Abstract
Salinity is nowadays considered one of the main factors that limit crop productivity and a threat to world’s food production. Hence, to breed salt tolerant varieties of crops and horticultural species is necessary to increase or at least maintain food production in order to feed the growing world’s population. Plant tolerance to salinity is a complex phenomenon at both cellular and plant level. Since salt causes several types of stresses, plants face salinity using different strategies, whose relative importance depends on the species and the growing conditions. Here we present an overview of the salt tolerance mechanisms to counteract osmotic, ionic and oxidative stress, as well as an update of the knowledge on the processes involved in salt tolerance gained in part through the new genomic approaches. To breed new cultivars able to grow and maintain crop productivity on saline conditions requires variability for some of the traits related to salinity tolerance, the discovery of quantitative trait loci (QTL) regulating those traits, a deep understanding of QTL interaction with other QTL and with the environment, and the transfer of QTL from donors to elite lines using phenotypic and marker assisted selection. We have summarised part of the information related to these four issues and some guidance is given to maximize the efficiency of the selection processes. Genetic transformation has become a powerful tool in plant breeding programs since it allows the introduction of gene(s) controlling traits without affecting the rest of the characteristics of an elite genotype. In this chapter we have reviewed the available information on several topics such as: salt tolerance improvement aided by genetic transformation, functional analysis of genes related salt-tolerance, the complexity of the trait and its evaluation method, the number of genes to be introduced, and the sources of genetic variability. Finally, the use of genomic tools like transcriptomic analysis, post-transcriptional gene silencing, insertional mutagenesis and gene traps, to perform the genetic dissection of this complex trait is discussed.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Over 6% of the world’s total land area (840 million hectares) is affected by salinity or sodicity (FAO 2005). This enormous area has only had small consequences for agriculture because there was enough good land for rain-fed crops: of the 1,500 million hectares dedicated to rain-fed agriculture only 32 millions were affected by secondary salinisation, the salinisation caused by agricultural practices (FAO 2005). This scenario is completely different when irrigated land is considered. The 230 million hectares of irrigated land produce about one third of world’s food and over 20% of this productive land is affected by secondary salinisation (FAO 2005) because salts dissolved in the irrigation water are deposited in the soil following evapotranspiration. And the greatest danger comes from the continuously increasing secondary salinisation in irrigated areas. In California for example, where irrigation started about 130 years ago, half of the irrigated area was affected by salinity more than 20 years ago (Lewis 1984). The risk of secondary salinisation is proportional to the amount of salts in the irrigation water. This risk will increase as population increases because cities (including gardens and sport premises) and industry will pay for the best quality water leaving the worst to agriculture.
Salinity is nowadays considered one of the main factors that limit crop productivity and a threat to world’s food production. In addition, world’s population is expected to increase about 50% by 2050. Hence, it is necessary to sustain agricultural productivity on the increasingly saline irrigated land by salt tolerant cultivars. However, it should be pointed out that salt-tolerant cultivars will only be part of the solution. Salinity in agricultural systems tends to increase rather than remain stable, and may rise in the 8–10 years period that takes to release a new cultivar. For this reason, plant breeding cannot be the full answer to the problem of salinisation but part of an integrated programme that would also include irrigation and drainage management (Flowers and Yeo 1995).
Higher plants do not have a salt-tolerant metabolism even if the plant itself thrives in seawater (Flowers 1972a, b). Salt tolerance in organisms other than the halobacteria do not depend on salt-tolerant proteins, but on keeping a defined micro-environment in the cytoplasm, regulated for the quantity and quality of inorganic ions. Single-celled aquatic organisms or cell cultures can excrete into the medium any dangerous substance. Animals have their cells designed to excrete into the bloodstream and special organs to deal with these toxic compounds. The plant root can only excrete into the solution being drawn towards it by transpiration, but the aboveground cells have nowhere to excrete apart from the small volume of the apoplast. The large central vacuole is the limited place where plant cells can store toxic materials, hence its importance in saline conditions. Glicophytes, where most of the crops are included, are not affected by the external salinity per se, but by growth fall below net ion import leading to ever-increasing internal concentrations and, eventually, to catastrophic failure (Munns and Termaat 1986). Contrarily, in halophytes, growth and net salt uptake are coupled, even if we do not know what controls what (Yeo and Flowers 1986). The targets to produce a plant tolerant to salinity are ion transport and compartmentalization and the synthesis of compatible solutes (osmolytes or osmoprotectants) to counteract osmotic and ionic stresses, oxidative protection, and the maintenance of ion homeostasis. But plant responses to salinity are even more complex at both the whole plant level and cellular level, and research efforts have been focused on understanding the physiological and molecular basis of salt tolerance in higher plants (Mansour and Salama 2004; Sairam and Tyagi 2004; Bartels and Dunkar 2005; Munns and Tester 2008).
In the first section of this chapter, we will describe the mechanisms developed by plants to cope with salinity and the relative importance of each one of them, which vary not only with the species but also with other factors such as stress level, exposure time to salinity, environmental conditions, etc. Advances in our knowledge of the physiological and molecular basis of salt tolerance so far achieved, with special mention to the emerging progress thanks to the new genomic approaches, are summarised. The achievement of salt tolerant cultivars is reviewed in the second part of the chapter, with special attention to the search for natural variation in phenotypic and physiological characters, the discovery of quantitative trait loci regulating salt tolerance, the interaction of these loci with the environment and other loci, and to design efficient breeding programmes based in marker assisted selection coupled with phenotypic selection. In the next section, the transfer and expression of genes related to salt tolerance into elite cultivars or parents of modern F1 hybrids through genetic transformation is a very attractive approach because, this way, susceptible but productive cultivars could be transformed into tolerant ones, while maintaining all the valuable traits today’s cultivars possess. Here we will review the progress in this field, raise the problems associated with the functional analysis of salt tolerance-related genes and discuss the adequacy of genomic tools to perform the genetic dissection of this complex trait.
2 Salt Tolerance Mechanisms at Physiological and Molecular Levels
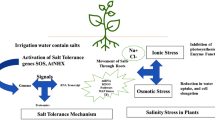
Salt causes several types of plant stress including osmotic stress and ionic stress due to the accumulation of toxic saline ions, nutritional stress due to the altered nutrient uptake, especially of ions such as K+ and Ca2+, and oxidative stress. However, the main physiological mechanism responsible of plant growth reduction and how environment and genotype modulate plant salinity response are not yet known.
To understand the physiological mechanisms responsible for salinity tolerance, it is important to bear in mind that plants respond to salinity by using two main tolerance mechanisms: mechanisms of osmotic tolerance to avoid the osmotic effect of the salt outside the roots, which occurs normally at low stress levels and over short periods of salt stress, and mechanisms of ionic tolerance to avoid the toxic effect of the salt within the plant, the main effect induced by long-term or high stress levels. However, the timescale over which ion-specific damage is manifested depends on the salt sensitivity of the species and the stress level. Thus, the ion-specific effect will start earlier in plants with a low ability to regulate the transport of saline ions to the shoot, or when high salt levels are applied. Although the two phases are generally separated in time for most plants, it is also possible for ion toxicity to take effect during the first phase itself and for osmotic effects to persist in the second phase (De Costa et al. 2007; Muñoz-Mayor et al. 2008).
2.1 Plant Response to Osmotic Stress Induced by Salinity
The osmotic effect starts immediately after the imposition of salt stress, before the saline ions are taken up by the roots. The salt concentration around the roots reduces the osmotic potential of the growing medium and thus the plant’s ability to take up water and nutrients by the roots, similar to the effect induced by drought. (Munns 2002). The plant needs to accumulate solutes to maintain cell volume and turgor, so the response to turgor reduction is osmotic adjustment, which is a major component of salt stress tolerance. Osmotic potential may be reduced either by the simple effect of solute concentration due to reduced water uptake by the plant or by the active solute accumulation. Osmotic adjustment is defined as the lowering of osmotic potential in plant tissue due to net accumulation of solute (Blum et al. 1996).
The main solutes contributing to osmotic adjustment are the inorganic solutes, which are taken up from the substrate and transported to the shoot, and the organic solutes, which are synthesised by the plant. Na+ and Cl− are energetically efficient osmolytes for osmotic adjustment, but must be compartmentalised into the vacuole to minimise cytotoxicity. Within the cytoplasm, osmotic adjustment is achieved by accumulation of compatible osmolytes. Some compatible osmolytes are essential elemental ions, such as K+, but the majority of these are organic solutes, especially sugars (mainly fructose and glucose) and organic acids. Other osmolytes are sugar alcohols (glycerol and methylated inositols), complex sugars (trehalose, raffinose and fructans), quaternary amino acid derivatives (proline, glycine betaine, β-alanine betaine, proline betaine), tertiary amines and sulfonium compounds (Zhu 2001). Another function of compatible osmolytes that may occur at lower concentrations is osmoprotection, which includes protection of thylakoid and plasma membrane integrity, stabilising proteins, a sink for energy or reducing power, a source of carbon and nitrogen for recovery, or scavenging of reactive oxygen species generated by salt stress (Bartels and Dunkar 2005). These beneficial impacts have been shown by overexpression of different genes involved in osmolyte accumulation (Ge et al. 2008; Yang et al. 2008). However, consensus has not been reached on the effectiveness of accumulation of osmolytes by genetic modification, as occurs in the case of proline. Therefore, further studies are required to give an insight into the role of the osmolytes and their effective utilisation for stress tolerance.
The use of organic solutes for osmotic adjustment is energetically much more expensive than the use of the saline ions proceeding from the substrate (Yeo 1983). Thus, the ATP requirement for the synthesis or accumulation of solutes in leaves was reported by Ravens (1985) as 3.5 for Na+, 34 for mannitol, 41 for proline, 50 for glycine betaine and about 52 for sucrose. In this respect, salt tolerance may not always be associated with low Na+ concentration in the leaves. While the more tolerant genotypes of many species are those better able to prevent excessive ion accumulation, the leaves of halophytes do contain high salt concentrations (Santa-Cruz et al. 1999), which are necessary to adjust the leaf water relations to low external potentials and plants use the cheapest solutes from an energetic point of view (Neumann 1997). Such a situation also seems to occur in species that are not very sensitive to salt, such as tomato grown at low-mid salinity levels, as the higher the fruit yield, the higher the contribution of inorganic solutes, including the saline ions, to the osmotic potential (Alarcon et al. 1993; Estañ et al. 2005). Thus, breeding for Na+ accumulation, rather than exclusion, could be a more effective strategy for improving salt tolerance of conventional crop plants.
2.2 Plant Response to Ionic Stress Induced by Salinity
Since NaCl is the major component of most saline soils, our usage of the terms salinity and salt stress here refers to stress caused by NaCl. The ionic stress starts generally later than osmotic stress, when the toxic saline ions are transported to the shoot and build up to toxic levels within the leaves. Thus, roots must exclude most of the Na+ and Cl− dissolved in the growing medium, or the salt in the shoot will gradually build up with time to toxic levels. The cause of injury is probably salt load exceeding the ability of the cells to compartmentalise salts in the vacuole. Salts would then build up rapidly in the cytoplasm and inhibit enzyme activity (Yokoi et al. 2002). Alternatively, they might build up in the cell walls and dehydrate the cell (Flowers et al. 1991). The rate of cell death is crucial for the survival of the plant. If new leaves are continually produced at a rate greater than that at which old leaves die, there will be enough photosynthesizing leaves on the plant to produce flowers and seeds, although reduced in number. However, if old leaves die more quickly than new ones develop, the plant may not survive.
In most studies on salinity it has not been possible to determine whether the toxic effects observed are due to Na+, Cl− or to a contribution of the two. In only a few species such as citrus (Moya et al. 2003) has there been conclusive evidence of greater sensitivity to Cl− than to Na+. Consequently, Na+ is considered the primary cause of ion-specific damage for many plants (Tester and Davenport 2003). However, similar relationships between fruit yield and leaf ionic concentrations for Na+ and Cl− were observed in tomato, which suggests that the toxic effects are due, at least in the long term, to the contribution of both ions (Estañ et al. 2005).
Salt tolerance of cultivated species is generally correlated to an efficient Na+ and Cl− exclusion mechanism and better maintenance of leaf K+ concentration at high external NaCl (Gorham et al. 1990; Matsushita and Matoh 1991). However, the higher salt tolerance of wild tomato species over cultivated forms has generally been associated with the halophytic character of Na+ accumulation in the wild relatives (Cuartero and Fernandez-Muñoz 1999). Tester and Davenport (2003) suggested that although halophytes accumulate Na+ in the shoot, it is unlikely that halophytic species have higher rates of Na+ transport at high salinities or over the long term than salt-sensitive species. Studies of halophytes at low salinities tend to obscure the true situation, because many halophytes show growth stimulation upon addition of NaCl to a growth medium when NaCl is rapidly accumulated and employed preferentially as an osmoticum (Alarcon et al. 1993; Glenn et al. 1999). Several recent reviews of this area of study have been carried out (Tester and Davenport 2003; Apse and Blumwald 2007; Munns and Tester 2008), and these describe in detail the transport of Na+ from the growing medium into the roots, the Na+ loading into and unloading from the xylem, and its redistribution within the plant.
2.3 Plant Response to Oxidative Stress Induced by Salinity
In addition to its known components of osmotic stress and ion toxicity, salt stress is also manifested as an oxidative stress, all of which contribute to its deleterious effects (Hernandez et al. 2000; Mittova et al. 2002). Oxidative stress is characterised by the overproduction of reactive oxygen species (ROS) represented predominantly by superoxide anion, hydrogen peroxide, hydroxyl radical, and singlet oxygen. Plants have defensive mechanisms and utilise several biochemical strategies to avoid damage caused by ROS. Plant enzymatic defences include antioxidant enzymes such as the phenol peroxidase, ascorbate peroxidase, glutathione peroxidase, superoxide dismutase, and catalase which, together with other enzymes of the ascorbate-glutathione cycle, promote the scavenging of ROS (Hernandez et al. 2001; Hong et al. 2007). The biochemical defence system also includes carotenoids, ascorbate, glutathione and tocopherols. Several authors have suggested that the function of sugars, poliols, glycine-betaine and proline could be to protect cells against the hydroxyl radical (Sickler et al. 2007).
The sources of ROS under stress, mechanisms of ROS detoxification and the role of ROS in stress signalling are all active areas of current research and have been extensively studied and reviewed (Ben Amor et al. 2007). However, more studies are necessary before a definitive conclusion can be reached about the role of the ROS production under stress. Thus, increased ROS production has long been known under the heading of ‘oxidative stress’, which in itself is a negative term implying a harmful process. In contrast to this negative term, implying a state to be avoided, Foyer and Noctor (2005) proposed that the syndrome would be more usefully described as ‘oxidative signalling’, that is, an important and critical function associated with the mechanisms by which plant cells sense the environment and make appropriate adjustments to gene expression, metabolism and physiology.
2.4 Homeostasis and Protection or Damage Repair Induced by Salinity
Responses to salt stress are often discussed in terms of homeostasis and protection or damage repair (Zhu 2001). Mechanisms of ion homeostasis and osmotic homeostasis attempt to restore the cellular ion or water content to levels similar to those present under unstressed conditions. Protection and damage repair mechanisms attempt to prevent or repair cellular damage caused by altered ion or water content under stress. To understand the molecular mechanisms that plants have developed to cope with salinity, key genes for salt stress should be identified. Present engineering strategies for enhanced salt tolerance rely on the transfer of one or several genes either involved in signalling and regulatory pathways or encoding enzymes present in pathways leading to the synthesis of functional and structural protectants, such as osmolytes and antioxidants, as is indicated in the next sections. Osmotic homeostasis probably depends on the action of genes for solute synthesis, and a number of channels and carriers for uptake and compartmentalisation of inorganic solutes, especially K+ (Rodriguez-Navarro 2000). Aquaporins may also have a role in osmotic homeostasis by facilitating water movement, but despite some progress achieved recently, not much is currently known about the levels of physiological relevant aquaporin function and regulation, and its effects on plant water balance (Kaldenhoff et al. 2008). Because of the most important effect induced by salinity to long-term is the toxic effect induced by the transport of saline ions to the shoot, most approaches have been directed to studying cation transporters and their regulation, especially the Na+ transporters genes, like SOS1 (Shi et al. 2002), AtHKT1 (Rus et al. 2004) and AtNHX1 (Apse and Blumwald 2002), whereas the Cl− transport mechanisms in plants have been rarely studied (Colmenero-Flores et al. 2007).
In spite of the important work carried out on the Na+ transport mechanism in plants in recent years, more advances are necessary to understand the role and regulation of genes involved in the re-establishment of Na+ homeostasis under salt stress (Maathuis 2006; Pardo et al. 2006; Apse and Blumwald 2007; Munns and Tester 2008). Moreover, these genes should be identified not only in model species like Arabidopsis but also in crop species, since their role may be different depending on the species, as seems to occur in the HKT1 genes. Recent studies on the role of AtHKT1 in Na+ transport have shown that this gene appears to control retrieval of Na+ from the xylem before it reaches the shoot (Davenport et al. 2007). However, the HKT gene family is quite diverse, and this diversity led to early reports of apparently contradictory properties (see ‘AtHKT1;1, A case study of confusion’, in Munns and Tester 2008). Increased clarity has been provided by dividing the HKT gene family into two distinct subfamilies (Platten et al. 2006). Recently, Nagata et al. (2008) did a comparative molecular-biological analysis of membrane transport genes in different organisms, ranging from bacteria to animals and plants. They compared the numbers of membrane transporter genes in Arabidopsis and rice. Although many transporter genes are similar in these plants, Arabidopsis has a more diverse array of genes for multi-efflux transport and for response to stress signals, and rice has more secondary transporter genes for carbohydrate and nutrient transport.
Another of the genes controlling long-distance transport in Arabidopsis thaliana is the SOS1 gene, which encodes a plasma membrane Na+/H+ antiporter that is essential for salt tolerance (Shi et al. 2003). Key intermediaries in the regulation of SOS1 are the protein kinase SOS2 and its associated Ca2+-sensor protein SOS3 (Pardo et al. 2006). Co-expression of the three proteins in a yeast strain lacking endogenous Na+ transporters restored salt tolerance to a much greater extent than SOS1 alone, whereas SOS2 or SOS3 individually failed to stimulate SOS1 activity (Quintero et al. 2002). Despite these advances, the function of SOS1 in plants needs to be clarified, as this transporter seems to function in retrieving Na+ from the xylem to prevent excess Na+ accumulation in the shoot under severe salt stress (Shi et al. 2003). On the contrary, SOS1 functions to load Na+ into the xylem for controlled delivery to the shoot when salinity is moderate (Shi et al. 2002).
Na+ has to be compartmentalised in the vacuole to prevent excess Na+ accumulation in the cytoplasm. Arabidopsis AtNHX1 protein was the first Na+/H+ exchanger identified in plants (Gaxiola et al. 1999). The presence of Na+/H+ antiporter activities has been physiologically characterised in tonoplast vesicles and it is molecularly represented by six Arabidopsis genes, AtNHX1–6 (Yokoi et al. 2002). AtNHX1 steady-state transcript levels were increased in response to NaCl, KCl, sorbitol, and ABA, suggesting that AtNHX1 transcript upregulation is not specific to ionic stress but is common to osmotic stress (Gaxiola et al. 1999; Shi and Zhu 2002). Overexpression of NHX antiporters has been used to improve salt tolerance in several plant species (Zhang and Blumwald 2001; Wu et al. 2004). Pardo et al. (2006) summarised the important work carried out in transformed plants with NHX antiporters and concluded that relevant information is still missing about the way NHX expression affects ion compartmentalisation within the cell, overall ion homeostasis, and osmotic adjustment, in order to fulfil the physiological premises basis that sustain the increase in salt tolerance.
With respect to the Na+ pumps, there are no classical pumps in higher plants. In fungi and mosses there is a Na+ pump, ENA1, which hydrolyses ATP to pump Na+ out of the cell (Benito and Rodriguez-Navarro 2003). Also in yeast are the HAL genes (Serrano et al. 1999). Overexpression of HAL1 conferred salt tolerance in yeast by facilitating intracellular K+ accumulation and decreasing intracellular Na+ (Rios et al. 1997). According to Munns (2005), the expression of these genes, which do not have orthologues in higher plants, may introduce new mechanisms for Na+ and/or K+ homeostasis. HAL1 was introduced into tomato and its overexpression led to higher salt tolerance in the progeny of different transgenic plants, and furthermore, a similar mechanism to that in yeast was observed, namely by facilitating K+/Na+ selectivity under salt stress (Gisbert et al. 2000; Rus et al. 2001). Afterwards, a transgenic line with a very high gene expression level was selected in order to corroborate the tolerance induced by HAL1 in tomato. However, the fruit yield of the homozygous plants was lower than that of the azygous plants, in spite of the lower Na+ uptake and Na+ translocation to the shoot that persisted over time in the homozygous line (Muñoz-Mayor et al. 2008). Moreover, the ability of HAL1 to reduce Na+ uptake in the long-term was shown by the low accumulation of Na+ in fruits. In-depth physiological characterisation of these plants showed that the greater ability for Na+ exclusion in the homozygous line caused another type of osmotic problem, as leaves required increased synthesis of organic solutes to maintain osmotic balance, which has a high energy cost (Balibrea et al. 2003), and hence a growth penalty that negatively impacted on fruit yield. These results demonstrate the importance of considering the osmotic component of salt stress, especially when overexpression is used as a tool to identify genes involved in salt tolerance. It may be concluded that the constitutive expression of a gene may induce improvements in a trait, e.g. Na+ exclusion, but this positive effect may provoke other physiological problems in the plant. In this sense, studies with AtNHX1-overexpressing tomato showed greater K+ uptake than control lines but nevertheless were prone to K+ deficiency symptoms at leaf K+ concentrations greater than the control line (Leidi et al. 2005). According to Pardo et al. (2006), this paradoxical phenotype is likely to be due to exacerbated activity of the AtNHX1 antiporter in the transgenic lines which could increase the vacuolar pool at the expense of the cytoplasmic K+ pool, thereby inducing a K+ starvation signal and eliciting greater K+ uptake by roots. Thus, proper modulation of gene expression in time and space may be more important than mere overexpression of the transgene (Tonsor et al. 2005), as is pointed out below.
Taken together, tolerance to salinity stress is a complex phenomenon at both the cellular and the whole plant level, and a considerable gap still exists between the knowledge gained by physiological and molecular studies in response to salt stress and the knowledge required to develop crop plants with enhanced tolerance to field saline conditions. A focus on comprehensive physiological, molecular and metabolic aspects of salt stress in crop plants is needed to advance in the knowledge of the salt tolerance basis and to facilitate the development of crop plants with enhanced stress tolerance. New tools for functional genomics emerged in recent years may enhance significantly the descriptive power of physiological analysis (Sanchez et al. 2008; Weckwertha 2008), although it is absolutely necessary to elucidate not only processes involved in ionic stress tolerance and compatible solute synthesis, but also other processes as mechanisms of osmotic tolerance, which remain unknown (Munns and Tester 2008). Furthermore, to benefit more from the new genomic approaches, molecular studies with plants grown in physiologically realistic conditions are needed. Finally, it should be mentioned that differences in salt tolerance mechanisms between salt-sensitive glycophytes and salt-tolerant halophytes may result from changes in regulation of the same basic set of genes involved in salt tolerance. Many genes encoding potential salt tolerance determinants have been identified in model plant species, but a comparative study of the expression of those genes in both halophytes and glycophytes has been hampered by the lack of genetic and molecular tools available for the halophytic species. An important step forward has been provided by the use of Arabidopsis molecular and genetic tools to characterise the halophyte T. halophila, which is allowing have shown that the two species exhibit both shared and divergent responses to salt stress (Gong et al. 2005a; Kant et al. 2006). Currently, we are trying to identify genes involved in salt tolerance in the wild salt-tolerant tomato species S. pennellii, using insertional mutagenesis as a genetic tool, as discussed in next sections.
3 Breeding for Salinity Tolerance
The first question that arises when breeding for salinity tolerance is the possibility to develop genotypes tolerant to salinity. Natural evolution has shaped flowering plant species and ecotypes to live under saline conditions, so that there is some compatibility between plant life and saline conditions and, therefore, it should be possible to artificially reproduce this process. However, the term ‘tolerant genotype’ depends upon the context. In natural environments a tolerant genotype is one that competes in the ecosystem and produce offspring. In an agricultural setting a tolerant genotype has to produce an economic yield (Yeo 2007). Breeders try to obtain genotypes not only able to live under saline conditions (biological tolerance) but also to grow and maintain agricultural productivity (agricultural tolerance) with the aid of appropriate cultivation methods. The success achieved in producing salt-tolerant varieties of crops has, however, been very limited. About 29 cultivars in only 12 species had been released for their salt tolerance until year 2000 (Flowers et al. 2000) and there has been little advance since then.
Obtaining salt-tolerant cultivars comprises three steps: (1) the presence of genetic variability for salinity tolerance in the species to be bred, in species that can be crossed with the target species, or even in organisms genetically far from the target species (2) to find out the gene or genes underlying the tolerance, and (3) the transmission of the gene(s) responsible of the tolerance to the cultivars.
3.1 Variability in Salinity Tolerance
All the crops where genetic variability for salt tolerance has been investigated have shown some degree of tolerance, either in the cultivated species or in closely related species that can be crossed with the cultivated one. Examples of species showing intra-specific variability are sorghum (Igartua and Garcia 1999), strawberry clover (Rumbaugh et al. 1993), rice (Alia et al. 2006), cotton (Ashraf 2002), lentil (Ashraf and Waheed 1990), etc. Inter-specific variability among genetically related species has been demonstrated in crops as different as tomato (Bolarin et al. 1991), durum wheat (Munns et al. 2000), eucalyptus (Niknam and McComb 2000) and many others.
Tolerance to salinity in higher plants affects numerous plant processes at all levels of organization (ion transport, osmotic adjustment, ion selectivity, nutrition, compartmentation, growth, water use, water use efficiency, etc.). Variability for osmotic adjustment has been demonstrated in wheat (Morgan 1992) barley (Blum 1989), rice (Lilley and Ludlow 1996), sorghum (Basnayake et al. 1993), maize (Bolaños et al. 1993), tomato (Borsani et al. 2002) etc.; for Na+ transport, in rice (Yeo 1992) and tomato (Cuartero et al. 2002). Cuartero et al. (2002) also demonstrated phenotypic variability in tomato for the relation [Na+]/leaf area reduction, for the relation [K+]/[Na+] in leaf, and for selective Na+ accumulation in old leaves.
It seems that the necessary variability for starting a breeding program is not hindering the development of salt tolerant cultivars in most crops. However “tolerance to salinity” is a very inaccurate term because it depends on the stage of plant development and where the tolerance has been measured. The easiest stage of development for determining tolerance to salinity is germination because experiments can be performed in the lab under controlled environmental conditions and with a high number of genotypes at the same time. But tolerance to salinity at germination stage has proved without relation to tolerance at adult stage in sorghum (Krishnamurthy et al. 2007), lentil (Ashraf and Waheed 1990), tomato (Foolad and Lin 1997), and rice (Moradi et al. 2003). Lack of relation between tolerance in vegetative stages and harvest has also been demonstrated (Greenway and Munns 1980; Shannon et al. 1987; Caro et al. 1991). Tolerance to salinity in one stage of development seems quite independent from tolerance to salinity in other stages what complicates indirect selection and comparison of results coming from different experiments and researchers.
Quantification of salt-tolerance is also difficult in the field, because (1) the stress may be experimentally uncontrollable because of variable rainfall, (2) the notoriously high field heterogeneity for salinity is liable to confound any planting plan designed for field experiments, and (3) salt uptake and sensitivity are modulated by environmental conditions that may affect each variety differently: any parameter which affects the transpiration rate (such as light intensity, temperature and humidity), can change dramatically a plant’s susceptibility to salinity (Yeo et al. 1990). To avoid field problems measuring tolerance, plants are usually grown on inert substrates supplemented with a nutrient solution salinized with NaCl or a known mixture of salts enriched in NaCl.
Salt concentrations at which plants are grown to measure tolerance to salinity deserve also attention. The degree of salt sensitivity of genotypes at the germination stage is generally maintained when different salt concentrations are tested (Cuartero and Fernandez-Muñoz 1999; Foolad 2004), but this behaviour is not general and Cruz (1990) points out that the most tolerant tomato cultivars at 5–7 dS m−1 do not correspond with the most tolerant ones at 13 dS m−1. It is necessary to define salinity conditions on the field and selection and experiments should be performed at those established salinity conditions (Cuartero et al. 2008).
3.2 Determinants Underlying Salinity Tolerance
It is known that most of the processes empirically determined to be important in plant tolerance to salinity exhibit quantitative inheritance (they show continuous variation) and a high degree of environmental influence (Cuartero and Fernandez-Muñoz 1999; Zhang et al. 1999; Mikiko et al. 2001).
Direct selection under field conditions for quantitative traits is difficult because fluctuating environmental factors adversely affect the accuracy and repeatability of such traits. Indirect selection for markers linked to quantitative traits has been suggested and put into practice for some time (Sax 1923; Falconer 1981). For an indirect selection marker to be useful in a breeding program, it has to exhibit both a significant genetic correlation with the trait and higher heritability than the trait itself (Falconer 1981). Molecular (DNA) and biochemical markers closely linked to quantitative trait loci (QTLs) affecting salt tolerance are good candidates because their expression are almost independent from the environment, and powerful biometric methods have been developed and applied to QTL mapping.
The main goal of QTL detection is marker-assisted selection (MAS) and QTL cloning (Asins 2002). The success in both of them depends on our ability to locate the QTL in chromosome regions as narrow as possible.
Salinity affects most of the biological processes in plants; hence tolerance to salinity is determined by many components. Some of them have been reviewed in the first part of this chapter, that is: accumulation of solutes (inorganic and organic) to maintain cell volume and turgor to counteract osmotic stress; ion exclusion from the root and ion compartmentalization to fight against ionic stress; preferential transport of water, K+ and Ca2+ to restore nutritional status; and over-production of antioxidant enzymes and antioxidant compounds to avoid oxidative stress. All of those characters are quantitatively inherited and will require the detection of the QTL governing their expression.
The basis of QTL detection is the identification of significant statistical association between phenotypes and specific genetic markers, established by the joint analysis of segregation of markers and phenotypes in individuals or lines. The most extensively segregating generation used has been the F2 because it can be rapidly obtained. Additive and dominant effects can be properly detected in F2 generations, but F2 have two important drawbacks: the genotypes cannot be replicated and evaluated several times and in several environments, and epistatic interactions are frequently undetected. According to Asins (2002) recombinant inbred lines (RIL) or double haploids (DH) can be reproduced independently and continuously evaluated with respect to additional quantitative traits and markers, with all the information being cumulative. Additive and epistatic interaction can be determined with RIL and DH but not dominant or over-dominance effects.
When dealing with a complex character as tolerance to salinity that depends on a number of quantitative traits, QTL analysis allows an integrative approach. Total phenotypic variation can be explained in three main ways: the identification of QTLs partially controlling the tolerance by analysing multiple traits in the same segregating population; the contribution of QTL × Environment (E) interaction; and the contribution of QTL × QTL interactions or epistastatic effects. Such analyses under different salinity levels and plant stage(s) can only be properly and efficiently carried out using populations of DHs or RILs (Cuartero et al. 2006).
3.2.1 QTL × E Interaction
Since salt concentration in the soil is highly variable, it is frequent to test plant tolerance to salinity in several salt concentrations applied to the root system. Genotype × salt treatment interaction has been found in several occasions and species (Bolarin et al. 1991; Asins et al. 1993; Igartua 1995; Lee et al. 2004). It is then not surprising that QTL also show interaction with the environment, which means that expression of particular chromosome regions differs across environments and markers identified would be significant in only one or some of the environments. However, genotype × salt interactions are detected with ANOVA in the numerous experiments including several genotypes and two or more environmental conditions, but to detect QTL × E interaction it is necessary to study segregant populations with several replicates of each genotype in two or more environments, and unfortunately this kind of studies are rare. In addition F2 population, the most frequent segregant population analyzed, is a poor plant material to detect QTL × E interactions and studies with RIL or DH populations in two or more environments are uncommon.
When appropriate segregant populations (RIL or DH) are grown in at least two salinity conditions (control and saline), more QTL have been identified in saline than in control conditions, and significant QTL × E interaction has been found in all the experiments designed to detect the interaction. Monforte et al. (1997) found only 6 QTL with expression in saline and control conditions, while 9 where expressed in the control and 18 specific for saline conditions. Takehisa et al. (2004) described 37 and 20 QTL in the fields irrigated with saline and fresh water respectively. Manneh et al. (2007) reported 8 markers expressed in control and saline conditions while 28 where expressed in only one of the conditions tested. Villalta et al. (2008) detected 4 QTL in control and 20 in saline conditions in one population, and 11 in saline, 2 in control and 2 in saline and control conditions in another population. Those examples have used only two experimental environments: control and saline conditions. It is necessary to asses that QTL detected under saline condition are expressed in different salt concentrations, otherwise QTL should be found for each specific salt concentration on which tolerant genotypes were to be grown.
QTL × E interaction, as genotype × environment interaction, has an evolutionary base because, according to Asins (2002), this sensitivity to the environment results in phenotypic plasticity (the possibility to take alternative development fates depending on environment) which is likely to be of particular importance in plants because they cannot move from one environment to another. But, from the viewpoint of the breeder, QTL × E interaction complicates the use of QTL in MAS for tolerance to salinity because salinity in the soils is not constant but variable, spatially in the field and also temporarily due to rain and irrigation variation from one year to another. To deal with this unpredictable situation breeders have to manage as many QTL related to tolerance to salinity as possible in order to breed salt tolerant genotypes. Some authors see the QTL × E interaction as lack of consistency of QTL effects in different environments and suggest using only QTL expressed in most of the environments in MAS programmes. However, as mentioned before, there are many more QTL related to tolerance to salinity expressed only under saline conditions and, according to their importance, they are key QTL to breed salt tolerant genotypes.
The number of QTL related to some characters important in salt tolerance is high: 33 in the case of Monforte et al. (1997), 57 detected by Takehisa et al. (2004), 38 by Manneh et al. (2007), and 17 or 21, depending on the population considered, by Villalta et al. (2008). This number will probably increase when more characters and environmental conditions (salt concentrations) are considered, making very difficult to handle all of them in a MAS programme. Fortunately some QTL associations have been found when they have been mapped (Villalta et al. 2008) which will assist in the introduction of those QTL clusters in elite lines.
3.2.2 QTL × QTL Interaction
QTL detection experiments are notoriously poor at detecting interactions between loci (Frankel and Schork 1996). For this reason, we do not have a good idea as to how common such epistatic effects are, but when experiments have been designed to test specifically for their presence, these interactions have been found (Flint and Mott 2001). The molecular dissection of variation in bristle number in Drosophila indicated that the combination of two QTL had much larger effect than predicted from their individual effects (Gurganus et al. 1999). Epistasis has also been documented in plants. Lark et al. (1995) demonstrated epistasis in soybean, Eshed and Zamir (1996) in tomato, Lukens and Doebley (1999) in maize, Maheswaran et al. (2000) in rice, etc. The detection of epistasis seems to be independent of the species and the lack of information about QTL × QTL interaction could be explained by the plant material employed in the experiments. According to Asins (2002), epistasis between QTL can hardly be detected in advanced backcrosses or F2 populations. The reason is that every backcross generation reduces the number of gene combinations while increasing genes from the recurrent genotype and in F2 populations, even if large, there are few individuals with two-locus double homozygotes. RIL or DH populations are, definitely, the appropriate plant material to detect epistasis.
MAS will lose efficiency if QTL × QTL interactions do exist and are ignored in the selection process because they have not been detected and quantified.
3.3 Transmission of Determinants Responsible of Salt Tolerance
The traits governing tolerance to salinity are quantitatively inherited with low heritability (Cuartero et al. 2006). In addition, these traits are difficult to measure in segregating populations, requiring meticulous control of environmental variables and replication of progenies over locations and seasons. MAS is an attractive approach when a few QTL control a significant portion of the variability for the traits under selection (Zhang et al. 1999).
Translating results from gene or QTL discovery to farm applications requires an agronomic approach rather than a purely academic perspective. We need not only an understanding of the genetic mechanisms underlying a particular trait but also a keen sense of the target environments and the appropriate germplasm to use as vehicles to deliver the traits (Leung 2008). It is important to note that cultivars bred for salt tolerance have to be not only salt tolerant, but also achieve the same desirable traits of productivity, quality, resistance to diseases, and adaptation to cultural techniques the current cultivars have. Elite lines, cultivars or parents of today’s hybrids would be the ideal candidates to be improved for tolerance to salinity by introducing the tolerant QTL from the donors.
Today cultivars are the result of many years of selection and incorporation of traits demanded by consumers and growers. The delicate equilibrium among productivity, quality, resistance to diseases, and other agronomic traits showed by those cultivars could be probably altered with the introduction of a substantial number of QTL as those required for tolerance to salinity. Even with the help of codominant molecular markers tightly linked to the QTL to be transmitted, MAS programmes will need the phenotypic selection of the breeder generation after generation. Phenotypic selection can only be properly practised in generations approaching homocigosity slowly. Accordingly, MAS programmes should be designed to slowly approach the recurrent parent genotype. Cuartero and Fernández-Muñoz (1999) described an example of such a programme. A partial solution would be to breed rootstocks tolerant to salinity, grafting onto them the shoot of today cultivars. Martinez-Rodriguez et al. (2008) have recently demonstrated the beneficial effects of some rootstocks chosen because of their tolerance to salinity on tomato yield irrigated with saline water.
Despite innovations like better marker systems and improved genetic mapping strategies, the success of MAS has been very limited even taking into account that tolerance of the breeding lines is not expected to be as high as that of the tolerant donors. Manneh et al. (2007) pointed out the reliability of MAS to identify superior yielding rice genotypes under stress in the field. Several rice lines have been bred and released in the Philippines, Bangladesh, and India demonstrating significant yield advantages over salt-sensitive varieties (Ismail et al. 2007). Those two examples of success in breeding for salinity tolerance have been performed with rice. Rice has two important advantages over other species, especially dicotyledonous species: (1) tolerance to salinity is controlled by a few QTL with large effects, and (2) it is especially sensitive to salinity only during early seedling and reproduction stages (Leung 2008). The partial success on breeding salt tolerant rice varieties, a feasible plant model, should encourage the work with other species.
Salinity increases steadily in agricultural soils because of the irrigation water employed and because inadequate irrigation practices. If a soil salinity target has been fixed at the beginning of a breeding programme, it can substantially increase in the about 10 year’s period needed to obtain a new cultivar with tolerance to salinity. So, breeding salinity tolerance cultivars cannot be alone a long term solution to grow in saline soils but it must be complemented by other cultural techniques to stabilize the soil salinity level (Cuartero et al. 2008).
4 Improving Salt Tolerance Trough Gene Transformation
Despite great efforts to increase the level of salinity tolerance in species of agronomic interest, the results obtained through conventional breeding methods, as well as by some biotechnological approaches (e.g. in vitro selection) have been rather scarce (Flowers 2004; Yamaguchi and Blumwald 2005; Ashraf et al. 2008). The difficulty in obtaining practical results through these and other approaches explains the expectations generated to obtain salt tolerant cultivars via genetic transformation.
In numerous papers published from early 1990s until the present time, several authors have claimed enhancement of salt tolerance through either overexpression of endogenous genes or, more frequently, heterologous expression of genes that supposedly act on different mechanisms involved in the process (Cuartero et al. 2006).
Genes that have proven somewhat effective in providing stress tolerance using a transgenic approach belong to different categories. Preliminary research in this field was mainly focused on the overproduction of metabolically compatible (organic) solutes in transgenic plants (Chen and Murata 2002; Penna 2003; Kavi Kishor et al. 2005). More recently, attention has been paid to the modification of the glycine betaine biosynthesis pathway by using genes isolated from different sources (Su et al. 2006; Shirasawa et al. 2006; Zhou et al. 2007; Waditee et al. 2007; Park et al. 2007), most probably because the quaternary ammonium compound glycine betaine is accumulated in numerous halophytes from several families (Rhodes and Hanson 1993; Flowers and Colmer 2008). In this respect, it has been shown that the accumulation of glycine betaine in genetically modified plants of tomato is more effective in the chloroplasts than in the cytosol (Park et al. 2007), in a similar way to that previously observed in rice (Sakamoto et al. 1998). Notably, the accumulation of glycine betaine in transplastomic plants of carrot led to high levels of salt tolerance (up to 400 mM NaCl; Kumar et al. 2004).
Another strategy to increase the level of salt tolerance has been the transfer of genes codifying different kinds of proteins functionally related to macromolecules protection (LEA proteins, osmotin, chaperons, mRNA binding proteins; Wang et al. 2003; Zhang et al. 2007) or the protection of key metabolic enzymes (Arrillaga et al. 1998).
The scavenging of reactive oxygen intermediates through the transfer and expression of genes encoding detoxification enzymes (e.g. glutathione S-transferase, superoxide dismutase, ascorbate peroxidase, catalase) is an alternative way to protect cells against oxidative stress, thus limiting the damage produced by salt treatments (Apel and Hirt 2004). Interestingly the co-expression of more than one gene involved in oxidative stress protection in both the chloroplasts and cytosol gave rise to plants with increased tolerance to different types of abiotic stress (Zhao and Zang 2006; Tseng et al. 2007; Lee et al. 2007).
Genetic manipulation with genes encoding membrane proteins involved in the uptake and transport of water and ions, such as water channel proteins and ion transporters, is an alternative approach (Yamaguchi and Blumwald 2005; Chinnusamy et al. 2005; Forrest and Bhave 2007). As ion transport across the tonoplast into vacuoles is energised by a proton moving force (Gaxiola et al. 2007), the strategy based on the use of antiporters has generated high expectations in recent years. By overexpressing the vacuolar Na+/H+ antiport from Arabidopsis thaliana (AtNHX1), a high level of salt tolerance was reported in genetically modified plants of arabidopsis (Apse et al. 1999), tomato (Zhang and Blumwald 2001) and canola (Zhang et al. 2001). Thereafter, the AtNHX1 gene was overexpressed in genetically modified plants of wheat (Xue et al. 2004), corn (Yin et al. 2004), beet (Yang et al. 2005), cotton (He et al. 2005) and tall fescue (Tian et al. 2006; Zhao et al. 2007), and in all the cases the authors indicated enhancement of salt tolerance. The greater tolerance conferred by the AtNHX1 gene has been attributed to a process of Na+ compartmentalisation into the vacuoles (Yamaguchi and Blumwald 2005). Despite being an attractive hypothesis, additional compelling evidence is needed before drawing a definitive conclusion. Since monovalent ions are judged toxic at the concentrations required for osmotic adjustment, it is generally assumed that Na+ and Cl− are compartmentalised in halophytes, predominantly in vacuoles, so that concentrations in the cytoplasm are maintained within tolerable limits. However, as stated by Flowers and Colmer (2008), experimental evidence for compartmentalisation of Na+ into vacuoles is still limited, even in halophytes.
New AtNHX genes have been cloned and characterised (Yokoi et al. 2002; Aharon et al. 2003; Wang et al. 2007; Liu et al. 2008) and much effort has been made to identify orthologous genes in different species and perform the functional analyses, usually by overexpression in genetically modified plants. Examples are AgNHX1 from Atriplex gmelini (Ohta et al. 2002); BnNHX1 from Brassica napus (Wang et al. 2004); GhNHX1 from Gossypium hirsutum (Wu et al. 2004); OsNHX1 from Oryza sativa (Fukuda et al. 2004; Wu et al. 2005; Chen et al. 2007); HbNHX1 from Hordeum brevisubulatum (Lu et al. 2005); GmNHX1 from Glycine max (Sun et al. 2006; Li et al. 2006); SsNHX1 from Suaeda salsa (Zhao et al. 2006b, c; Li et al. 2007); PgNHX1 from Pennisetum glaucum (Verma et al. 2007; Rajagopal et al. 2007); TNHX1 from Triticum aestivum (Brini et al. 2007); and AeNHX1 from Agropirum elongatum (Qiao et al. 2007).
Overexpression of a vacuolar H+-pyrophosphatase (AVP1) from Arabidopsis thaliana in transgenic plants of the same species increases the level of salt tolerance (Gaxiola et al. 2001). Similar results have been achieved by overexpressing the homologues from Thellungiella halophila (TsVP) in tobacco (Gao et al. 2006) and cotton (Lv et al. 2008), Suaeda salsa (SsVP) in arabidopsis (Guo et al. 2006), and Triticum aestivum (TVP1) also in arabidopsis (Brini et al. 2007). Interestingly, TsVP and SsVP genes have been cloned from halophytes (Thellungiella halophila and Suaeda salsa, respectively).
Likewise, a higher level of salt tolerance has been described through the overexpression of genes that codify plasma membrane Na+/H+ antiports cloned from different sources. For example, AtSOS1 from Arabidopsis thaliana (Shi et al. 2003); SOD2 from Schizosaccharomyces pombe (Gao et al. 2003; Zhao et al. 2006a); nhaA from Escherichia coli (Wu et al. 2005); and OsSOS1 from Oryza sativa (Martínez-Atienza et al. 2007).
Regulatory genes such as transcription factors and those codifying signal transduction components or receptor-related proteins are another target (Kaur and Gupta 2005; Agarwal et al. 2006). Cloning of genes codifying transcription factors is a promising field as they lie upstream of many other genes. Recent research has allowed the identification of several transcription factors that are important in regulating plant stress responses, including not only different kinds of abiotic stress but also pathogen-induced defence responses, various physiological processes, hormonal signalling pathways and several developmental processes (Agarwal et al. 2006; Ham et al. 2006; Sohn et al. 2006; Seong et al. 2007; Ogawa et al. 2007; Liu et al. 2007; Dai et al. 2007; Nakashima et al. 2007).
It has also been suggested that genes codifying calcium sensors (Cheong et al. 2003) or even DNA helicases (pea DNA helicase 45, PDH45, Sanan-Mishra et al. 2005) and RNA helicases (DEAD-box helicase, Gong et al. 2005b; Owttrim 2006) could be involved in the process of salt tolerance. The role of siRNAs in stress conditions is also under study (Sunkar and Zhu 2004; Borsani et al. 2005; Jung and Kang 2007). Finally, knowledge of the processes related to DNA/RNA metabolism and G-protein signalling pathways could be useful in elucidating the less known stress signalling networks and thereby be helpful for engineering salinity-tolerance in crop plants (Tuteja 2007).
5 Functional Analysis of Salt Tolerance-Related Genes
Overall, the results obtained in this field show that the expression of different kinds of genes in transgenic plants can increase salinity tolerance, at least to some extent. Unfortunately, it is not possible to conclude for the moment that true halotolerant cultivars (i.e. with a sufficient tolerance level from an agronomic point of view) have been obtained via transformation. In fact, it would be best to avoid excessive optimism when drawing conclusions on the current state of this topic (Flowers 2004). Furthermore, when performing the functional analysis of a salt tolerance-related gene it would be advisable to take into consideration aspects such as the species used in the transformation, the procedure for evaluating the tolerance to salinity, and the complexity of the trait.
With respect to the first issue, the majority of transformation experiments have been carried out with the model species arabidopsis and tobacco (Vij and Tyagi 2007) which means we should be cautious when drawing conclusions. It would be best to perform such experiments in crops (Grover et al. 2003; Yamaguchi and Blumwald 2005; Cuartero et al. 2006; Bhatnagar-Mathur et al. 2008), as it is not certain what will occur when the genes that have given a positive result in models are expressed in cultivated species. The suitability of tobacco as a model in this field has been seriously questioned (Murthy and Tester 1996) and results from the evaluation of salt tolerance in transgenic arabidopsis plants cannot easily be extrapolated to a crop species, as the important trait in the latter is the maintenance of production under stress. Extrapolations between different crop species cannot even be made, since the effects of salinity can be very different between species. If true advances are sought, the best approach is to focus efforts on cultivated species where the transformation technology is already available. Without doubt, the difficulties will be greater and the advances slower in crops than in model species, but the results will indicate the true importance of a certain transgene in the genetic context in which the tolerant phenotype will supposedly occur. Last but not least, any results could be of practical interest (Cuartero et al. 2006).
Regarding the procedure for evaluating the tolerance to salinity, if the published results are scrutinised, some of the methods of evaluation of transgenic materials appear of doubtful value. Results of a descriptive type or those based upon photographic evidence of the performance of plants may lead to confusing or erroneous conclusions (Flowers 2004). Responses to salt are frequently studied using small samples, in the very short-term, by using shock treatments and, furthermore, the data are collected during very specific growth periods (Yamaguchi and Blumwald 2005), in spite of the fact that in most crop species salt sensitivity depends on the growth stage (Perez-Alfocea et al. 1993; Khatum and Flowers 1995). Moreover, tolerance estimated on the basis of seed germination is not correlated with tolerance at later growth stages (Foolad and Lin 1997; Cuartero and Fernandez-Muñoz 1999). The usefulness of in vitro tests, frequently used for the evaluation of salt tolerance, could also be questioned because transpiring conditions have a major influence on Na+ transport and tolerance (Moller and Tester 2007). However, a clear relationship between tolerance to salinity in vitro (callus) and in vivo (plants grown in greenhouse) has been observed for cultivated and wild tomato species (Perez-Alfocea et al. 1994; Cano et al. 1996) and similar results were obtained for cultivated species of Cucumis and Citrullus (melon, cucumber, and watermelon) and related wild species (Barage 2002). In vitro tests can provide complementary information on the effect of some transgenes (e.g. genes involved in ionic homeostasis) and can be useful for the pre-selection of transgenic lines (if an in vitro and in vivo correlation has previously been shown), but they should not be used as the only criterion to determine the degree of salt tolerance. In evaluating the tolerance of transgenic crops, it is important to perform long-term experiments, focus on growth and yield, and provide quantitative data (Flowers 2004; Munns and Tester 2008).
Bressan et al. (2008) have discussed the convenience of defining a minimum set of criteria for establishing unambiguously that transgenic plants do indeed show tolerance that is attributable to the transgene. In this respect, an important aspect in the functional analysis of a salt tolerance-related gene is the plant material to be used. The use of TG1 plants (primary transformants) is questionable because epigenetic effects (which are very important in some cases) may lead to erroneous conclusions. The evaluation in TG2 avoids the above problem, but it is necessary to take into account that this is a segregant progeny. In the authors’ opinion, the best materials are the homozygous and azygous lines obtained in TG3. Thus, each homozygous line should be compared with two controls: the wild type and the corresponding azygous line without the transgene. Positional effects can generate great differences in the expression of a given transgene in independent transgenic lines, indicating the necessity for selecting those with the best expression for the trait (Pineda 2005). Dose effects of the transgene can be estimated by comparing the behaviour of homozygous versus hemizygous lines (i.e. those derived from the sexual crossing between the homozygous and azygous lines). The relative tolerance of these lines can be estimated in the short- and mid-term, although, ultimately, the long-term response (estimating yield with quantitative data) must also be reported.
5.1 Complexity of the Trait and Sources of Genetic Variation
Salt tolerance is a complex trait (Bohnert et al. 1996; Tuteja 2007; Munns and Tester 2008). If one takes into account the diversity of mechanisms involved, the question that immediately arises is whether the introduction of a single gene can produce a sufficient level of tolerance or whether it is necessary to introduce several genes involved in different processes (e.g. osmotic adjustment, osmoprotection, ionic homeostasis, oxygen free radical scavengeing, stress response, restoration of enzymatic activity, photorespiration; Bohnert et al. 1996). Of course a particular gene (e.g. one that codes for a transcription factor) can have a cascade effect, modifying the expression of many genes. Alternatively, the expression of a gene involved in the compartmentalisation of ions in the vacuoles may alleviate toxic effects. Even so, it seems unlikely that a single gene could affect all the processes influenced by salinity. What is most likely is that the transference and expression, in a co-ordinated way, of a series of genes, each of which would affect one of the principal mechanisms of the process, would produce tolerant plants. The problem is that there is still not a clear idea of which genes have to be transferred.
When considering the future targets in this field, one can argue that rather than looking for salt tolerant-related genes in salt sensitive species, like arabidopsis, it would be better to focus on halotolerant plants. Flowers and Colmer (2008) have recently reviewed the mechanisms of tolerance in halophytes, plants that are able to survive and reproduce in environments where the salt concentration is around 200 mM NaCl or more. In this respect, the authors have proposed that research should be concentrated on a number of ‘model’ (halotolerant) species that are representative of the various mechanisms that might be involved in tolerance. As these halophytes are evolutionarily distant from the main crop species, from a breeding point of view it would perhaps be better to take advantage of the existence of halotolerant accesions of wild species related to a given crop, as occurs in the genus Solanum (Cuartero et al. 2006), Citrullus (Barage 2002), Cucumis (Barage et al. 2002) and many others. Unfortunately, despite the wealth of sources of variation, it is still not known which are the key genes determining the high level of salt tolerance in those plants.
6 Genomic Approaches for Dissection of Salinity Tolerance
The use of functional genomic approaches may serve to overcome the above mentioned problems. Transcriptomic analysis provides the expression profiles of hundreds or thousands of genes. At present, this kind of approach is being used to identify those genes that are expressed or deactivated in response to saline or other types of abiotic stress (Cuartero et al. 2006, and references therein). Although these methods might lead to an overestimation of the number of genes supposedly involved, which would make the identification of relevant genes among an enormous number of other genes with purely secondary or irrelevant functions more difficult, it is foreseeable that transcriptomic analysis will become a valuable tool in the near future. However, in order to fulfil the expectations created in this field, it would be sensible to take into account the stage of development at which the saline treatment is applied, to perform tissue-specific studies and to avoid traumatic or unnatural treatments (Munns 2005). In fact, transcriptomics studies can produce different answers depending on the tissue examined and whether the plant is growing or dying (Munns and Tester 2008). The relationship between the intensity of saline treatment and the degree of salt tolerance is another point to be considered. A high-salt treatment for a sensitive plant like arabidopsis will induce changes predominantly associated with senescence; however, a low-salt treatment may not result in discernable changes in gene expression (Munns and Tester 2008). The opposite situation should also be taken into account, as a high-salt treatment for a salt-tolerant plant (e.g. halophyte) may not produce any remarkable change in gene expression (as those plants grow normally in that situation and tolerate salt concentrations that kill or inhibit the growth of the majority of glycophytes) and a low-salt treatment may just reflect an abnormal situation for that species. Rather than apply these approaches either to model (salt-sensitive) species or halophytes, it would be better to apply them in both crop species and halotolerant accessions of related wild species and thus, by comparison, try to identify the genes responsible for tolerance (Bohnert et al. 2006; Cuartero et al. 2006).
Other genomic approaches should provide very useful information. For example, major advances have been achieved in the study of mechanisms of post-transcriptional gene silencing and high throughput systems are available to infer gene function (Baulcombe 2004; Herr et al. 2005; Cherian et al. 2006). We foresee that, if systematically applied in a large-scale program using halotolerant plants, this approach would be particularly valuable for the identification of genes involved in different mechanisms related to salt tolerance.
Overexpression has hitherto been the most widely used strategy for both the functional analysis of candidate genes as well as for the increase in salt tolerance in transgenic plants. The underlying idea is that by overexpressing a certain gene or by expressing it in a constitutive way it would always have a positive effect on the phenotype. But increasing evidence supports the idea that sometimes strong and constitutive promoters (e.g. CaMV-35S, mostly for dicots, or actin and ubiquitin1, for monocot species) involve a high energetic cost and yield a penalty in transgenic plants (Rus et al. 2001; Grover et al. 2003; Pineda 2005; Muñoz-Mayor et al. 2008) and, in other cases, the beneficial effects of the transgene are masked by pleiotropic effects derived from the use of strong promoters (Romero et al. 1997; Capell et al. 1998; Kasuga et al. 1999). Yield penalty and/or pleiotropy not only make the interpretation of the results on the functional analysis of a given transgene difficult but also hamper any practical application. In fact, evidence from research in this field supports the advantages of using inducible promoters (Kasuga et al. 1999; Garg et al. 2002; Rohila et al. 2002; Lee et al. 2003; Su and Wu 2004; Nakashima et al. 2007). Moreover, the use of inducible or specific promoters will be essential when tackling the co-transference and co-expression of several genes to avoid homology-based gene silencing (Grover et al. 2003; Cuartero et al. 2006). It is expected that the identification of new cis regulatory elements, which allow proper expression in time and space, will be a major target in the near future (Yamaguchi and Blumwald 2005; Cherian et al. 2006; Cuartero et al. 2006; Bhatnagar-Mathur et al. 2008).
The use of mutants as genomic tools should also be one of the main research areas in the coming years. One of the key factors explaining our present knowledge in several areas of plant development lies in the detection and characterisation of mutants that have altered developmental traits, for example those affected in tomato fruit development and maturation (Emmanuel and Levy 2002; Tanksley 2004; Giovannoni 2007; Lozano et al. 2009). By comparison, the number of mutants with the level of salt tolerance affected in other species than arabidopsis which are already available for the scientific community is rather scarce, perhaps due to the difficulty in performing proper evaluation of the trait. Occasional spontaneous mutants or, alternatively, those generated by chemical (e.g. EMS) or physical (e.g. fast neutrons) methods could provide the basis for advancing the knowledge of physiological processes related to salt tolerance. However, in the absence of obvious candidate genes, the isolation of a gene altered in the mutant through a positional cloning strategy requires huge effort. Fortunately, at present we can overcome these problems by using alternative approaches.
Insertional mutagenesis with T-DNA or transposable elements constitutes a basic tool for the identification of genes and the analysis of their function. With respect to insertional mutagenesis with T-DNA, we can approach the tagging of genes by using a simple construction with a marker gene. In this way, the integration of T-DNA within the structural sequence or the controlling elements of a given gene will lead to its disruption and the consequent loss-of-function or, depending of the characteristics of the T-DNA-insert, gain-of-function or change in its level of expression (Krysan et al. 1999). Upon detecting the mutant phenotype in TG1 (in the case of dominant, semidominant or additive effects) or TG2 (recessive), its cloning can be easier as the gene is tagged by the T-DNA.
By comparison with classical insertional mutagenesis, trapping systems (Springer 2000) can be particularly useful for the identification of genes related to salt tolerance. The advantage of using enhancer, promoter or gene traps resides in its self dual nature. Like any other T-DNA, those traps act as insertion mutagens, but also, when T-DNA is integrated inside an endogenous gene in the appropriate orientation, the reporter gene lies under the control of the regulatory elements of the tagged gene. By analysing the reporter gene expression one can obtain a precise picture of the spatial and temporal expression pattern of the endogenous gene tagged by the trap. In this respect, trapping strategies bring great advantages over insertional mutagenesis by allowing identification of functionally redundant genes, those expressed at multiple developmental stages (generating confusion during phenotyping), genes whose disruption causes early lethality, and genes whose disruption causes a soft phenotype that may not be detected (in this case, the reporter expression gives a clue to identify the phenotype during evaluation). In addition, gene identification is independent of the expression level of the gene, avoiding the risk of rejecting genes that are expressed at low levels, even though they have major effects on the phenotype. Finally, this is the best way to identify genes that are activated or repressed in response either to an external stimulus or biotic and abiotic stress situations.
Using an enhancer trap (kindly provided by Dr. Thomas Jack, Department of Biological Sciences, Dartmouth College, USA) and a promoter trap (developed in the laboratory of Drs. Rafael Lozano and Trinidad Angosto, Universidad de Almería, Spain), a collection of more than 2,000 T-DNA lines of halotolerant accessions of the wild tomato-related species Solanum pennellii has been generated (Anton et al 2009). Scrutiny of this collection to identify insertion mutants with altered levels of saline stress (mainly hypersensitive) is under way. Given that the collection of T-DNA lines is going to enlarge progressively, identification of new salt tolerance related-genes is expected in the near future. It is foreseeable that the use of these genomic approaches will allow the genetic dissection of the trait and, thereafter, the proper design of a breeding program.
7 Conclusions
A significant gap still exists between the knowledge gained in physiological and molecular studies of responses to salt stress, and the knowledge required to develop crop plants with enhanced tolerance to saline conditions. A focus on physiological, molecular and metabolic aspects of salt stress in crop plants would help to bridge this gap. It is still necessary to elucidate some processes involved in ionic stress tolerance, synthesis of compatible solute and also other processes as the mechanisms of osmotic tolerance. New tools arising from functional genomics may significantly enhance the descriptive power of physiological analysis and molecular studies with plants grown under realistic saline conditions.
It seems then that the necessary variability for starting breeding programs is not hindering the development of salt tolerant cultivars in most crops, although it is necessary to look for variability to some key physiological traits related to osmotic and ionic stresses, in specifically salt concentrations and plant development stages. QTL detection and mapping requires the development of large RIL or DH generations from parents with a wide variation in a number of traits related to tolerance to salinity. These populations will help to: (1) locate the QTL in chromosome regions as narrow as possible, (2) accumulate QTL information gained in different experiments and growing seasons, and (3) quantify QTL × E and QTL × QTL interactions. Elite cultivars or elite lines, parents of today’s hybrids, are the ideal candidates to receive the salt tolerance QTL from donor genotypes. Introduction of QTL should be made by combining marker assisted and phenotypic selection in breeding programs approaching slowly the recurrent elite lines. This will allow breeders to maintain the delicate equilibrium among productivity, quality, resistance to diseases, and other agronomic traits showed by elite cultivars and lines, that could be otherwise be altered with the introduction of a substantial number of QTL as those required to obtain tolerance to salinity.
Expression of some transgenes promotes a high level of salt tolerance in different species. Despite these interesting results, it is not possible to conclude yet that cultivars tolerant enough from an agronomic point of view have been obtained via transformation. To fulfil the expectations created by the plant transformation technique, it is necessary to advance on different aspects: (1) identification of genes actually involved in the process of salt tolerance, (2) isolation of genes from adequate sources of variation (wild species related to a given crop), (3) design vectors that allow the transfer and coordinate expression of several genes (since salt tolerance is a complex trait), and (4) identification of regulatory elements modulating spatially and temporarily the expression level of the transgenes. Despite present limitations, it is foreseeable that breeding programmes will benefit from ongoing functional genomics projects that could allow the use of genetic transformation as a regular breeding tool.
References
Agarwal PK, Agarwal P, Reddy MK et al (2006) Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep 25:1263–1274
Aharon GS, Apse MP, Duan S et al (2003) Characterization of a family of vacuolar Na+/H+ antiporters in Arabidopsis thaliana. Plant Soil 253:245–256
Alarcon JJ, Sanchez-Blanco MJ, Bolarin MC et al (1993) Water relations and osmotic adjustment in Lycopersicon esculentum and L. pennellii during short-term salt exposure and recovery. Physiol Plant 89:441–447
Alia AJ, Xua JL, Ismail AM et al (2006) Hidden diversity for abiotic and biotic stress tolerances in the primary gene pool of rice revealed by a large backcross breeding program. Field Crops Res 97:66–76
Anton T, Garcia-Abellan JO, Perez F et al (2009) Identification of genes related to salt and drought tolerance in T-DNA tagging lines of cultivated and wild tomato species. In: Plant abiotic stress tolerance. Proc of Internat Conferen, Vienna feb 2009:161
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399
Apse MP, Blumwald E (2002) Engineering salt tolerance in plants. Curr Opin Biotechnol 13:146–150
Apse MP, Blumwald E (2007) Na+ transport in plants. FEBS Lett 581:2247–2254
Apse MP, Aharon GS, Snedden WA et al (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis thaliana. Science 285:1256–1258
Arrillaga I, Gil-Mascarell R, Gisbert C et al (1998) Expression of the yeast HAL2 gene in tomato increases the in vitro ssalt tolerance of transgenic progenies. Plant Sci 136:219–226
Ashraf M (2002) Salt tolerance of cotton: some new advances. Crit Rev Plant Sci 21:1–30
Ashraf M, Waheed A (1990) Screening of local/exotic accessions of lentil (Lens culinaris Medic.) for salt tolerance at two growth stages. Plant Soil 128:167–176
Ashraf M, Athar HR, Harris RJC et al (2008) Some prospective strategies for improving crop salt tolerance. Adv Agron 97:45–110
Asins MJ (2002) Present and future of quantitative trait locus analysis in plant breeding. Plant Breed 121:281–291
Asins MJ, Bretó MP, Cambra M et al (1993) Salt tolerante in Lycopersicon species. II. Genetics effects and a search for associated traits. Theor Appl Genet 86:769–774
Balibrea ME, Cuartero J, Bolarin MC et al (2003) Sucrolytic activities during fruit development of Lycopersicon genotypes differing in tolerance to salinity. Physiol Plant 118:38–46
Barage M (2002) Identificación de fuentes de tolerancia a la salinidad y al estrés hídrico en especies silvestres de la familia Cucurbitaceae. Ph.D. thesis. Universidad Politécnica de Valencia, Valencia, Spain
Barage M, Pineda B, Atares A et al (2002) Identificación de fuentes de tolerancia a la salinidad en especies silvestres de la familia Cucurbitaceae sobre la base de parámetros de crecimiento vegetativo. Actas de Hrticultura 34:133–138
Bartels D, Dunkar R (2005) Drought and salt tolerance in plants. Crit Rev Plant Sci 24:23–58
Basnayake J, Ludlow MM, Cooper M et al (1993) Genotypic variation among adjustment and desiccation tolerance in contrasting sorghum inbred lines. Field Crops Res 35:51–62
Baulcombe D (2004) RNA silencing in plants. Nature 431:356–363
Ben Amor N, Jimenez A, Megdiche W et al (2007) Kinetics of antioxidant response to salinity in the halophyte Cakile maritima. J Integr Plant Biol 49:1–11
Benito B, Rodriguez-Navarro A (2003) Molecular cloning and characterization of a sodium-pump ATPase of the moss Physcomitrella patens. Plant J 36:382–389
Bhatnagar-Mathur P, Vadez V, Sharma KK (2008) Transgenic approaches for abiotic stress tolerance in plants: retrospect and prospects. Plant Cell Rep 27:411–424
Blum A (1989) Osmotic adjustment and growth of barley genotypes under drought stress. Crop Sci 29:230–233
Blum A, Munns R, Passioura JB et al (1996) Genetically engineered plants resistant to soil drying and salt stress: How to interpret osmotic relations? Plant Physiol 110:1051
Bohnert HJ, Gong Q, Li P et al (2006) Unraveling abiotic stress tolerance mechanisms - getting genomics going. Curr Opin Plant Biol 9:180–188
Bohnert HJ, Golldack D, Ishitani M et al (1996) Salt tolerance engineering requires multiple gene transfer. In: Collins GB, Shepherd RJ (eds) Engineering plants for commercial products and application. Ann New York Acad Sci 729:115–125
Bolaños J, Edmeades GO, Martinez L (1993) Eight cycles of selection for drought tolerance in lowland tropical maize. III. Responses in drought-adaptive physiological and morphological traits. Field Crops Res 31:269–286
Bolarin MC, Fernández FG, Cuartero J et al (1991) Salinity tolerance in four wild tomato species using vegetative yield-salinity response curves. J Am Soc Hort Sci 116:286–290
Borsani O, Cuartero J, Valpuesta V et al (2002) Tomato tos1 mutation identifies a gene essential for osmotic tolerance and abscisic acid sensitivity. Plant J 32:905–914
Borsani O, Zhu J, Verslues PE et al (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123:1279–1291
Bressan RA, Bohner HJ, Hasegawa PM (2008) Genetic engineering for salinity stres tolerance. Adv Plant Biochem Mol Biol 1:347–384
Brini F, Hanin M, Mezghani I et al (2007) Overexpression of wheat Na+/H+ antiporter TNHX1 and H+-pyrophosphatase TVP1 improve salt- and drought-stress tolerance in Arabidopsis thaliana plants. J Exp Bot 58:301–308
Cano EA, Perez-Alfocea F, Moreno V et al (1996) Responses to NaCl stress of cultivated and wild species and their hybrids in callus culture. Plant Cell Rep 15:791–794
Capell T, Escobar C, Lui H et al (1998) Overexpression of the oat arginine decarboxylase cDNA in transgenic rice (Oryza sativa L.) affects normal development patterns in vitro and results in putrescine accumulation in transgenic plants. Theor Appl Genet 97:246–254
Caro M, Cruz V, Cuartero J et al (1991) Salinity tolerance of normal-fruited and cherry tomato cultivars. Plant Soil 136:249–255
Chen H, An R, Tang JH et al (2007) Over-expression of a vacuolar Na+/H+ antiporter gene improves salt tolerance in an upland rice. Mol Breed 19:215–225
Chen THH, Murata N (2002) Enhancement of tolerance of abiotic stress by metabolic engineering of betaines and other compatible solutes. Curr Opin Plant Biol 5:250–257
Cheong YH, Kim KN, Pandey GK et al (2003) CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell 15:1833–1845
Cherian S, Reddy MP, Ferreira RB (2006) Transgenic plants with improved dehydration-stress tolerance: progress and future prospects. Biol Plant 50:481–495
Chinnusamy V, Jagendorf A, Zhu JK (2005) Understanding and improving salt tolerance in plants. Crop Sci 45:437–448
Colmenero-Flores JM, Martinez G, Gamba G et al (2007) Identification and functional characterization of cation-chloride cotransporters in plants. Plant J 50:278–292
Cruz V (1990) Tolerancia a la salinidad y criterios de selección en Lycopersicon Mill. spp. Tesis doctoral. Universidad de Málaga, 484 p
Cuartero J, Fernández-Muñoz R (1999) Tomato and salinity. Sci Hort 78:83–125
Cuartero J, Bolarin MC, Asíns MJ et al (2006) Increasing salt tolerance in the tomato. J Exp Bot 57:1045–1058
Cuartero J, Bolarin MC, Moreno V et al (2008) Tolerancia a la salinidad. In: Moreno MT, Cubero JI, Atienza S, et al (eds) El ambiente y los estreses abióticos en mejora vegetal. IFAPA-Junta de Andalucía:231–262
Cuartero J, Romero-Aranda R, Yeo AR et al (2002) Variability for some physiological characters affecting salt tolerance in tomato. Acta Hort 573:435–441
Dai X, Xu Y, Ma Q et al (2007) Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol 143:1739–1751
Davenport RJ, Muñoz-Mayor A, Jha D et al (2007) The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ 30:497–507
De Costa W, Zorb C, Hartung W et al (2007) Salt resistance is determined by osmotic adjustment and abscisic acid in newly developed maize hybrids in the first phase of salt stress. Physiol Plant 131:311–321
Emmanuel E, Levy AA (2002) Tomato mutants as genomic tools. Curr Opin Plant Biol 5:112–117
Eshed Y, Zamir D (1996) Less-than-additive epistatic interactions of quantitative trait loci in tomato. Genetics 143:1807–1817
Estañ MT, Martinez-Rodriguez MM, Perez-Alfocea F et al (2005) Grafting raises the salt tolerance of tomato through limiting the transport of sodium and chloride to the shoot. J Exp Bot 56:703–712
Falconer DS (1981) Introduction to quantitative genetics. Longman, New York
FAO (2005) Global Network of Integrated Soil Management for Sustainability Use of Salt-Affected Soils. Rome FAO Land and Plant Nutrition Management Service http://www.fao.org/ag/agl/agll/spush
Flint J, Mott R (2001) Finding the molecular basis of quantitative traits: successes and pitfalls. Nat Genet Rev 2:437–445
Flowers TJ (1972a) Salt tolerance in Suaeda maritima L. Dum. The effect of sodium chloride on growth, respiration and soluble enzymes in a comparative study with Pisum sativum. J Exp Bot 23:310–321
Flowers TJ (1972b) The effect of sodium chloride on enzyme activities from four halophyte species of Chenopodiaceae. Phytochem 11:1881–1886
Flowers TJ (2004) Improving crop salt tolerance. J Exp Bot 55:307–319
Flowers TJ, Colmer TD (2008) Salinity tolerance in halophytes. New Phytol 179:945–963
Flowers TJ, Yeo AR (1995) Breeding for salinity tolerance in crop plants: Where next? Aust J Plant Physiol 22:875–884
Flowers TJ, Flowers SA, Hajibagheri MA et al (1991) Ion accumulation in the cell walls of rice plants growing under saline conditions: evidence for the Oertli hypothesis. Plant Cell Environ 14:319–325
Flowers TJ, Koyama ML, Flowers SA et al (2000) QTL: their place in engineering tolerance of rice to salinity. J Exp Bot 51:99–106
Foolad MR (2004) Recent advances in genetics of salt tolerance in tomato. Plant Cell Tissue Organ Cult 76:101–119
Foolad MR, Lin GY (1997) Absence of a genetic relationship between salt tolerance during seed germination and vegetative growth in tomato. Plant Breed 116:363–367
Forrest KL, Bhave M (2007) Major intrinsic proteins (MIPs) in plants: a complex gene family with major impacts on plant phenotype. Funct Integr Genomics 7:263–289
Foyer CH, Noctor G (2005) Oxidant and antioxidant signalling in plants: a re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ 28:1056–1071
Frankel WN, Schork NJ (1996) Who’s afraid of epistasis? Nat Genet 14:371–373
Fukuda A, Nakamura A, Tagiri A et al (2004) Function, intracellular localization and the importance in salt tolerance of a vacuolar Na+/H+ antiporter from rice. Plant Cell Physiol 45:146–159
Gao F, Gao Q, Duan XG et al (2006) Cloning of an H+-PPase gene from Thellungiella halophila and its heterologous expression to improve tobacco salt tolerance. J Exp Bot 57:3259–3270
Gao XH, Ren ZH, Zhao YX et al (2003) Overexpression of SOD2 increases salt tolerance of Arabidopsis. Plant Physiol 133:1873–1881
Garg AK, Kim JK, Owens TG et al (2002) Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci USA 99:15898–15903
Gaxiola RA, Li JL, Undurraga S et al (2001) Drought and salt tolerant plants result from overexpression of the AVP1 H+ pump. Proc Natl Acad Sci USA 98:11444–11449
Gaxiola RA, Palmgren MG, Schumacher K (2007) Plant proton pumps. FEBS Lett 581:2204–2214
Gaxiola RA, Rao R, Sherman A et al (1999) The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. PNAS-USA 96:1480–1485
Ge LF, Chao DY, Shi M et al (2008) Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 228:191–201
Giovannoni JJ (2007) Fruit ripening mutants yield insights into ripening control. Curr Opin Plant Biol 10:283–289
Gisbert C, Rus AM, Bolarin MC et al (2000) The yeast HAL1 gene improves salt tolerance of transgenic tomato. Plant Physiol 123:393–402
Glenn EP, Brown JJ, Blumwald E (1999) Salt tolerance and crop potential of halophytes. Crit Rev Plant Sci 18:227–255
Gong Q, Li P, Ma S et al (2005a) Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J 44:826–839
Gong Z, Dong CH, Lee H et al (2005b) A DEAD box RNA helicase is essential form RNA export and important for development and stress responses in Arabidopsis. Plant Cell 17:256–267
Gorham J, Bristol A, Young EM et al (1990) Salt tolerance in the Triticeae: K/Na discrimination in barley. J Exp Bot 41:1095–1101
Greenway H, Munns R (1980) Mechanisms of salt tolerance in non-halophytes. Annu Rev Plant Physiol Plant Mol Biol 31:149–190
Grover A, Aggarwal PK, Kapoor A et al (2003) Addressing abiotic stresses in agriculture through transgenic technology. Curr Sci 84:355–367
Guo S, Yin H, Zhang X et al (2006) Molecular cloning and characterization of a vacuolar H+-pyrophosphatase gene, SsVP, from the halophyte Suaeda salsa and its overexpression increases salt and drought tolerance of Arabidopsis. Plant Mol Biol 60:41–50
Gurganus MC, Nuzhdin SV, Leips JW et al (1999) High-resolution mapping of quantitative trait loci for sternopleural bristle number in Drosophila melanogaster. Genetics 152:1585–1604
Ham BK, Park JM, Lee SB et al (2006) Tobacco Tsip1, a DnaJ-Type Zn finger protein, is recruited to and potentiates Tsi1-mediated transcriptional activation. Plant Cell 18:2005–2020
He CX, Yan JQ, Shen GX et al (2005) Expression of an arabidopsis vacuolar sodium/proton antiporter gene in cotton improves photosynthetic performance under salt conditions and increases fibre yield in the field. Plant Cell Physiol 46:1848–1854
Hernandez JA, Jimenez A, Mullineaux P et al (2000) Tolerance of pea (Pisum sativum L.) to long-term salt stress is associated with induction of antioxidant defences. Plant Cell Environ 23:853–862
Hernandez JA, Ferrer MA, Jimenez A et al (2001) Antioxidant systems and O-2(.-)/H2O2 production in the apoplast of pea leaves. Its relation with salt-induced necrotic lesions in minor veins. Plant Physiol 127:817–831
Herr AJ, Jensen MB, Dalmay T et al (2005) RNA polymerase IV directs silencing of endogenous DNA. Science 308:118–120
Hong C, Hsu YT, Tsai YH et al (2007) Expression of ASCORBATE PEROXIDASE 8 in roots of rice (Oryza sativa L.) seedlings in response to NaCl. J Exp Bot 58:3273–3283
Igartua E (1995) Choice of selection environments for improving crops yields in saline areas. Theor Appl Genet 91:1061–1021
Igartua E, Gracia P (1999) Divergent selection for salinity tolerance at germination-emergence stage in grain sorghum. Maydica 43:161–168
Ismail AM, Heuer S, Thomson MJ et al (2007) Genetic and genomics approaches to develop rice germplasm for problem soils. Plant Mol Biol 65:547–550
Jung HJ, Kang H (2007) Expression and functional analyses of microRNA417 in Arabidopsis thaliana under stress conditions. Plant Physiol Biochem 45:805–811
Kaldenhoff R, Ribas-Carbo M, Sans JF et al (2008) Aquaporins and plant water balance. Plant Cell Environ 31:658–666
Kant S, Kant P, Raveh E et al (2006) Evidence that differential gene expression between the halophyte Thellungiella halophila and Arabidopsis thaliana is responsible for higher levels of the compatible osmolyte proline and tight control of Na+ uptake in T. halophila. Plant Cell Environ 29:120–134
Kasuga M, Liu Q, Miura S et al (1999) Improving plant drought, salt and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17:287–291
Kaur N, Gupta AK (2005) Signal transduction pathways under abiotic stresses in plants. Curr Sci 88:1771–1780
Kavi Kishor PB, Sangam S, Amrutha RN et al (2005) Regulation of proline biosynthesis, degradation, uptake and transport in higher plants: its implications in plant growth and abiotic stress tolerance. Curr Sci 88:424–438
Khatum S, Flowers TJ (1995) Effects of salinity on seed set in rice. Plant Cell Environ 18:61–67
Krishnamurthy L, Serraj R, Hash CT et al (2007) Screening sorghum genotypes for salinity tolerant biomass production. Euphytica 156:15–24
Krysan PJ, Young JC, Sussman MR (1999) T-DNA as an insertional mutagen in arabidopsis. Plant Cell 11:2283–2290
Kumar S, Dhingra A, Daniell H (2004) Plastid-expressed betaine aldehyde dehydrogenase gene in carrot cultured cells, roots, and leaves confers enhanced salt tolerance. Plant Physiol 136:2843–2854
Lark KG, Chase K, Adler F et al (1995) Interactions between quantitative trait loci in soybean in which trait variation at one locus is conditional upon a specific allele at another. Proc Natl Acad Sci USA 92:4656–4660
Lee GJ, Boerma HR, Villagarcia MR et al (2004) A major QTL conditioning salt tolerance in S-100 soybean and descendent cultivars. Theor Appl Genet 109:1610–1619
Lee JT, Prasad V, Yang PT et al (2003) Expression of Arabidopsis CBF1 regulated by an ABA/stress inducible promoter in transgenic tomato confers stress tolerance without affecting yield. Plant Cell Environ 26:1181–1190
Lee YP, Kim SH, Bang JW et al (2007) Enhanced tolerance to oxidative stress in transgenic tobacco plants expressing three antioxidant enzymes in chloroplasts. Plant Cell Rep 26:591–598
Leidi EO, Barragán V, Cubero B et al (2005) Function of endosomal NHX antiporters on plant K nutrition. Society for Experimental Biology Annual Main Meeting. Comp Biochem Physiol-Part A 141:S342
Leung H (2008) Stressed genomics - bringing relief to rice fields. Curr Opin Plant Biol 11:201–208
Lewis IN (1984) A vital resource in danger. Calif Agric 38:2
Li J, Jiang G, Huang P et al (2007) Overexpression of the Na+/H+ antiporter gene from Suaeda salsa confers cold and salt tolerance to transgenic Arabidopsis thaliana. Plant Cell Tissue Organ Cult 90:41–48
Li WYF, Wong FL, Tsai SN et al (2006) Tonoplast-located GmCLC1 and GmNHX1 from soybean enhance NaCl tolerance in transgenic bright yellow (BY)-2 cells. Plant Cell Environ 29:1122–1137
Lilley JM, Ludlow MM (1996) Expression of osmotic adjustment and dehydration tolerance in diverse rice lines. Field Crops Res 48:185–197
Liu H, Wang QQ, Yu MM et al (2008) Transgenic salt-tolerant sugar beet (Beta vulgaris L.) constitutively expressing an Arabidopsis thaliana vacuolar Na+/H+ antiporter gene, AtNHX3, accumulates more soluble sugar but less salt in storage roots. Plant Cell Environ 31:1325–1334
Liu K, Wang L, Xu Y et al (2007) Overexpression of OsCOIN, a putative cold inducible zinc finger protein, increased tolerance to chilling, salt and drought, and enhanced proline level in rice. Planta 226:1007–1016
Liu N, Zhong NQ, Wang GL et al (2007) Cloning and functional characterization of PpDBF1 gene encoding a DRE-binding transcription factor from Physcomitrella patens. Planta 226:827–838
Lozano R, Giménez E, Cara B et al (2009) Developmental genetics of tomato, a model species in plant biology. Int J Dev Biol (in press)
Lu SY, Jing YX, Shen SH et al (2005) Antiporter gene from Hordum brevisubulatum (Trin.) Link and its overexpression in transgenic tobacco. J Integr Plant Biol 47:343–349
Lukens LN, Doebley J (1999) Epistatic and environmental interactions for quantitative trait loci involved in maize evolution. Genet Res 74:291–302
Lv S, Zhang KW, Gao Q et al (2008) Overexpression of an H+-PPase gene from Thellungiella halophila in cotton enhances salt tolerance and improves growth and photosynthetic performance. Plant Cell Physiol 49:1150–1164
Maathuis FJM (2006) The role of monovalent cation transporters in plant responses to salinity. J Exp Bot 57:1137–1147
Maheswaran M, Huang N, Sreerangasamy SR et al (2000) Mapping QTL associated with days to flowering and photoperiod sensitivity in rice (Oryza sativa L.). Mol Breed 6:145–155
Manneh B, Stam P, Struik PC et al (2007) QTL-based analysis of genotype-by-environment interaction for grain yield of rice in stress and non-stress environments. Euphytica 156:213–226
Mansour MMF, Salama KHA (2004) Cellular basis of salinity tolerance in plants. Environ Exp Bot 52:113–122
Martínez-Atienza J, Jiang X, Garciadeblas B et al (2007) Conservation of the salt overly sensitive pathway in rice. Plant Physiol 143:1001–1012
Martinez-Rodriguez MM, Estañ MT, Moyano E et al (2008) The effectiveness of grafting to improve salt tolerance in tomato when an ‘excluder’ genotype is used as scion. Environ Exp Bot 63:392–401
Matsushita N, Matoh T (1991) Characterization of Na+ exclusion mechanisms of salt-tolerant reed plants in comparison with salt-sensitive rice plants. Physiol Plant 83:170–176
Mikiko LK, Levesley A, Koebner RMD et al (2001) Quantitative trait loci for component physiological traits determining salt tolerance in rice. Plant Physiol 125:406–422
Mittova V, Tal M, Volokita M et al (2002) Salt stress induces up-regulation of an efficient chloroplast antioxidant system in the salt-tolerant wild tomato species Lycopersicon pennellii but not in the cultivated species. Physiol Plant 115:393–400
Moller IS, Tester M (2007) Salinity tolerance of Arabidopsis: a good model for cereals?. Trends Plant Sci 12:534–540
Monforte AJ, Asins MJ, Carbonell E (1997) Salt tolerance in Lycopersicon species. VI. Genotype-by-salinity interaction in quntitaive trait loci detection: constitutive and response QTLs. Theor Appl Genet 95:706–713
Moradi F, Ismail AM, Gregorio GB et al (2003) Salinity tolerance of rice during reproductive development and association with tolerance at the seedling stage. Indian J Plant Physiol 8:105–116
Morgan JM (1992) Osmotic components and properties associated with genotypic differences in osmorregulation in wheat. Aust J Plant Physiol 19:67–7676
Moya JL, Gomez-Cadenas A, Primo-Millo E et al (2003) Chloride absorption in salt-sensitive Carrizo citrange and salt-tolerant Cleopatra mandarin citrus rootstocks is linked to water use. J Exp Bot 54:825–833
Munns R, Hare RA, James RA, Rebetzke GJ (2000) Genetic variation for improving the salt tolerance of durum wheat. Aust J Agric Res 51:69–74.
Munns R (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25:239–250
Munns R (2005) Genes and salt tolerance: bringing them together. New Phytol 167:645–663
Munns R, Termaat A (1986) Whole-plant responses to salinity. Aust J Plant Physiol 13:143–160
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Muñoz-Mayor A, Pineda B, Garcia-Abellán JO et al (2008) The HAL1 function on Na+ homeostasis is maintained over time in salt-treated transgenic tomato plants, but the high reduction of Na+ in leaf is not associated with salt tolerance. Physiol Plant 133:288–297
Murthy M, Tester M (1996) Compatible solutes and salt tolerance: misuse of transgenic tobacco. Trends Plant Sci 1:294–295
Nagata T, Iizumi S, Satoh K et al (2008) Comparative molecular biological analysis of membrane transport genes in organisms. Plant Mol Biol 66:565–585
Nakashima K, Tran LSP, Nguyen DV et al (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51:617–630
Neumann P (1997) Salinity resistance and plant growth revisited. Plant Cell Environ 20:1193–1198
Niknam SR, McComb J (2000) Salt tolerance screening of selected Australian woody species - a review. Forest Ecol Manage 139:1–19
Ogawa D, Yamaguchi K, Nishiuchi T (2007) High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased thermotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J Exp Bot 58:3373–3383
Ohta M, Hayashi Y, Nakashima A et al (2002) Introduction of a Na+/H+ antiporter gene from Atriplex gmelini confers salt tolerance to rice. FEBS Lett 532:279–282
Owttrim GW (2006) RNA helicase and abiotic stress. Nucleic Acids Res 34:3220–3230
Pardo JM, Cubero B, Leidi EO et al (2006) Alkali cation exchangers: roles in cellular homeostasis and stress tolerance. J Exp Bot 57:1181–1199
Park EJ, Jeknic Z, Pino MT et al (2007) Glycinebetaine accumulation is more effective in chloroplasts than in the cytosol for protecting transgenic tomato plants against abiotic stress. Plant Cell Environ 30:994–1005
Penna S (2003) Building stress tolerance through over-producing trehalose in transgenic plants. Trends Plant Sci 8:355–357
Perez-Alfocea F, Estañ MT, Santa Cruz A et al (1993) Effects of salinity on nitrate, total nitrogen, soluble protein and free amino acid levels in tomato plants. J Hort Sci 68:1021–1027
Perez-Alfocea F, Santa Cruz A, Guerrier G et al (1994) NaCl stress-induced organic solute changes on leaves and calli of Lycopersicon esculentum, L. pennellii and their interspecific hybrid. J Plant Physiol 143:106–111
Pineda B (2005) Análisis functional de diversos genes relacionados con la tolerancia a la salinidad y el estrés hídrico en plantas transgénicas de tomate (Lycopersicon esculentum Mill). Ph.D. thesis. Universidad Politécnica de Valencia, Valencia, Spain
Platten JD, Cotsaftis O, Berthomieu P et al (2006) Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci 11:372–374
Qiao WH, Zhao XY, Li W et al (2007) Overexpression of AeNHX1, a root-specific vacuolar Na+/H+ antiporter from Agropyron elongatum, confers salt tolerance to Arabidopsis and Festuca plants. Plant Cell Rep 26:1663–1672
Quintero FJ, Ohta M, Shi H et al (2002) Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA 99:9061–9066
Rajagopal D, Agarwal P, Tyagi W et al (2007) Pennisetum glaucum Na+/H+ antiporter confers high level of salinity tolerance in transgenic Brassica juncea. Mol Breed 19:137–151
Ravens JA (1985) Regulation of pH and generation of osmolarity in vascular plants - a cost-benefit analysis in relation to efficiency of use of energy, nitrogen and water. New Phytol 101:25–77
Rhodes D, Hanson AD (1993) Quaternary ammonium and tertiary sulfonium compounds in higher-plants. Annu Rev Plant Physiol Plant Mol Biol 44:357–384
Rios G, Ferrando A, Serrano R (1997) Mechanisms of salt tolerance conferred by overexpression of the HAL1 gene in Saccharomyces cerevisiae. Yeast 13:515–528
Rodriguez-Navarro A (2000) Potassium transport in fungi and plants. Biochim Biophys Acta 1469:1–30
Rohila JS, Jainb RK, Wu R (2002) Genetic improvement of Basmati rice for salt and drought tolerance by regulated expression of a barley Hva1 cDNA. Plant Sci 163:525–532
Romero C, Belles JM, Vaya JL et al (1997) Expression of the yeast trehalose-6-phosphate synthase gene in transgenic tobacco plants: pleiotropic phenotypes include drought tolerance. Planta 201:293–297
Rumbaugh MD, Pendery BM, James DW (1993) Variation in the salinity tolerance of strawberry clover (Trifolium fragiferum L.). Plant Soil 153:265–271
Rus A, Lee BH, Muñoz-Mayor A et al (2004) AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol 136:2500–2511
Rus AM, Estañ MT, Gisbert C et al (2001) Expressing the yeast HAL1 gene in tomato increases fruit yield and enhances K+/Na+ selectivity under salt stress. Plant Cell Environ 24:875–880
Sairam RK, Tyagi A (2004) Physiology and molecular biology of salinity stress tolerance in plants. Curr Sci 86:407–421
Sakamoto A, Alia, Murata N (1998) Metabolic engineering of rice leading to biosynthesis of glycinebetaine and tolerance to salt and cold. Plant Mol Biol 38:1011–1019
Sanan-Mishra N, Phan XH, Sopory SK et al (2005) Pea DNA helicase 45 overexpression in tobacco confers high salinity tolerance without affecting yield. Proc Natl Acad Sci USA 102:509–514
Sanchez DH, Siahpoosh MR, Roessner U et al (2008) Plant metabolomics reveals conserved and divergent metabolic responses to salinity. Physiol Plant 132:209–219
Santa-Cruz A, Acosta M, Rus A et al (1999) Short-term salt tolerance mechanisms in differentially salt tolerant tomato species. Plant Physiol Biochem 37:65–71
Sax K (1923) The association of size differences with seed coat pattern and pigmentation in Phaseolus vulgaris. Genetics 8:552–560
Seong ES, Cho HS, Choi D et al (2007) Tomato plants overexpressing CaKR1 enhanced tolerance to salt and oxidative stress. Biochem Biophys Res Commun 363:983–988
Serrano R, Culiañez-Macia FA, Moreno V (1999) Genetic engineering of salt and drought tolerance with yeast regulatory genes. Sci Hort 78:261–269
Shannon MC, Gronwald JW, Tal M (1987) Effects of salinity on growth and accumulation of inorganic ions in cultivated and wild tomato species. J Am Soc Hort Sci 112:416–423
Shi HZ, Zhu JK (2002) Regulation of expression of the vacuolar Na+/H+ antiporter gene AtNHX1 by salt stress and abscisic acid. Plant Mol Biol 50:543–550
Shi HZ, Lee BH, Wu SJ et al (2003) Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol 21:81–85
Shi HZ, Quintero FJ, Pardo JM et al (2002) The putative plasma membrane Na1/H1 antiporter SOS1 controls long-distance Na1 transport in plants. Plant Cell 14:465–477
Shirasawa K, Takabe T, Takabe T et al (2006) Accumulation of glycinebetaine in rice plants that overexpress choline monooxygenase from spinach and evaluation of their tolerance to abiotic stress. Ann Bot 98:565–571
Sickler CM, Edwards GE, Kiirats O et al (2007) Response of mannitol-producing Arabidopsis thaliana to abiotic stress. Funct Plant Biol 34:382–391
Sohn KH, Lee SC, Jung HW et al (2006) Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol Biol 61:897–915
Springer PS (2000) Gene traps: tools for plant development and genomics. Plant Cell 12:1007–1020
Su J, Wu R (2004) Stress-inducible synthesis of proline in transgenic rice confers faster growth under stress conditions than that with constitutive synthesis. Plant Sci 166:941–948
Su J, Hirji R, Zhang L et al (2006) Evaluation of the stress-inducible production of choline oxidase in transgenic rice as a strategy for producing the stress-protectant glycine betaine. J Exp Bot 57:1129–1135
Sun YX, Wang D, Bai YL et al (2006) Studies on the overexpression of the soybean GmNHX1 in Lotus corniculatus: the reduced Na+ level is the basis of the increased salt tolerance. Chin Sci Bull 51:1306–1315
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019
Takehisa H, Shimodate T, Fukuta Y et al (2004) Identification of quantitative trait loci for plant growth of rice in paddy field flooded with salt water. Field Crops Res 89:85–95
Tanksley SD (2004) The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell 16:181–189
Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot 91:503–527
Tian L, Huang C, Yu R et al (2006) Overexpression AtNHX1 confers salt-tolerance of transgenic tall fescue. African J Biotechnol 5:1041–1044
Tonsor SJ, Alonso-Blanco C, Koornneef M (2005) Gene function beyond the single trait: natural variation, gene effects, and evolutionary ecology in Arabidopsis thaliana. Plant Cell Environ 28:2–20
Tseng MJ, Liu CW, Yiu JC (2007) Enhanced tolerance to sulfur dioxide and salt stress of transgenic Chinese cabbage plants expressing both superoxide dismutase and catalase in chloroplasts. Plant Physiol Biochem 45:822–833
Tuteja N (2007) Mechanisms of high salinity tolerance in plants. Meth Enzymol 428:419–438
Verma D, Singla-Pareek SL, Rajagopal D et al (2007). Functional validation of a novel isoform of Na+/H+ antiporter from Pennisetum glaucum for enhancing salinity tolerance in rice. J Biosci 32:621–628
Vij S, Tyagi AK (2007) Emerging trends in the functional genomics of the abiotic stress response in crop plants. Plant Biotechnol J 5:361–380
Villalta I, Reina-Sánchez A, Bolarín MC et al (2008) Genetic analysis of Na+ and K+ concentrations in leaf and stem as physiological components of salt tolerance in tomato. Theor Appl Genet 116:869–880
Waditee R, Bhuiyan NH, Hirata E et al (2007) Metabolic engineering for betaine accumulation in microbes and plants. J Biol Chem 282:34185–34193
Wang J, Zuo K, Wu W et al (2004) Expression of a novel antiporter gene from Brassica napus resulted in enhanced salt tolerance in transgenic tobacco plants. Biol Plant 48:509–515
Wang W, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218:1–14
Wang WQ, Li Y, Zhang YY et al (2007) Comparative expression analysis of three genes from the Arabidopsis vacuolar Na+/H+ antiporter (AtNHX) family in relation to abiotic stresses. Chin Sci Bull 52:1754–1763
Weckwertha W (2008) Integration of metabolomics and proteomics in molecular plant physiology – coping with the complexity by data-dimensionality reduction. Physiol Plant 132:176–189
Wu CA, Yang GD, Meng QW et al (2004) The cotton GhNHX1 gene encoding a novel putative tonoplast Na+/H+ antiporter plays an important role in salt stress. Plant Cell Physiol 45:600–607
Wu YY, Chen QJ, Chen M et al (2005) Salt-tolerant transgenic perennial ryegrass (Lolium perenne L.) obtained by Agrobacterium tumefaciens-mediated transformation of the vacuolar Na+/H+ antiporter gene. Plant Sci 169:65–73
Xue ZY, Zhi DY, Xue GP et al (2004) Enhanced salt tolerance of transgenic wheat (Tritivum aestivum L.) expressing a vacuolar Na+/H+ antiporter gene with improved grain yields in saline soils in the field and a reduced level of leaf Na+. Plant Sci 167:849–859
Yamaguchi T, Blumwald E (2005) Developing salt-tolerant crop plants: challenges and opportunities. Trends Plant Sci 10:615–620
Yang AF, Duan XG, Gu XF et al (2005) Efficient transformation of beet (Beta vulgaris) and production of plants with improved salt-tolerance. Plant Cell Tissue Organ Cult 83:259–270
Yang L, Tang R, Zhu J et al (2008) Enhancement of stress tolerance in transgenic tobacco plants constitutively expressing AtIpk2b, an inositol polyphosphate 6-/3-kinase from Arabidopsis thaliana. Plant Mol Biol 66:329–343
Yeo AR (1983) Salinity resistance: physiologies and prices. Physiol Plant 58:214–222
Yeo AR (1992) Variation and inheritance of sodium transport in rice. Plant Soil 146:109–116
Yeo AR (2007) Salinity. In: Yeo AR, Flowers TJ (eds) Plant solute transport. Blckwell, Oxford
Yeo AR, Flowers TJ (1986) Ion transport in Suaeda maritima: its relation to growth, and implications for the pathway of radial transport of ions across the root. J Exp Bot 37:143–159
Yeo AR, Yeo, ME, Flowers SA et al (1990) Screening of rice (Oryza sativa L.) genotypes for physiological characters contributing to salinity resistance, and their relationship to overall performance. Theor Appl Genet 79:377–384
Yin XY, Yang AF, Zhang KW et al (2004) Production and analysis of transgenic maize with improved salt tolerance by the introduction of AtNHX1 gene. Acta Bot Sin 46:854–861
Yokoi S, Quintero FJ, Cubero B et al (2002) Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J 30:529–539
Zhang HX, Blumwald E (2001) Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol 19:765–768
Zhang HX, Hodson JN, Williams JP et al (2001) Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc Natl Acad Sci USA 98:12832–12836
Zhang J, Nguyen HT, Blum A (1999) Genetic analysis of osmotic adjustment in crop plants. J Exp Bot 50:291–302
Zhang Y, Wang Z, Xu J (2007) Molecular mechanism of dehydrin in response to environmental stress in plant. Prog Nat Sci 17:237–246
Zhao F, Zhang H (2006) Salt and paraquat stress tolerance results from co-expression of the Suaeda salsa glutathione S-transferase and catalase in transgenic rice. Plant Cell Tissue Organ Cult 86:349–358
Zhao F, Guo S, Zhang H et al (2006a) Expression of yeast SOD2 in transgenic rice results in increased salt tolerance. Plant Sci 170:216–224
Zhao F, Wang Z, Zhang Q et al (2006b) Analysis of the physiological mechanism of salt-tolerant transgenic rice carrying a vacuolar Na+/H+ antiporter gene from Suaeda salsa. J Plant Res 119:95–104
Zhao F, Zhang XJ, Li PH et al (2006c) Co-expression of the Suaeda salsa SsNHX1 and Arabidopsis AVP1 confer greater salt tolerance to transgenic rice than the single SsNHX1. Mol Breed 17:341–353
Zhao J, Zhi D, Xue Z et al (2007) Enhanced salt tolerance of transgenic progeny of tall fescue (Festuca arundinacea) expressing a vacuolar Na+/H+ antiporter gene from Arabidopsis. J Plant Physiol 164:1377–1383
Zhou SF, Chen XY, Xue XN et al (2007) Physiological and growth responses of tomato progenies harboring the betaine alhyde dehydrogenase gene to salt stress. J Integr Plant Biol 49:628–637
Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6:66–71
Acknowledgments
The authors like to thank Dr. Eva Domínguez, Estación Experimental La Mayora, CSIC, for valuable suggestions while editing the manuscript. V. Moreno, B. Pineda and M.C. Bolarín would like to thank the CICYT (Ministerio de Ciencia e Innovación, Spain) for financial support (project AGL2006 15290). V. Moreno would also like to thank Dr. Thomas Jack, Department of Biological Sciences, Dartmouth College, USA for providing the pD991 vector.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2010 Springer Science+Business Media B.V.
About this chapter
Cite this chapter
Cuartero, J., Bolarin, M.C., Moreno, V., Pineda, B. (2010). Molecular Tools for Enhancing Salinity Tolerance in Plants. In: Jain, S., Brar, D. (eds) Molecular Techniques in Crop Improvement. Springer, Dordrecht. https://doi.org/10.1007/978-90-481-2967-6_16
Download citation
DOI: https://doi.org/10.1007/978-90-481-2967-6_16
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-90-481-2966-9
Online ISBN: 978-90-481-2967-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)