Abstract
Use of DNA-based markers can accelerate cultivar development in variable cultivation environments since, in contrast to phenotype, DNA markers are environment-independent. In an effort to elucidate the genetic basis of genotype-by-environment interaction (G × E) for yield of rice (Oryza sativa L.), the associations between 139 AFLP markers and grain yield were determined for rice grown in fresh water (EC of 0.65 dS m−1) and saline conditions (EC of 4–8 dS m−1) with 0 kg ha−1 or 100 kg ha−1 nitrogen fertilizer in the years 2000 and 2001. A population of recombinant inbred lines of rice, developed from an IR29 × Pokkali cross, was used in the study. Both genotype × salinity and genotype × nitrogen level interactions were significant, with the genotype × salinity interaction being stronger. Through multiple regression analysis using a stepwise procedure for selecting markers, 36 markers were detected for grain yield in the four test conditions and of these 28 were detected in only one test condition implying strong environmental specificity for yield QTL expression. However, the fact that eight QTLs were detected in more than one test condition points to the existence of wide-adaptability genes in this cross. Markers with significant associations with yield explained between 37% and 48% of the yield variation in each test condition. Superior genotypes of rice were identified in all four test conditions based on their marker signatures. Furthermore, across N fertilizer regimes, yield predicted from summed additive effects of QTLs were significantly correlated with observed yield in the same year and across years. Thus marker-assisted selection can help breeders overcome the problem of low selection efficiency encountered during phenotypic selection for yield in stress environments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The bulk of rice in many African countries is produced under traditional production systems where field conditions are often variable and yield is low. This is due to the preponderance of abiotic stresses such as poor soil nutrient supply, soil salinity, droughts and flooding in these systems. Improving the cultivation environment through fertilizer application, liming or irrigation is not feasible for many of these farmers due to the high costs of these technologies relative to their incomes, or due to unavailability or lack of sufficient knowledge of the technologies. The improvement of the genetic yield potential of rice for marginal environments remains the cheapest and most accessible option for most of these low resource farmers.
Crop yield is quantitative in nature and under the control of many genes or gene complexes that are described as quantitative trait loci (QTLs). Improving such quantitative traits through conventional breeding is slow and difficult due to the often large genotype-by-environment interaction associated with these traits. Therefore, the use of molecular techniques that can accurately predict crop performance in diverse environments holds much promise for crop breeding programmes.
The basis of QTL detection is the identification of significant statistical association between phenotypes and specific genetic markers (Yin et al. 2002). Once such an association is established, indirect selection can be targeted at the presence or absence of markers of interest in breeding lines. Some success has already been reported in rice breeding employing this method of selection, which is called marker-assisted selection (MAS). For example, Cho et al. (1994) used molecular markers to select for the semi-dwarf characteristic in rice. Singh et al. (2001) pyramided three genes conferring resistance to bacterial leaf blight of rice through marker-assisted selection. Wang et al. (2005) also successfully introgressed three QTLs with large effects on spikelet fertility into near-isogenic lines using marker-assisted selection. Introgression of osmotic adjustment, associated with drought tolerance, has been achieved in rice by Robin et al. (2003). These authors detected 14 QTLs that explained altogether 58% of phenotypic variation in a re-hydration experiment.
The objectives of this paper were (i) to identify molecular markers that can significantly account for variation in yield of rice under diverse environmental conditions, characteristic for the rice cultivation in Africa, and (ii) to determine the efficacy of marker-assisted selection to identify superior genotypes of rice in stress and non-stress environments.
Materials and methods
Plant material
A subset of 98 RILs randomly selected from a segregating population of rice (Oryza sativa L.) comprising 276 recombinant inbred lines (RILs) in the ninth filial generation (F9) was used. The RIL population was developed at the International Rice Research Institute (IRRI) in the Philippines, from the cross IR29 × Pokkali (both indica varieties) by single seed descent. IR29 is a short, high yielding modern cultivar released by IRRI and is known to be very sensitive to salinity (Gregorio 1997). Pokkali is a tall, traditional variety from India known to be tolerant to salinity (Yeo and Flowers 1986; Garcia et al. 1995; Gregorio 1997). As one of the two parents is a high-yielding, modern cultivar and the other is a traditional variety, it is expected that the RIL population will also be segregating for response to nitrogen fertilizer. This is due to the fact that modern rice cultivars have been bred for response to high levels of mineral fertilizer application while traditional varieties have usually been selected by farmers in environments with sub-optimal levels of nutrient supply.
The RIL population, together with the parents, was grown during the rainy season (June–October) of 2000 and 2001 at Sapu (13.55° N latitude), in The Gambia. Two rice cultivars were introduced into the trial in 2001—ROK5 and ITA212. ROK5 is a tall, moderately salt-tolerant cultivar bred in Sierra Leone (WARDA, 1994) and has been used in the saline swamps of West Africa for many years. ITA212 is a semi-dwarf, high-yielding lowland rice cultivar bred by researchers at the International Institute of Tropical Agriculture (IITA), in Ibadan, Nigeria.
Experimental design
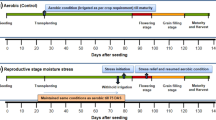
In both years 2000 and 2001, a split-split plot experimental design was used with salinity as the main plot factor, rate of nitrogen fertilizer application as the sub-plot factor and genotype as the sub-subplot factor. Each sub-subplot measured 2.6 m × 3.0 m and a spacing of 20 cm × 20 cm within and between rows was used. Two levels of salinity and two levels of nitrogen fertilizer application, giving a total of four test conditions, were applied. Three replications were maintained in each year of the trials. Additional information on the trials is given in Table 1. The following test conditions were maintained:
-
S1N1—Fresh water (river water) at an electrical conductivity (EC) of 0.65 dS m−1 and 0 kg N ha−1;
-
S1N2—Fresh water (river water) at an EC of 0.65 dS m−1 and 100 kg N ha−1 as urea;
-
S2N1—Salt water at an EC of 4–8 dS m−1 and 0 kg N ha−1;
-
S2N2—Salt water at an EC of 4–8 dS m−1 with 100 kg N ha−1 as urea.
Selection scheme
For our study, 98 RILs were randomly selected from the 276 RILs available and grown together with their parents in 2000. In an earlier work by Gregorio (1997) 80 RILs were selectively genotyped from this RIL population (276 lines) based on their response to salt stress. The 98 RILs selected by us comprised 25, which were earlier on fingerprinted by Gregorio and another 73 that were not fingerprinted.
As the polyacrylamide gels used to visualize the amplified DNA fragments had 80 lanes we needed to select an extra 7 RILs which when added to the 73 mentioned above would total 80. These 7 RILs were randomly selected from the 123 RILs not included in the earlier selections. This new set of 80 RILs was fingerprinted for our purpose and then the marker dataset was combined with that of the 80 RILs selectively genotyped by Gregorio giving a total of 160 fingerprinted RILs.
After analyses of yield data from year 2000, 38 RILs were selected from the 160 genotyped RILs for high-, medium- and low-yielding ability in the different test conditions. These selections were made from yield data predicted from the molecular marker signatures of the different RILs. Of these 38 RILs, 22 were present in the 98 RILs grown in year 2000 while the remaining 16 RILs had not been grown before in our trial.
Cultural practices
Pre-germinated rice seeds were sown in a nursery that was well supplied with fresh water and regularly hand-weeded. At 22–27 days after sowing the seedlings were transplanted to the field following the experimental design described above. After transplanting the trial plots were kept continuously flooded by irrigating with river water until all RILs and cultivars were close to physiological maturity.
Salinity was imposed by manually broadcasting measured amounts of granular table salt in standing water to attain the required salinity. Salinity of the ponded-water was measured 2 days after every significant rainfall or after a protracted period without rains (more than four consecutive days). An ES-421 digital salt meter (Atago Co. Ltd., Japan) was used to measure salt concentration of standing water in % (g/100 g). This was converted into dS m−1 by using the ratio of 1 dS m−1/727.273 mg/l (Primary Industries, SA 1994). When the salinity level was too low (<4 dS m−1) more salt was added to raise the salinity and when the salinity was too high (>8 dS m−1) the saline plots were irrigated with fresh water to reduce the salinity to the desired level.
Data collection
Yield data were collected by harvesting plants from plots after removing border rows as recommended in the Standard Evaluation System of IRRI (IRRI 1996). The harvested plants were threshed, dried, cleaned and weighed. The yield (in g per plot) from each net plot (2.2 m × 2.6 m) was converted into yield in kg ha−1 by multiplying with a factor of 1.7482.
Statistical analysis
Yield data were analyzed per year for 97 RILs and their parents in year 2000 and for 38 RILs, two parents and two cultivars in year 2001. An analysis of yield across years was done using pooled yield data for the 22 RILs and their parents common to both years. In each of these analyses, an analysis of variance model was fitted to yield using the Mixed Procedure (Proc MIXED) of SAS (1999) and to generate least squares means (LSMeans) for the line × salinity level × nitrogen combinations for yield, i.e., for each of the four test conditions a vector of line means was produced. In year 2000 one RIL was dropped from the data set due to ambiguous labelling of the RIL. Subsequent QTL analysis was performed using the LSMeans for 97 RILs.
The following mixed model was used to analyze the data per year (random terms are underlined):
where
-
yijkl is the observation on the k-th replication, j-th salinity level, i-th nitrogen level and l-th genotype
-
μ is the grand mean of grain yield
-
rk is the effect of the k-th replication
-
sj is the effect of the j-th salinity level
-
fjk is the main plot error term
-
ni is the effect of the i-th nitrogen level
-
(n × s)ij is the effect of the ij-th salinity-by-nitrogen level interaction
-
bijk is the split plot error term
-
gl is the effect of the l-th genotype
-
(g × s)jl is the effect of the jl-th genotype-by-salinity level interaction
-
(g × n)il is the effect of the il-th genotype-by-nitrogen level interaction
-
(g × s × n)ijl is the effect of the ijl-th genotype-by-salinity-by-nitrogen level interaction
-
\( \underline \varepsilon\)ijkl is the split-split plot error term.
The pooled yield data for the 22 RILs and their parents were analyzed using the mixed model below (random terms underlined):
where
-
y, μ, sj, f, ni, (n × s)ij, bijk, gl, (g × s)jl, (g × n)il, (g × n × s)ijl and \( \underline \varepsilon \), represent the corresponding parameters as specified in the one year ANOVA model above;
-
am is the effect of the m-th year;
-
tkm is the effect of the k-th replicate within the m-th year;
-
(a × s)jm is the effect of the jm-th year-by-salinity interaction;
-
(a × n)im is the effect of the im-th year-by-nitrogen interaction;
-
(a × n × s)ijm is the effect of the ijm-th year-by-nitrogen-by-salinity interaction;
-
(a × g)lm is the effect of the lm-th genotype-by-year interaction;
-
(a × g × s)jlm is the effect of the jlm-th genotype-by-salinity-by-year interaction;
-
(a × g × n)ilm is the effect of the ilm-th genotype-by-nitrogen-by-year interaction;
-
(a × g × n × s)ijlm is the effect of the ijlm-th genotype salinity-by-nitrogen-by-year interaction;
Molecular marker analysis
A molecular marker map was developed at IRRI with the same RIL population to map QTLs for salt tolerance. The map contained 205 AFLP markers, 3 STS (sequence tagged sites) markers and one phenotypic marker (for salt tolerance) distributed over all the 12 chromosomes of rice and was generated using 32 primer combinations (Gregorio 1997). The AFLP map was prepared using DNA collected from 80 RILs selected through selective genotyping for salinity tolerance (38 very tolerant and 42 very sensitive).
For this study, 25 primer combinations were selected from the 32 used by Gregorio and these were used to generate 139 polymorphic AFLP markers from the 80 RILs randomly selected by us. This new marker dataset was combined with the marker dataset for the 80 selectively genotyped RILs to give a total of 160 fingerprinted RILs with 209 markers. However, out of these 209 markers only 139 were present for all 160 RILs and the other 70 were missing amongst the 80 RILs genotyped specifically for our purpose. These 70 markers included the 3 STS markers and one phenotypic marker (for salt tolerance).
The marker dataset containing 139 markers for the combined 160 RILs was analyzed using JoinMap (Stam and van Ooijen 1995) with the aim of producing a linkage map. Sixty-one of the 139 markers showed highly skewed segregation ratios and estimated recombination frequencies between certain markers were unrealistically high. Only 78 markers could be assigned to 11 of the 12 linkage groups of rice. Linkage group 4 could not be constructed, since of the five markers located on this linkage group by Gregorio (1997), in our tentative map only one had a segregation ratio within the range (40:60) expected for RILs. As a result of these deficiencies in the tentative map, a linkage map was not constructed for QTL mapping.
Amongst the 160 RILs, the missing scores for the 139 markers were estimated using information about correlations of the affected markers with other markers. A new data file was generated in which all 160 RILs had scores for the 139 markers.
QTL detection
For each test condition, line means for yield, as produced from the earlier described analysis of variance model, were regressed on marker data, where markers were represented by a value of 1 for band presence and 0 otherwise. Stepwise regression as implemented in Genstat (Genstat 6.1, 2002) was used to identify sets of markers within multiple regression, i.e., multiple QTL models. For the yield data from the trials of year 2000, markers were included when the F-value for inclusion exceeded 4, and were dropped from the model when the F-value for exclusion was less than 1 (Montgomery and Peck 1982).
Marker-assisted selection (MAS)
For each test condition in year 2000, marker based predicted values for yield were obtained from the final selected multiple regression models. In addition, for 38 RILs grown in year 2001, marker based predictions were created from estimated marker effects in year 2000. The proportion of phenotypic variance for yield described by identified QTLs was estimated by the squared correlation, R2, for fitted regression models.
To determine the feasibility of MAS in the presence of significant genotype-by-environment interaction, for each test condition, the Pearson correlation coefficient was computed between the observed yield and QTL based predicted yield as obtained under the alternative test conditions.
Results
Genotype × Environment interaction for yield
ANOVA revealed a significant (P < 0.001) genotype × salinity × nitrogen fertilizer interaction for grain yield in both 2000, 2001 and for the pooled yield data for the 2 years (Table 2). Salinity caused a reduction in mean yield in both years while N fertilizer application increased mean yield. Mean yield of Pokkali and ROK5 were reduced in fresh water conditions with high N fertilizer application.
The RIL population used in this study exhibited larger genotype × salinity interaction than genotype × nitrogen fertilizer interaction (Table 2). Nonetheless, the level of genotype × nitrogen interaction was also significant in both 2000 and 2001 and for the pooled data.
The year effect on the three-way interaction of genotype × salinity × nitrogen fertilizer for the pooled yield data of the 22 RILs and their parents grown both in 2000 and 2001 was not significant (P > 0.05). Nonetheless the diluting effect of the higher rainfall in year 2000 compared to year 2001, on applied salt and nitrogen fertilizer, resulted in a smaller yield gain from applied fertilizer and less yield loss from salinity stress in year 2000 relative to year 2001 (Table 3).
There was a stronger linear relationship between grain yield of rice genotypes when test conditions differed only in the level of N fertilizer application than when test conditions differed in their level of salinity (Fig. 1).
Linear relations between yield in different test conditions for RILs and their parents, IR29 and Pokkali (POK) in years 2000 and 2001. S1N1, S1N2—fresh water with 0 kg ha−1 or 100 kg ha−1 N fertilizer, respectively; S2N1, S2N2—saline water with 0 kg ha−1 or 100 kg ha−1 N fertilizer, respectively; r2-proportion of yield variation along one axis that is explained by a linear regression of yield in the other axis; ** P < 0.0001; * P < 0.05; n.s. P > 0.05
Segregation of AFLP markers
One hundred and thirty-nine AFLP markers were generated for our study. Sixty-one of these 139 markers had segregation ratios significantly deviating from the theoretical 50:50 ratio. The segregation was skewed in favour of Pokkali at 33 of these marker loci (>60% of offspring) and in favour of IR29 at another 28 marker loci (>60% of offspring).
Molecular markers significantly associated with grain yield
Through the use of multiple regression of grain yield on molecular marker scores, 36 markers were identified that had significant associations with grain yield in the four test conditions (Table 4). The proportion of yield variation explained by all markers significantly associated with yield was higher in fresh water treatments than in saline water treatments. Consequently, marker-based yield predictions were better correlated with observed yield in fresh water treatments than in saline water treatments (Fig. 2).
Comparison between observed and predicted values of grain yield in year 2000 (a(i)–a(iv)) and 2001 (b(i)–b(iv)). Yield predicted based on sums of estimates of additive effects of markers for grain yield (n = 97 in 2000 and n = 38 in 2001). The thick line represents simple linear regression of predicted on observed yield. The highest and lowest yielding RILs in each test condition are indicated. Note: S1N1, S1N2—fresh water with 0 kg ha−1 or 100 kg ha−1 N fertilizer, respectively; S2N1, S2N2—saline water with 0 kg h−1 or 100 kg h−1 N fertilizer, respectively; ** P < 0.0001; * P < 0.05
In all four test conditions the tall, salt-tolerant parent, Pokkali, supplied the yield-increasing alleles for most of the markers found to be significantly associated with grain yield (Table 4). Between the fresh and saline water treatments, the proportion of marker loci with superior IR29 alleles was higher in saline than in fresh water treatments, with zero N fertilizer application. However, with high N fertilizer application this proportion was higher in fresh water than in saline water treatments.
QTL × Environment interaction
When there is significant QTL × E it would be expected that for the same trait evaluated in different environments, markers identified would be significant in only a subset of the environments. In this study, 28 of the 36 markers significantly associated with yield were expressed in one test condition. Only 8 markers were expressed in more than one test condition (Table 4). Of these 8 markers, more markers showed consistency across fresh water conditions than across saline conditions.
Marker-assisted selection (MAS)
Within and across years, markers detected for grain yield could significantly explain variation in observed yield across N fertilizer levels but not so well (in year 2000) or not at all (in year 2001 or across years) across salt and fresh water treatments (Tables 5 and 6). However, the accuracy of MAS was greater in 2000 than in 2001. This was reflected in the stronger correlations between observed yield and yield predicted from molecular marker signatures in 2000 than in 2001 (Table 5) in most cases.
Discussion
The larger genotype × salinity interaction compared to the genotype × nitrogen fertilizer interaction exhibited by the RIL population used in this study was expected. This was due to the fact that this population was developed from a cross of a salt-sensitive (IR29) and a salt-tolerant parent (Pokkali). However, the significant genotype × nitrogen fertilizer interaction for grain yield in both 2000 and 2001 implied that this population can be used for studying response of rice to nitrogen fertilizer application.
The year × genotype × salinity × nitrogen effect was not significant. Yet, Table 1 shows that the amount of rainfall received in 2000 was almost twice that of 2001. This would dilute applied salt in saline treatments and wash away applied N fertilizer quicker. Thus the yield increment due to N fertilizer application would be smaller in 2000 than in 2001. Consequently mean RIL yield in fresh water with high N fertilizer application was lower in 2000 (with more rainfall) than in 2001 (with less rainfall). Likewise, yield reduction due to salt stress was smaller in 2000 than in 2001 resulting in higher mean yield in saline test conditions in 2000 than in 2001.
Fewer polymorphic markers were generated in our study than in the research of Gregorio (1997) using the same RIL population. The two parents, IR29 and Pokkali, are both indica varieties. Common indica varieties belong to the same group (group I) on the basis of genetic affinity following the isozyme classification system of Glaszmann (Khush 1997). This close genetic relationship between many rice cultivars leads to the generation of relatively low numbers of polymorphic markers per primer combination in segregating populations of rice. In the earlier work of Gregorio (1997) 32 primer combinations were used to generate 205 AFLP markers. We used 25 primer combinations and were able to produce only 139 polymorphic markers. In other crops such as barley (Schut 1998; Qi et al. 1998) and tomato (Haanstra 2000) relatively fewer primer combinations are sufficient to generate many polymorphic AFLP markers.
Skewed segregation was detected in 44% of the 139 markers generated for our study. This distorted segregation in molecular markers is common in rice (Gregorio 1997). Distorted segregation in segregating populations of rice is attributed to the possible association between distorted markers and genes responsible for gametophytic incompatibility (He et al. 2001) or selection in earlier generations against traits such as shattering (Thomson et al. 2003). The RIL population used in this study was developed through single seed descent without selection. Thus the distorted segregation we observed in some of the markers could be due to association of the concerned markers with genes for gametophytic incompatibility.
There was a high degree of environmental specificity with regards to expression of markers with significant associations to grain yield. Due to the lack of a molecular linkage map it was difficult to ascertain whether some of the identified markers were actually linked to the same QTLs or not. Stepwise regression has the deficiency that only the ‘best’ subset regression model is identified while there could be several equally good models (Montgomery and Peck 1982). The selection of markers through stepwise regression suffers from slight changes with respect to included markers as a result of chance variations in the response variable. Hence when two or more markers are closely linked and all are significantly related to a trait under study, one marker may be selected in one environment and not in another environment. This is due to the fact that in the second environment another closely linked marker may be selected first and it would account for most of the observed variation in the trait.
Total variation in yield accounted for by markers was lower in treatments where salt was applied. Soil salinity under natural field conditions is quite variable (Russell 1978; Sylla 1994). This variability would be increased when salinity was imposed manually as it was done in our trial. Gomez and Gomez (1984) pointed out that non-uniform application of treatments may result in higher variability in treated plots than in untreated plots. This may have caused the lower proportion of variance accounted for by markers in the N fertilizer regime in the saline test conditions.
Markers were detected for yield that showed consistent effects in more than one test condition. Such markers would be good candidates for inclusion in breeding programmes aimed at wide adaptation. Furthermore, MAS can be useful in identifying superior genotypes at different N fertilizer levels. However, most markers associated with yield in this study, were detected in only one test condition. As noted earlier, due to the lack of a molecular marker map it could not be confirmed whether the expression of markers in only one test condition was the result of QTL × Environment (QTL × E) interaction or was caused by stepwise regression picking up different markers linked to the same QTL under different test conditions. Nonetheless, proper characterization of cultivation environments would help determine the extent to which MAS can be used in rice breeding programmes. In environments differentiated solely by level of N fertilizer application MAS can be used to identify high-yielding genotypes of rice across different N fertilizer application levels. However, when cultivation environments comprise both saline and fresh water conditions, then MAS should be used to identify high-yielding genotypes specifically for either fresh water or saline conditions.
Differential expression of QTLs, also known as QTL × E interaction has been reported in other studies, for example, for plant type traits of rice at two different locations (Yan et al. 1999), root characteristics of rice in contrasting water-deficit regimes (Price et al. 2002), for growth and grain yield-related traits of rice across nine locations (Hittalmani et al. 2003) and for grain quality traits of rice across different environments (Wan et al. 2005). In these studies, QTLs that were expressed across a broad range of environments have also been reported. Hence in breeding programmes employing the technique of MAS to increase rice yield, the best approach would be to select QTLs for wide adaptability supplemented with QTLs specifically expressed in the range of environments targeted by the breeding programme. Such an approach is also advocated for by other researchers such as Li et al. (2003) and Yang and Zhu (2005).
Yield increasing alleles appeared to be dispersed over the two parents indicating that this cross offers the opportunity for improvement in yield by pyramiding the relevant QTL alleles. However, the tall traditional cultivar, Pokkali, supplied most of the superior alleles for grain yield in all test conditions. This was actually unexpected for it is commonly believed that high-yielding modern cultivars would have accumulated more superior alleles for yield through selection than traditional cultivars. Near isogenic lines could be developed in an IR29 background in order to determine the full benefit of the superior Pokkali alleles for grain yield. In a cross between O. sativa and wild O. rufipogon, Moncada et al. (2001) also found that 56% of QTLs with positive effects on yield and yield components were supplied by O. rufipogon. Thus QTL studies are very useful in uncovering new alleles that can be introgressed into modern high yielding cultivars raising the possibility of increasing the yield of modern cultivars still further (through better adaptation) in diverse environments. Introgression of new alleles from wild or traditional varieties would help to broaden the genetic base of rice cultivars leading to potential improvements in pest and disease resistance.
Conclusions
In this study it was seen that MAS can successfully be used to identify superior genotypes of rice in different test conditions. However, MAS with yield QTLs was generally more successful in fresh water than in saline conditions. Nonetheless, the reliability of MAS to identify superior-yielding genotypes under stress would help breeders overcome the reported problem of low selection efficiency through phenotypic selection for yield in stress environments. In addition, use of MAS can accelerate breeding programmes by enabling breeders to reliably predict performance of rice genotypes from DNA sequence information only.
Both parents possessed superior alleles for grain yield. This offers the opportunity to improve grain yield of rice in different environments through a pyramiding approach. Pokkali contributed many favourable alleles to the cross and could therefore be a source of alleles in rice breeding programmes.
Abbreviations
- AFLP:
-
Amplified Fragment Length Polymorphism
- CM:
-
Centi-Morgan
- DS:
-
Deci-Siemens
- EC:
-
Electrical Conductivity
- QTL:
-
Quantitative Trait Locus
- RIL:
-
Recombinant Inbred Line
- STS:
-
Sequence Tagged Sites
References
Cho YG, Eun MY, McCouch SR, Chae YA (1994) The semidwarf gene, sd-1, of rice (Oryza sativa L.). II. Molecular mapping and marker-assisted selection. Theor Appl Genet 89:54–59
Garcia A, Senadhira D, Flowers TJ, Yeo AR (1995) The effects of selection for sodium transport and of selection for agronomic characteristics upon salt resistance in rice (Oryza sativa L.). Theor Appl Genet 90:1106–1111
Genstat 6.1, (2002) Genstat 6 Release 1. Oxford Sci. Publ., Oxford
Gomez KA, Gomez AA (1984) Statistical procedures for agricultural research, 2nd edn. John Wiley & Sons Inc., Singapore, 680 pp
Gregorio GB (1997) Tagging salinity tolerance genes in rice using amplified fragment length polymorphism (AFLP). PhD thesis, University of the Philippines, Los Baños, Philippines, 118 pp
Haanstra JPW (2000) Characterization of resistance genes to Cladosporium fulvum on the short arm of chromosome 1 of tomato. PhD thesis, Wageningen Agricultural University, Wageningen, The Netherlands, 119 pp
He P, Li JZ, Zheng XW, Shen LS, Lu CF, Chen Y, Zhu LH (2001) Comparison of molecular linkage maps and agronomic trait loci between DH and RIL populations derived from the same rice cross. Crop Sci 41:1240–1246
Hittalmani S, Huang N, Courtois B, Venuprasad R, Shashidhar HE, Zhuang JY, Zheng KL, Liu GF, Wang GC, Sidhu JS, Srivantaneeyakul S, Singh VP, Bagali PG, Prasanna HC, McLaren G, Khush G (2003) Identification of QTL for growth and grain yield-related traits in rice across nine locations in Asia. Theor Appl Genet 107:679–690
IRRI (1996) Standard evaluation system for rice, 4th edn. IRRI, Manila, Philippines, 52 pp
Khush GA (1997) Origin, dispersal, cultivation and variation of rice. Plant Mol Biol 35:25–34
Li ZK, Yu SB, Lafitte HR, Huang N, Courtois B, Hittalmani S, Vijayakumar CHM, Liu GF, Wang GC, Shashidar HE, Zhuang JY, Zheng KL, Singh VP, Sidhu JS, Srivantaneeyakul S, Khush GS (2003) QTL × environment interactions in rice. I. Heading date and plant height. Theor Appl Genet 108:141–153
Moncada P, Martinez CP, Borrero J, Chatel M, Gauch Jr H, Guimaraes E, Tohme J, McCouch SR (2001) Quantitative trait loci for yield and yield components in an Oryza sativa × Oryza rufipogon BC2F2 population evaluated in an upland environment. Theor Appl Genet 102:41–52
Montgomery DC, Peck EA (1982) An introduction to linear regression. Wiley, New York, 504 pp
Price AH, Steele KA, Moore BJ, Jones RGW (2002) Upland rice grown in soil-filled chambers and exposed to contrasting water-deficit regimes II. Mapping quantitative trait loci for root morphology and distribution. Field Crops Res 76:25–43
Primary Industries, SA (1994) Testing for soil and water salinity. Government of South Australia Fact Sheet No. 66/00. http://www.pir.sa.gov.au/factsheets
Qi X, Stam P, Lindhout P (1998) Use of locus-specific AFLP markers to construct a high-density molecular map in barley. Theor Appl Genet 96:376–384
Robin S, Pathan B, Courtois B, Lafitte R, Carandang S, Lanceras S, Amante M, Nguyen HT, Li Z (2003) Mapping osmotic adjustment in an advanced back-cross inbred population of rice. Theor Appl Genet 107:1288–1296
Russell EW (1978) Soil conditions and plant growth. Longman Group Limited, London, 849 pp
SAS Inst. (1999) SAS/STAT User’s Guide. SAS Institute, Cary, NC
Schut JW (1998) Prediction of cross performance in barley. PhD thesis, Wageningen Agricultural University, Wageningen, The Netherlands, 121 pp
Singh S, Sidhu JS, Huang N, Vikal Y, Li Z, Brar DS, Dhaliwal HS, Khush GS (2001) Pyramiding three bacterial blight resistance genes (xa5, xa13 and Xa21) using marker-assisted selection into indica rice cultivar PR106. Theor Appl Genet 102:1011–1015
Stam P, van Ooijen JW (1995) JoinMap(tm) version 2.0: software for the calculation of genetic linkage maps. CPRO-DLO, Wageningen
Sylla M (1994) Soil salinity and acidity: spatial variability and effects on rice production in West Africa’s mangrove zone. PhD thesis, Wageningen Agricultural University, Wageningen, The Netherlands, 175 pp
Thomson MJ, Tai TH, McClung AM, Lai X-H, Hinga ME, Lobos KB, Xu Y, Martinez CP, McCouch SR (2003) Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor Appl Genet 107:479–493
Wan XY, Wan JM, Weng JF, Jiang L, Bi JC, Wang CM, Zhai HQ (2005) Stability of QTLs for rice grain dimension and endosperm chalkiness characteristics across eight environments. Theor Appl Genet 110:1334–1346
Wang GW, He YQ, Xu CG, Zhang Q (2005) Identification and confirmation of three neutral alleles conferring wide compatibility in inter-subspecific hybrids of rice (Oryza sativa L.) using near-isogenic lines. Theor Appl Genet 111:702–710
WARDA (1994) Training in mangrove rice production: Instructors’ manual. WARDA, Bouake, Ivory Coast, 254 pp
Yan J, Zhu J, He C, Benmoussa M, Wu P (1999) Molecular marker-assisted dissection of genotype × environment interaction for plant type traits in rice (Oryza sativa L.). Crop Sci 39:538–544
Yang J, Zhu J (2005) Methods for predicting superior genotypes under multiple environments based on QTL effects. Theor Appl Genet 110:1268–1274
Yeo AR, Flowers TJ (1986) Salinity resistance in rice (Oryza sativa L.) and a pyramiding approach to breeding varieties for saline soils. Aust J Plant Physiol 13:161–173
Yin X, Chasalow SD, Stam P, Kropff MJ, Dourleijn CJ, Bos I, Bindraban PS (2002) Use of components analysis in QTL mapping of complex crop traits: a case study on yield in barley. Plant Breed 121:314–319
Acknowledgments
The authors would like to acknowledge the support of the Netherlands Foundation for the Advancement of Research in the Tropics (WOTRO), the International Foundation for Science, Sweden, Wageningen University, the International Rice Research Institute (IRRI), Philippines and the National Agricultural Research Institute (NARI) of The Gambia. The help of Marcos Malosetti at Wageningen University with the data analyses is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Manneh, B., Stam, P., Struik, P.C. et al. QTL-based analysis of genotype-by-environment interaction for grain yield of rice in stress and non-stress environments. Euphytica 156, 213–226 (2007). https://doi.org/10.1007/s10681-007-9368-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-007-9368-8