles assayed after cold conditioning show that the transformant with two disrupted PEP4 copies (S-CSK10) did not observably affect the aromatic compound content of beer compared with the host strain (S-6) (Table 3). However, the Ilv5p (acetohydroxyacid reductoisomerase) overexpression transformant (S-CSIK12) had increased fusel alcohols, esters, and acetaldehyde (Table 3). Considering fusel alcohols and esters are the products of amino acid catabolism in the ILV biosynthetic pathway, their increased amounts may have been caused by increased flux in the ILV biosynthetic pathway [41–43]. Although the fusel alcohols and acetaldehyde, which negatively affect beer quality, were increased when S-CSIK12 was used, their concentrations in the beer were within the normal sensory thresholds.
The sensorial analysis results indicated that the beer brewed with the host strain (S-6) and that brewed with the two PEP4 allele disruption strain (S-CSK10) did not differ in flavor or taste. The tasting experts considered the beers brewed with the two PEP4 allele disruption and ILV5 overexpression strain (S-CSIK12) better tasting than those brewed with S-6 and S-CSK10. The flavor component profiles of the finished beer products also showed the values determined for the beers brewed with S-6, S-CSIK12 and S-CSK10, as shown in Table 4. In addition, the resulting beers produced by microscale fermentation were comparable with the pilot brews of beers.
The wort apparent extract was detected, and a slight reduction in wort sugar consumption during fermentation was observed in the two PEP4 allele disruption strains (S-CSK10 and S-CSIK12) compared with the host strain (S-6) (Fig. 6). This phenomenon should be interpreted as follows: First, PrA is essential to the Saccharomyces cerevisiae vacuolar proteolytic system during nutritional stress and vegetative growth [44], especially during beer fermentation. Second, Chen et al. [39] reported that in industrial S. cerevisiae, PrA may control glycolytic enzyme expression (HK: hexokinase, Pfk: phosphofructokinase, and PYKi: pyruvate kinase) directly or indirectly, which delays cell metabolism in PrA-modified strains, and that glycolytic flux direction and rate may be regulated by vacuolar PrA in industrial S. cerevisiae [39]. Mutations at the PEP4 locus may exhibit a dosage effect on sugar consumption. PrA deficiency in the yeast may affect the physiological and metabolic functions of cells, including sugar consumption.
Conclusions
Brewing with two PEP4 allele disruption and ILV5 overexpression recombinant industrial brewing yeast strains significantly decreased the PrA activity of beer, improves foam stability, and decreases diacetyl production in beer. Our investigations show that the PrA activity of the beer brewed with strains with two disrupted PEP4 copies (S-CSIK12 and S-CSK10) was decreased compared with that using the host strain. Head retention was also improved. In addition, the diacetyl production of ILV5 overexpression strain (S-CSIK12) was always lower than that of the host strain at all stages of beer fermentation. Brewing with lower diacetyl production strain optimizes the timing of yeast cropping, decreases the PrA activity of beer, and improves head retention. This discussed the relationship between diacetyl production and beer head retention, two aspects of beer quality. The current study would help in maintaining beer foam performance, especially in stabilizing the foam in unpasteurized beer. The KanMX markers of all transformants were excised such that the beers brewed with the transformants were considered safe. However, genetically modified foods produced by the fermentation of recombinant microorganisms are not commercially available due to the lack of public acceptance. Thus, the recombinant yeast strains in this study could not be applied in the commercial brewing industry at present because of the controversy over genetically modified organisms (GMOs) in Europe. Nevertheless, the present study provides reference for the brewing industry as well as research on the diacetyl reduction and foam stability of beer. With stronger understanding of and further research into genetically modified organisms, these will likely be widely accepted in the future.
Abbreviations
- PrA:
-
Proteinase A
- PEP4:
-
A kind of gene that encodes proteinase A
- ILV5:
-
A kind of gene that encodes reductoisomerase
- RI:
-
Acetohydroxyacid reductoisomerase
- ILV pathway:
-
Isoleucine–valine pathway
- ALDC:
-
α-Acetolactate decarboxylase
- ILV2:
-
A kind of gene that encodes acetolactate synthase I/II/III large subunit
- ILV3:
-
A kind of gene that encodes dihydroxy-acid dehydratase
- ILV6:
-
A kind of gene that encodes acetolactate synthase I/III small subunit
- LTP1:
-
Lipid transfer protein 1
- GPD1:
-
A kind of gene that encodes glycerol-3-phosphate dehydrogenase
- ROX:
-
Reference dye for real-time PCR
- HK:
-
Hexokinase
- Pfk:
-
Phosphofructokinase
- PYKi:
-
Pyruvate kinase
- S-6:
-
Wild-type industrial brewer’s yeast
- S-CSK10:
-
Two PEP4 allele disruption strain
- S-CSIK12:
-
Two PEP4 allele disruption and ILV5 overexpression strain
References
Saison D, De Schutter DP, Uyttenhove B, Delvaux F, Delvaux FR (2009) Contribution of staling compounds to the aged flavour of lager beer by studying their flavour thresholds. Food Chem 114(4):1206–1215
Meilgaard M (1975) Flavor chemistry of beer. II. Flavor and threshold of 239 aroma volatiles. Tech Q Master Brew Assoc Am 12(3):151–168
Haukeli A, Lie S (1978) Conversion of α-acetolactate and removal of diacetyl: a kinetic study. J Inst Brew 84:85–89
Wang Z-Y, He X-P, Liu N, Zhang B-R (2008) Construction of self-cloning industrial brewing yeast with high-glutathione and low-diacetyl production. Int J Food Sci Technol 43(6):989–994
Yamano S, Tanaka J, Inoue T (1994) Cloning and expression of the gene encoding [alpha]-acetolactate decarboxylase from Acetobacter aceti ssp. xylinum in brewer’s yeast. J Biotechnol 32(2):165–171
Blomqvist K, Suihko ML, Knowles J, Penttilä M (1991) Chromosomal integration and expression of two bacterial α-acetolactate decarboxylase genes in brewer’s yeast. Appl Environ Microbiol 57(10):2796–2803
Fujii T, Kondo K, Shimizu F, Sone H, Tanaka J, Inoue T (1990) Application of a ribosomal DNA integration vector in the construction of a brewer’s yeast having alpha-acetolactate decarboxylase activity. Appl Environ Microbiol 56(4):997–1003
Sone H, Fujii T, Kondo K, Shimizu F, Tanaka J, Inoue T (1988) Nucleotide sequence and expression of the Enterobacter aerogenes alpha-acetolactate decarboxylase gene in brewer’s yeast. Appl Environ Microbiol 54(1):38–42
Duong CT, Strack L, Futschik M, Katou Y, Nakao Y, Fujimura T, Shirahige K, Kodama Y, Nevoigt E (2011) Identification of Sc-type ILV6 as a target to reduce diacetyl formation in lager brewers’ yeast. Metab Eng 13(6):638–647
Villanueba K, Goossens E, Masschelein C (1990) Subthreshold vicinal diketone levels in lager brewing yeast fermentations by means of ILV5 gene amplification. J Am Soc Brew Chem 48:111–114
Villa K, Lee S, Goossens E, Debourg A, Masschelein C (1995) Control of vicinal diketone production by brewer’s yeast. I. Effects of ILV5 and ILV3 gene amplification on vicinal diketone production and ILV enzyme activity. J Am Soc Brew Chem 53:49–53
Brey SE, de Costa S, Rogers PJ, Bryce JH, Morris PC, Mitchell WJ, Stewart GG (2003) The effect of proteinase A on foam-active polypeptides during high and low gravity fermentation. J Inst Brew 109(3):194–202
Leisegang R, Stahl U (2005) Degradation of a foam-promoting barley protein by a proteinase from brewing yeast. J Inst Brew 111(2):112–117
Evans DE, Sheehan M, Stewart D (1999) The impact of malt derived proteins on beer foam quality. Part II. The influence of malt foam-positive proteins and non-starch polysaccharides on beer foam quality. J Inst Brew 105:171–178
Sorensen S, Bech L, Muldbjerg M, Beenfeldt T, Breddam K (1993) Barley lipid transfer protein 1 is involved in beer foam formation. Tech Quart 30:136–145
Evans DE, Bamforth CW (2008) Beer foam: achieving a suitable head: beer: a quality perspective. Academic Press, Burlington, pp 7–66
Lynch D, Bamforth C (2002) Measurement and characterization of bubble nucleation in beer. J Food Sci 67(7):2696–2701
Yokoi S, Shigyo T, Tamaki T (1996) A fluorometric assay for proteinase A in beer and its application for the investigation of enzymatic effects on foam stability. J Inst Brew 102(1):33–37
Cooper D, Stewart G, Bryce J (2000) Yeast proteolytic activity during high and low gravity wort fermentations and its effect on head retention. J Inst Brew 106(4):197–201
Wang ZY, He GQ, Ruan H, Liu ZS, Yang LF, Zhang BR (2006) Construction of proteinase A deficient transformant of industrial brewing yeast. Eur Food Res Technol 225(5–6):831–835
Wang ZY, He XP, Zhang BR (2007) Over-expression of GSH1 gene and disruption of PEP4 gene in self-cloning industrial brewer’s yeast. Int J Food Microbiol 119(3):192–199
Westphal V, Marcusson EG, Winther JR, Emr SD, van den Hazel HB (1996) Multiple pathways for vacuolar sorting of yeast proteinase A. J Biol Chem 271(20):11865
Woolford CA, Noble J, Garman JD, Tam MF, Innis M, Jones EW (1993) Phenotypic analysis of proteinase A mutants. Implications for autoactivation and the maturation pathway of the vacuolar hydrolases of Saccharomyces cerevisiae. J Biol Chem 268(12):8990–8998
Maddox I, Hough J (1970) Proteolytic enzymes of Saccharomyces carlsbergensis. Biochem J 117(5):843
Quilliam W, Hulse G, Cameron-Clarke A (2000) Yeast management and fermentation performance: a brewer’s perspective. Brewing yeast fermentation performance. Blackwell Science, Oxford, pp 189–200
Zhang HB, Ruan H, Li WF, Zhang W, Su ZR, He GQ, Chen QH (2011) Construction of recombinant industrial S. cerevisiae strain with barley lipid-transfer protein 1 secretion capability and lower PrA activity. Eur Food Res Technol 233(4):707–716
Liu XF, Wang ZY, Wang JJ, Lu Y, He XP, Zhang BR (2009) Expression of GAI gene and disruption of PEP4 gene in an industrial brewer’s yeast strain. Lett Appl Microbiol 49(1):117–123
Dillemans M, Goossens E, Goffin O, Masschelein C (1987) The amplification effects of the ILV5 gene on the production of vicinal diketones in Saccharomyces cerevisiae. J Am Soc Brew Chem 45(3):81–84
Gjermansen C, Nilsson-Tillgren T, Petersen JGL, Kielland-Brandt MC, Sigsgaard P, Holmberg S (1988) Towards diacetyl-less brewers’ yeast. Influence of ilv2 and ilv5 mutations. J Basic Microb 28(3):175–183
Kondo H, Yomo H, Furukubo S, Fukui N, Nakatani K, Kawasaki Y (1999) Advanced method for measuring proteinase A in beer and application to brewing. J Inst Brew 105(5):293–300
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, vol 2. CSHL press, New York
Lilly M, Lambrechts M, Pretorius I (2000) Effect of increased yeast alcohol acetyltransferase activity on flavor profiles of wine and distillates. Appl Environ Microbiol 66(2):744–753
Gueldener U, Heinisch J, Koehler G, Voss D, Hegemann J (2002) A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucl Acid Res 30(6):e23–e23
Hao JG, Dong JJ, Speers RA, Shen W, Shan LJ, Fan W, Li Q, Gu GX, Chen J (2008) Construction of a single PEP 4 allele deletion in Saccharomyces carlsbergensis and a preliminary evaluation of its brewing performance. J Inst Brew 114(4):322–328
Daniel Gietz R, Woods RA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Meth Enzymol 350:87–96
Cha HJ, Chae HJ, Choi SS, Yoo YJ (2000) Production and secretion patterns of cloned glucoamylase in plasmid-harboring and chromosome-integrated recombinant yeasts employing an SUC2 promoter. Appl Biochem Biotech 87(2):81–93
Saerens SMG, Verbelen P, Vanbeneden N, Thevelein J, Delvaux F (2008) Monitoring the influence of high-gravity brewing and fermentation temperature on flavour formation by analysis of gene expression levels in brewing yeast. Appl Microbiol Biotechnol 80(6):1039–1051
Zhang HB, Zhang HF, Chen QH, Ruan H, Fu ML, He GQ (2009) Effects of proteinase a on cultivation and viability characteristics of industrial Saccharomyces cerevisiae WZ65. J Zhejiang Univ Sci B 10(10):769–776
Chen QH, Liu XJ, Fu ML, Zhang HB (2010) Effect of PrA encoding gene-PEP4 deletion in industrial S. cerevisiae WZ65 on key enzymes in relation to the glycolytic pathway. Eur Food Res Technol 231(6):943–950
Bond U, Neal C, Donnelly D, James TC (2004) Aneuploidy and copy number breakpoints in the genome of lager yeasts mapped by microarray hybridisation. Curr Genet 45(6):360–370
Dickinson JR, Harrison SJ, Hewlins MJE (1998) An investigation of the metabolism of valine to isobutyl alcohol in Saccharomyces cerevisiae. J Biol Chem 273(40):25751–25756
Dickinson JR, Harrison SJ, Dickinson JA, Hewlins MJE (2000) An investigation of the metabolism of isoleucine to active amyl alcohol in Saccharomyces cerevisiae. J Biol Chem 275(15):10937–10942
Omura F (2008) Targeting of mitochondrial Saccharomyces cerevisiae Ilv5p to the cytosol and its effect on vicinal diketone formation in brewing. Appl Microbiol Biotechnol 78(3):503–513
Parr CL, Keates RAB, Bryksa BC, Ogawa M, Yada RY (2007) The structure and function of Saccharomyces cerevisiae proteinase A. Yeast 24(6):467–480
Hao X, Xiao DG, Zhang CY, Chen YF (2010) Influence of nutrients on proteinase A activity in draft beer during fermentation. Int J Food Sci Tech 45(6):1169–1174
Sampaio TL, Kennedy JA, Vasconcelos MC (2007) Use of microscale fermentations in grape and wine research. Am J Enol Vitic 58(4):534–539
Acknowledgments
The current study was financially supported by the program for Changjiang Scholars and Innovative Research Team in University (IRT1166), the National Natural Science Foundation of China (No.31271916).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lu, J., Dong, J., Wu, D. et al. Construction of recombinant industrial brewer’s yeast with lower diacetyl production and proteinase A activity. Eur Food Res Technol 235, 951–961 (2012). https://doi.org/10.1007/s00217-012-1821-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-012-1821-9