Abstract
In order to evaluate the potential responses of deep-burying and mulching on soil bacteria community abundance and soil fertility in the same cropping systems, we designed a field experiment as follows: (1) no straw residue (CK), (2) straw mulching (M); (3) straw residue placed in 20–40 cm (DS); and (4) straw residue placed in 20–40 cm with decomposing agent plus (DSP). The results showed that straw deep placement could increase soil organic matter and improve soil microbial community by Illumina HiSeq high-throughput sequencing approach and real-time quantitative PCR (qPCR). Straw deep-burying reduced soil pH (2%) and increased soil total organic carbon (TOC), microbial biomass carbon (MBC), microbial biomass nitrogen (MBN) by 27–339%, as well as soil enzyme activities (urease, dehydrogenase, and cellulase) by over 7%. Thereafter, we found that the deep buried straw residue increased the soil bacteria abundance by 175%, especially Proteobacteria, Bacteroidetes, and Acidobacteria. The family of Xanthomonadaceae and Chitinophagaceae had achieved remarkable growth. In canonical correspondence analysis (CAA), soil organic matter increasing and pH reducing were the main reasons to shift soil bacterial community. A positive correlation was found between straw deep-burying and soil bacteria community (P < 0.01). Therefore, our results highlight that deep-buried maize straw residue changes soil bacteria community abundance and improves soil fertility. Deep placement could potentially be an effective use of straw residues for the future sustainable cropping systems.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
As a side product of an increasing worldwide production of corn to meet the ever-growing human demands, a large number of straw residues are expected. According to China Ministry of Agriculture, China produces more than 0.8 billion tons of crop residue per year, of which only 14% are returned to the soil. How to effectively manage those corn residues has become an important issue for the government. In order to resource reuse the increasing crop straw, now we are used to place them in field by mulching or deep-burying. However, straw is difficult to decompose in cold winters in the northeast of China; the soil microbial community develop into a limiting factor in straw retention farming land. Straw mulching and deep-burying play different roles in affecting soil bacteria community abundance and soil fertility. We should evaluate the effect to soil bacteria between straw mulching and deep-burying, and then pick the better method to support sustainable farming systems. But the information of effect to soil bacteria between straw mulching and deep-burying is lacked. The aim of this study was therefore to evaluate the effects of straw deep-burying on the soil fertility and the potential responses of bacteria community to such a residue addition.
Based on past practice experience, we hypothesize that deep-burying is the better way to return straw. Firstly, straw is difficult to decompose in cold winters in the northeast of China. Mulching straw as a kind of solid obstacle would disturb next spring’s seeding. In addition, some pests and diseases would remain in the leaving straw and contract the plants. The production after leaving stubble or mulching were always dropped (Thorburn et al. 2012). Secondly, straw buried in 0–20-m layer soil would harm plant root growth. Straw mulching or returning in a shallow layer may form obstructer to scramble for soil N (Ndegwa and Thompson 2000). In addition, the fermented residue could release some small molecular weight organic acids, like oxalate (Li et al. 2011). So the soil pH would be altered around plant root when we are mulching or burying straw in 0–20-cm layer. It is unfavorable for crop farming (Ma et al. 2016). Thus, aiming to separate seed and root from straw residue in soil space, burying straw in a depth of 20–40 cm becomes a vital way of straw returning to the field (Wang et al. 2015a). Some straw-decomposing agents were used to accelerate residue decomposition when deep-burying straw, so the yield after straw deep-burying was always higher than mulching (Li et al. 2014).
The main reason of corn yield increased after straw returning was the soil fertility improvement (Lueders et al. 2006). The nutrients from straw decomposed by soil microbial increased soil total organic carbon (TOC) and total nitrogen (TN), which control soil degradation and promoted crop yield (Wang et al. 2019; Yin et al. 2018). In this straw retention farming biochemical process, soil bacteria is pivotal constituent of soil characteristics (Berg 2009; Sharma et al. 2011). The more rich soil microbial biomass and more active soil enzymes reflect the better function of soil microorganism in organic matters transformation (Burns et al. 2013; Dariusz et al. 2004). The previous studies of microbiology mechanism on straw mulching have been conducted widely, while straw deep-burying was short. Straw mulching could increase soil enzyme activities and growth of soil bacteria (Grandy et al. 2013; Zhao et al. 2016). It is soil bacteria which dominates in the initial phase of crop residues decomposition and increases soil carbon stocks (Hao et al. 2019). The abundances of Proteobacteria were increased after straw mulching, especially Actinobacteria, Betaproteobacteria, and Gammaproteobacteria (Li et al. 2015). In straw deep-burying studies, we have preliminary knowledge that straw deep-burying could obtain higher soil organic carbon and microbial biomass (Wang et al. 2015b). But the information on how deep-buried straw residue affects soil microorganisms is limited, the shift of soil bacteria after straw deep-burying is a confusion.
Up to now, real-time quantitative PCR (qPCR) and high-throughput sequencing techniques take a study for soil microbial species, community, function, and genetic diversity to elucidate the microbiology mechanism of biogeochemical cycling (Lou et al. 2014). We inferred residue deep-burying could shift soil microbial community abundance by soil microbial biomass carbon (MBC) increased in a deep-buried straw experiment (Zou et al. 2016). qPCR and high-throughput sequencing techniques can bring insight into soil bacteria community. We suspected straw deep-burying may promote soil enzyme activities and bacteria abundance. It will be important to decrease straw wasting and inorganic fertilization using to sustain the development of ecological agriculture.
2 Materials and Methods
2.1 Experiment Design
This straw placement experiment was conducted in the Agricultural Science and Technology Test Station, in the northeast of China (119° 32′ E, 41° 20′ N). The soil typical there was carbonate cinnamon soil of loam, pH 7.8, contain TOC 7.06 g kg−1, TN 108 g kg−1, available phosphorus (AP) 17.3 mg kg−1, and available potassium (AK) 130 mg kg−1. The annually average rainfall is 450 mm, frost-free period is 135 days, and effective accumulated temperature is 3200 °C.
Maize variety of Liaodan565 was plant in blocks, keeping 25-cm row spacing and 66,000–67,500 plants per hectare. Every block designed 4.8 m in width and 24 m in length with management as normal. Four treatments and three replicates were designed: (1) No straw residue control (CK); (2) Straw mulching (M); (3) Straw residue placed under 40 cm in soil (DS); and (4) Straw residue and decomposing agent (30 Kg ha−1) placed under 40 cm in soil together (DSP). The amount of returned straw was 12,000 Kg ha−1. We evenly spread 31 Kg ha−1 urea in the blocks of M, DS, and DSP to control straw residue C/N ratio of 25:1 at the same time of straw returning (Ndegwa and Thompson 2000). When seedlings are planted, every treatments contain the same fertilization amount of total N 3600 Kg ha−1, total P (P2O5) 1800 Kg ha−1, and total K (K2O) 1350 Kg ha−1.
The decomposing agent was granular materials as the promotion to accelerate straw decomposition. Thirty kilograms per hectare of decomposing agent was sprinkled evenly on straw of DSP block by manual when we put straw in ditch and then bury soil. We select a production of Henglongtai Biological Engineering Company in Henan Province named HM Straw Decomposing Agent. It contains a lot of bacteria to decompose protein, cellulose, hemicellulose, and lignin. The effective number of viable cells of the decomposing agent was more than 0.5 × 109 N g−1 with standard plate counting method (r ≥ 0.99). The detailed information was in Appendix.
After the harvest in 2013, we furrowed and buried straw residue by a self-developed agricultural straw return machinery (Zou et al. 2014). Two rows corn as a team, we put two rows straw into the medial of their growth land, as shown in Fig. 1a. In 2014 spring, we took no-till farming, and in autumn returned the obtained straw into the other side of their growth land, as shown in Fig. 1b. Farming repeated as this cycle. It takes a long-term farm management as this method. Soil samples and corncob of this paper were collected after harvest in 2015 by “S” method with 5 places of 0–40-cm layer to further analyze. Dry weight of maize grain was used to calculate yield.
2.2 Soil Analyses
Soil pH was measured with a glass electrode using a soil-to-water ratio of 1:2.5. Soil TOC, TN, MBN, and MBC were determined by routine methods (Xiangui Lin 2009).
In this study, we determined soil cellulase, dehydrogenase, invertase, and urease activities respectively by using 3,5-dinitrosalicylic acid method (Zhao et al. 2008), the reduction of 2,3,5-triphenyltetrazolium (Serra-Wittling et al. 1995), 3,5-dinitrosalicylic acid method (Frankenberger and Johanson 1983), and indophenol blue colorimetric method (Gosewinkel and Broadbent 1984).
2.3 DNA Extraction
DNA was extracted from 0.5 g of each individual replicate soil using a FastDNA® SPIN Kit (MP Biomedicals, Santa Ana, CA) following the manufacturer’s instructions.
2.4 Real-Time Quantitative PCR of Bacterial 16 S rDNA Genes
SYBR Premix Ex Tap Perfect Real-Time Kit of TaKaRa with Bio-Rad CFX96 Real-Time PCR System (Bio-Rad, CA, USA) recommended primer sets 519F/907R (519F: 5′-GTGCCAGCMGCCGCGG-3′, 907R: 5′-CCGTCAATTCMTTTRAGTTT-3′) in qPCR of bacterial 16 s rDNA genes analysis (Biddle et al. 2008). In brief, 25-μl reaction mixtures contained 12.5-μl of SYBR® Premix ExTaq™, 0.5 μM of each primer, and 1.0 μl template containing 2–9 ng DNA, and the details were observed in the study of Xu et al. (2013).
2.5 PCR and Preparation of the Amplicon Libraries
The brief DNA of purification was the template for PCR copy 16SV4 gene fragment. And then, common bacteria primer with Barcode (519F/907R) could identify bacteria diversity by Phusion® High-Fidelity PCR Master Mix with GC Buffer of New England Biolabs Company. The production of PCR was detected by lipid sugar gel electrophoresis in 2% and construct the library with TruSeq® DNA PCR-Free Sample Preparation Kit. The qualified library was sequenced by Qubit and qPCR with HiSeq2500 PE250.
2.6 Processing of Pyrosequencing Data
The 16-s data, Raw Tags was PE Reads jointed after split through FLASH (Lozupone et al. 2011) and filtered by Qiime (Caporaso et al. 2010) according to quality. At last, we got the effective tags through length filtering and chimera deleting. (Bokulich et al. 2013). Using a 97% identity threshold, the most abundant sequence from each operational taxonomic units (OUTs) was selected as a representative sequence for that OTUs. Simpson and Shannon index has been figured out by Qiime (Version 1.7.0), as well as Rarefaction Curve, ACE, and Chao1 index to richness between different treatments (Amato et al. 2013; Sun et al. 2013).
2.7 Statistics Method
IBM SPSS Statistic 22, R, and Origin Lab 9.0 were carried out to calculate standard deviation, Tukey’s multiple range test and do some figures. The differences of each mean at P < 0.05 were considered statistically significant.
3 Results
3.1 The Soil Fertility and Crop Yield
Compared to the no-straw control (CK), straw residue significantly got an increased yield of maize crop (P < 0.05, Table 1). It is the phenomenon that the straw has been hidden underground in a valid way. The yield of straw deep application (DS and DSP) is higher than mulching; DSP was higher than DS by 8%.
From the soil characters, straw addition could reduce soil pH; mulching dropped 1%; DS and DSP dropped 2%. Straw deep application (DS and DSP) got a significant increased value of TN (27%), TOC (23%), and MBN (48%), MBC (> 200%). There is no difference between CK and M. Further statistical tests revealed a positive correlation between soil characters and crop yield. Especially the differences among M, DS, and DSP were highlighted in MBC and MBN; the MBC of DSP and DSP were higher than M by 36% and 182%; MBN was higher than by 17% and 33%. It is a strong evidence of soil microbial community changed.
3.2 The Effect to Soil Enzyme Activity
Among the four tested soil enzyme activities, except invertase, activities of urease, dehydrogenase, and cellulase were generally significantly increased under the addition of either mulching or deep placement of straw residue (P < 0.05, Fig. 2), compared to the no-straw control. The activities of invertase decreased after straw amendment.
The trend of dehydrogenase and urease activity is consistent. Mulching increased urease by 12%, dehydrogenase by 360%, and cellulose 66%. DS and DSP were more active than M, urease 7%, dehydrogenase 16%, and cellulose more than 58%. Activities of urease and dehydrogenase were similar between DS and DSP. It showed the same trend as maize production and soil TOC. The urease activity significantly correlates with MBN (P < 0.05, R2 = 0.93) and qPCR (P < 0.01, R2 = 0.98); cellulose activity has a linear correlation with MBC (P < 0.01, R2 = 0.99). Therefore, this nutrition came from straw decomposition can stimulate soil bacteria growth to promote more enzyme release. Straw cellulose decomposing and organics release must be the result of the more active enzyme. It was further corroborated the responses of soil enzyme activities that straw deep-burying developed richer bacteria biomass than mulching.
3.3 Copy Number of Bacterial 16 s rDNA Genes
Gene copy number refers to a gene or a specific segment of the DNA sequence in the haploid genome. The result of copy number of dry weight (DW) soil bacteria 16 s rDNA is showed as Fig. 3 (P < 0.05). Compared to the CK, the gene copy number in soil samples from the straw return by deep ditching treatments, DS (142%) and DSP (208%) increased significantly. DSP was significantly higher than M by 56%. By contrast, the copy number of mulching (M) was no difference greater than CK. From the copy number, soil enzyme activities were enhanced along with bacteria growth in quantity.
3.4 Pyrosequencing Information and Overall Taxonomic Richness
The rationality of sequencing data would be reflected as reasonable directly when the curve tends to flat, and the richness of simple species as well (Appendix). As in Fig. 4a, the distribution of 16S sequence number at each classification level obtained is shown in Fig. 4b. Compared with the CK, sequence numbers decreased for all levels of classification in M; by contrast, numbers increased for all levels in DSP.
In phylum, the maximum abundance of the top 10 in species relative abundance cylindrical cumulative graph was shown in Fig. 4c. Compared to the CK, Proteobacteria, Bacteroidetes, and Acidobacteria have an obvious shift and increased by fluctuation in deep burying treatments (DS and DSP). Simultaneously, the abundance of Bacteroidetes, Crenarchaeota, and Firmicutes also shifted.
In Venn diagrams (Fig. 5), a mean of 5530 bacterial OTUs was observed in the group of CK, M, DS, and DSP; the number of shared OTUs was 1955 (35.35% of the total). Straw deep-burying either DS or DSP owned the relative lower unique OTUs numbers in contrast to CK and M treatments. From Table 2, bacterial α-diversity of DS and DSP waslower than M and CK; this further evidenced that the bacteria community after straw deep-burying will not be increased species number or richness. Some local dominant bacteria may develop well and increased in quantity. There was no significant difference among different treatments in Simpson index. The diversity index indicated that no significant differences were potentially related to straw mulching, deep-burying, and decomposition agent.
3.5 The Relative Abundance of Bacteroidetes, Acidobacteria, and Proteobacteria
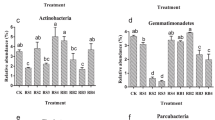
The abundance of each sample in Class classification level was showed as (Fig. 6) Acidobacteria, Acidobateria-6, and [Chloracidobacteria] got obviously changes. Straw mulching stay a same abundance with CK. Acidobacteria increased in the treatments of straw deep placement. For DS, sequences decreased significantly but an equal relative abundance to CK. Within the Proteobacteria, straw deep applied (DS and DSP) increased sequences of Alphaproteobacteria, Deltaproteobacteria, and Gammaproteobacteria, while relative abundance in contrast. Mulching did not get a significant shift and almost was same as CK.
In further analysis, the family level classification within Proteobacteria and Bacteroidetes was conducted to determine whether sequences were different among treatments (Fig. 7). Within the Proteobacteria in the CK, the families Xanthomonadaceae and Sinobacteraceae in the order Xanthomonadales contained the dominant indigenous bacteria. The sequence number and relative abundance of Xanthomonadaceae increased significantly in straw deep placement. The treatment with the decomposing agent (DSP) supported a homogeneous propagation rate of Xanthomonadaceae and Sinobacteraceae. The shift within Bacteroidetes was more complex than that within Proteobacteria.
The shift of Bacteroidetes was more complex than Proteobacteria. First, straw amendment (M, DS, and DSP) got an increased sequence number of Cyclobacteriaceae. Second, mulching (M) decreased the sequences and relative abundance of Chitinophagaceae and Sphingobacteriaceae. However, deep placement (DS) increased Sphingobacteriaceae and supported a homogeneous propagation rate of Chitinophagaceae. Third, the decomposing agent (DSP) got a significant stimulated to S24–7 compared to others.
3.6 Relationship Between Soil Environmental Factors and Bacterial Community
Maize yield was positively correlated with bacterial α-diversity (Shannon index, R2 = 0.737, P < 0.01), richness (Chao1 index, R2 = 0.811, P < 0.01) as well as gene abundance (R2 = 0.858, P < 0.01), as Fig. 8. This suggests that the bacterial community plays an important role in improving yield after straw amendment.
CCA was used to reveal what environmental factors shifted bacteria assemblages in soils (Fig. 9). The four treatments resulted in different distributions. Dehydrogenase activity was positively associated with straw deep-burying and soil C or N. The shifts in OTUs of bacteria in different treatments were correlated with environmental variables. Compared to CK, straw amendment led to clear differences. The bacteria shift of straw deep placement treatments shows positive correlation with soil MBC, MBN, TOC, and TN. Straw mulching and no - straw tillage could not promote soil bacteria growth via increasing soil TOC and microbial biomass, whereas the dominant change was in pH. DSP had a significant difference with DS in more obvious positive correlation with MBC, TOC, and dehydrogenase activity. The straw decomposition agent accelerated residue carbon translation into soil microorganisms.
4 Discussion
Soil organic matter is the representative of soil fertility and it could be stocked by straw returning (Laird and Chang 2013). Straw residue retains crude fiber, trace element, and essential amino acid like lysine, methionine, and cysteine. These residues as a kind of organic fertilization could enhance soil TN, TOC, MBC, and MBN; reduced soil pH; and promote plant growth (Sun et al. 2015; Wang et al. 2015a, b). According to some reports, straw deep-burying could more effectively modify soil physical and increase soil organic carbon stock, among other ways of returning straw to the field (Ludwig et al. 2011; Malhi et al. 2011; Schneider et al. 2017). On the one hand, straw deep-burying increases the plant availability of subsoil nutrients, which does not come at the cost of impaired topsoil fertility (Schneider et al. 2017). On the other hand, the 40-cm depth was conducive to the straw anaerobic decomposition and fixed more C in soil than straw mulching (Olk et al. 2007). In the same experiment condition, straw deep-burying would keep the better soil situation than straw mulching, where the soil temperature is more constant, soil moisture is more sufficient. Straw mineralization is thus increased (Dan et al. 2015; Li et al. 2006; Wang et al. 2011). In this paper, straw deep-burying got the highest TN and TOC among mulching and no-straw, so it got the biggest gains.
The affluent bacteria community was the key factor to release straw nutrient and then promote corn growth. After returning straw, soil organic matter and pH were changed. Soil pH is a powerful driving factor for bacterial diversity cation (Griffths et al. 2011; Bartram et al. 2014), and straw became the culture medium of bacteria (Chaudhry et al. 2012). This study found that straw deep-burying with lowest pH value could promote bacterial growth. Furthermore, the straw of mulching was on the dry surface of soil, while the straw of deep-burying was in the wet soil. Microbial activity of straw mulching was limited by temperature and moisture (Cao et al. 2015; Cao et al. 2016). The increased soil enzyme activities after straw deep-burying further evidenced a more active soil bacteria community. Dehydrogenase activity has been considered as an indicator of the overall soil microbial activity, urease and cellulase activities characterize soil microorganism’s ecological function involved in soil N, and C cycles (Xu et al. 2013). Cellulase and urease are hydrolytic enzymes, which are responsible for the acquisition of C and N by degrading cellulose (Bowles et al. 2014). More cellulose from straw can promote fiber hydrolization into reducing sugars, and combined with other production, like protein and amino acids, to be consumed by microbes easily. However, the invertase is the enzyme to hydrolyze sucrose. It was measured as potential enzyme activity rather than real enzyme activity (Nannipieri et al. 2012). In addition, the application of chemical fertilizer can decrease the enzyme activity for retarding the enzyme synthesis (Nannipieri et al. 2008).
Straw amendment has been able to enhance soil bacteria community (Huang et al. 2015; Sun et al. 2015) by the changes in nutrient substrates (Yuan et al. 2013; Zhao et al. 2014). Usually, straw made bacterial richness increased without new bacterial species emerging (He et al. 2008; Börjesson et al. 2012). Based on the further results from high-throughput sequencing, clear shifts were observed in Proteobacteria, Bacteroidetes, and Acidobacteria. Proteobacteria is the bacteria which includes many aerobic or facultative bacteria, and usually has a strong degradation capacity (Timothy et al. 2011). Proteobacteria relative abundance has been significant increased after straw deep-burying (Fig. 4). The phylum of Proteobacteria contains a variety of metabolic types; the class Gamma-Proteobacteria currently contains the best known types of bacteria and includes important model organisms, such as Colibacillus. Most nitrogen-fixing bacteria are within the Proteobacteria, and with soil microbial activity, soil nutrition is promoted by the processes of nitrification and oxidation and increases in the concentrations of nitrogen, sulfide, and ammonia (Coleman et al. 1996). Many species within Bacteroidetes can degrade cellulose, especially Chitinophaga, which can efficiently degrade organic matters (Weilandbräuer et al. 2017). Straw deep-burying creates better conditions for bacteria to decompose organic matter than straw mulching or no straw. Within Acidobacteria, returned straw decreased the abundance of Acidobacteria-6 and [Chloracidobacteria], unlike the treatment with the decomposing agent. It is similar to a study who displayed a negative correlation between Acidobacteria and soil total carbon (Sun et al. 2015). Because Acidobacteria was negatively related with soil carbon availability (Fierer et al. 2007), we always found that the highest abundances are present in the lowest pH soils (Lauber et al. 2009; Chu et al. 2010; Shen et al. 2013). The effects of decomposing agents are compounded by the inclusion of some complex bacteria like Acidobacteria; it may help to maintain a stable and balanced increasing in bacteria abundance rather than affecting diversity. But the further effect of decomposing agents to soil bacteria remained to be solved.
5 Conclusions
In this study, straw mulching almost did not change the bacteria community of soil. Straw deep-burying could not affect the soil bacterial community. The decomposing agents maintain a balance in the abundance of soil bacteria. Therefore, straw deep-burying with decomposing agents is the better straw retention or returning method than mulching, from the perspective of soil bacteria. It will help the problem of too much straw wasted in north of China.
References
Amato KR, Yeoman CJ, Kent A, Righini N, Carbonero F, Estrada A, Gaskins HR, Stump RM, Yildirim S, Torralba M, Gillis M, Wilson BA, Nelson KE, White BA, Leigh SR (2013) Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. The ISME Journal 7(7):1344–1353. https://doi.org/10.1038/ismej.2013.16
Bartram AK, Jiang X, Lynch MD, Masella AP, Nicol GW, Dushoff J, Neufeld JD (2014) Exploring links between pH and bacterial community composition in soils from the Craibstone experimental farm. FEMS Microbiol Ecol 87:403–415. https://doi.org/10.1111/1574-6941.12231
Berg G (2009) Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl Microbiol Biotechnol 84:11–18. https://doi.org/10.1007/s00253-009-2092-7
Biddle JF, Fitz-Gibbon S, Schuster SC, Brenchley JE, House CH (2008) Metagenomic signatures of the Peru Margin subsea floor biosphere show a genetically distinct environment. Proceedings of the National Academy of Sciences of the United States of America (PNAS), 105: 10583-10588. https://doi.org/10.1073/pnas.0709942105
Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10(1):57–59. https://doi.org/10.1038/nmeth.2276
Börjesson G, Menichetti L, Kirchmann H, Kätterer T (2012) Soil microbial community structure affected by 53 years of nitrogen fertilisation and different organic amendments. Biol Fertil Soils 48(3):245–257. https://doi.org/10.1007/s00374-011-0623-8
Bowles TM, Acosta-Martinez V, Calderon F, Jackson LE (2014) Soil enzyme activities, microbial communities, and carbon and nitrogen availability in organic agroecosystems across an intensively-managed agricultural landscape. Soil Biol Biochem 68:252–262. https://doi.org/10.1016/j.soilbio.2013.10.004
Burns RG, DeForest JL, Marxsen J, Sinsabaugh RL, Stromberger ME, Wallenstein MD, Weintraub MN, Zoppini A (2013) Soil enzymes in a changing environment: Current knowledge and future directions. Soil Biol Biochem 58:216–234. https://doi.org/10.1016/j.soilbio.2012.11.009
Cao YF, Zhang H, Liu K, Dai YC, Lv JL (2015) Organic acids variation in plant residues and soils among agricultural treatments. Agron J 107(6):2171–2180. https://doi.org/10.2134/agronj15.0137
Cao YF, Zhang H, Liu K, Lv JL (2016) Decomposition characteristics of crop residues among different agricultural treatments. Transactions of the Chinese Society of Agricultural Machinery 47(9):212–219. https://doi.org/10.6041/j.issn.1000-1298.2016.09.030
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336. https://doi.org/10.1038/nmeth.f.303
Chaudhry V, Rehman A, Mishra A, Chauhan PS, Nautiyal CS (2012) Changes in bacterial community structure of agricultural land due to long-teen organic and chemical amendments. Microb Ecol 64:450–460. https://doi.org/10.1007/s00248-012-0025-y
Chu H, Fierer N, Lauber CL, Caporaso JG, Knight R, Grogan P (2010) Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ Microbiol 12:2998–3006. https://doi.org/10.1111/j.1462-2920.2010.02277.x
Coleman DC, Crossley DA, Hendrix PF (1996) Fundamentals of soil ecology. Academic Press, London, pp 48–77
Dan T, Lynne CB, Christian T, John PR (2015) Crop production and soil water management in conservation agriculture, no-till, and conventional tillage systems in Malawi, agriculture. Ecosystem and Environment 212:285–296. https://doi.org/10.1016/j.agee.2015.07.011
Dariusz K, Christopher M, Hubert H, Marcos G, Bernhard F, Ludwig B, Paul V (2004) Litter decomposition, microbial biomass and activity of soil organisms in three agroforestry sites in central Amazonia. Nutr Cycl Agroecosyst 69:257–267. https://doi.org/10.1023/B:FRES.0000035196.19804.13
Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88:1354–1364. https://doi.org/10.1890/05-1839
Frankenberger WT, Johanson JB (1983) Method of measuring invertase activity in soils. Plant Soil 74:301–311. https://doi.org/10.1007/BF02181348
Gosewinkel U, Broadbent FE (1984) Conducti metric determination of soil urease activity. Commun Soil Sci Plant Anal 15:1377–1389. https://doi.org/10.1080/00103628409367564
Grandy AS, Salam DS, Wickings K, McDaniel M, Culman SW, Snapp SS (2013) Soil respiration and litter decomposition responses to N fertilization rate in no-till corn systems. Agric Ecosyst Environ 179:35–40. https://doi.org/10.1016/j.agee.2013.04.020
Griffths RI, Thomson BC, James P, Bell T, Bailey M, Whiteley AS (2011) The bacterial biogeography of British soils. Environ Microbiol 13:1642–1654. https://doi.org/10.1111/j.1462-2920.2011.02480.x
Hao M, Hu H, Liu Z, Dong Q, Sun K, Feng Y, Li G, Ning T (2019) Shifts in microbial community and carbon sequestration in farmland soil under long-term conservation tillage and straw returning. Appl Soil Ecol 136:43–54. https://doi.org/10.1016/j.apsoil.2018.12.016
He JZ, Zheng Y, Chen CR, He YQ, Zhang LM (2008) Microbial composition and diversity of an upland red soil under long-term fertilization treatments as revealed by culture-dependent and culture-independent approaches. Journal of Soil Sediments 8:349–358. https://doi.org/10.1007/s11368-008-0025-1
Huang XQ, Liu LL, Wen T, Zhu R, Zhang JB (2015) Illumina MiSeq investigations on the changes of microbial community in the Fusarium oxysporum f.sp. cubense infected soil during and after reductive soil disinfestation. Microbiol Res 181:33–42. https://doi.org/10.1016/j.micres.2015.08.004
Laird DA, Chang CW (2013) Long-term impacts of residue harvesting on soil quality. Soil Tillage Res 134:33–40. https://doi.org/10.1016/j.still.2013.07.001
Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75:5111–5120. https://doi.org/10.1128/AEM.00335-09
Li XG, Li FM, Bhupinderpal S, Cui ZJ, Rengel Z (2006) Decomposition of maize straw in saline soil. Biol Fertil Soils 42:366–370. https://doi.org/10.1007/s00374-005-0042-9
Li NJ, Zeng GM, Huang DL, Hu S, Feng CL, Zhao MH, Lai C, Huang C, Wei Z, Xie GX (2011) Oxalate production at different initial Pb2+ concentrations and the influence of oxalate during solid-state fermentation of straw with Phanerochaete chrysosporium. Bioresour Technol 102:8137–8142. https://doi.org/10.1016/j.biortech.2011.05.092
Li XZ, Rui JP, Xiong JB, Li JB, He ZL, Zhou JZ, Anthony C (2014) Functional potential of soil microbial communities in the maize rhizosphere. PLoS One 9(11):e11609. https://doi.org/10.1371/journal.pone.0112609
Li J, Cooper JM, Lin ZN, Li YT, Yang XD (2015) Soil microbial community structure and function are significantly affected by long-term organic and mineral fertilization regimes in north China plain. Appl Soil Ecol 96:75–87. https://doi.org/10.1016/j.apsoil.2015.07.001
Lin XG (2009) Principles and methods of soil microbiology research. Beijing, China
Lou J, Liu Y, Li Y (2014) Review of high-throughput sequencing techniques in studies of soil microbial diversity. Chinese Agricultural Science Bulletin 30(15):256–260 (in Chinese, with English abstract)
Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R (2011) UniFrac: an effective distance metric for microbial community comparison. The ISME journal 5(2):169–172. https://doi.org/10.1038/ismej.2010.133
Ludwig B, Geisseler D, Michel K, Joergensen RG, Schulz E, Merbach L, Raupp J, Rauber R, Hu K, Niu L, Liu X (2011) Effects of fertilization and soil management on crop yields and carbon stabilization in soils. A review. Agronomy for Sustainable Development 31:361–372. https://doi.org/10.1051/agro/2010030
Lueders T, Kindler R, Miltner A, Friedrich MW, Kaestner M (2006) Identification of bacterial micropredators distinctively active in a soil microbial food web. Appl Environ Microbiol 72:5342–5348. https://doi.org/10.1128/AEM.00400-06
Ma ZG, Frédéric M et al (2016) Glyoxylate cycle and metabolism of organic acids in the scutellum of barley seeds during germination. Plant Sci 248:37–44. https://doi.org/10.1016/j.plantsci.2016.04.007
Malhi SS, Nyhorg M, Goddard T, Puurveen D (2011) Long-term tillage, straw management and N fertilization effects on quantity and quality of organic C and N in a Black Chernozem soil. Nutr Cycl Agroecosyst 90:227–241. https://doi.org/10.1007/s10705-011-9424-6
Nannipieri P, Ascher J, Ceccherini MT, Landi L, Pietramellara G, Renella G, Valori F (2008) Effects of root exudates in microbial diversity and activity in rhizosphere soils. In: Nautiyal CS, Dion P (eds) Molecular mechanisms of plant and microbe coexistence. Soil biology, vol 15. Springer, Berlin. https://doi.org/10.1007/978-3-540-75575-3_14
Nannipieri P, Giagnoni L, Renella G, Puglisi E, Ceccanti B, Masciandaro G, Fornasier F, Moscatelli M, Marinari S (2012) Soil enzymology: classical and molecular approaches. Biol Fertil Soils 48:743–762. https://doi.org/10.1007/s00374-012-0723-0
Ndegwa PM, Thompson SA (2000) Effects of C-to-N ratio on vermicomposting of biosolids. Bioresour Technol 75:7–12. https://doi.org/10.1016/S0960-8524(00)00038-9
Olk DC, Samson MI, Gapas P (2007) Inhibition of nitrogen mineralization in young humic fractions by anaerobic decomposition of rice crop residues. Eur J Soil Sci 58(1):270–281. https://doi.org/10.1111/j.1365-2389.2006.00836.x
Schneider F, Don A, Hennings I, Schmittmann O, Seidel SJ (2017) The effect of deep tillage on crop yield - what do we really know? Soil Tillage Res 174:193–204. https://doi.org/10.1016/j.still.2017.07.005
Serra-Wittling C, Houot S, Barriuso E (1995) Soil enzymatic response to addition of municipal solid-waste compost. Biol Fertil Soils 20(4):226–236. https://doi.org/10.1007/BF00336082
Sharma SK, Ramesh A, Sharma MP, Joshi OP, Govaerts B, Steenwerth K, Karlen DL (2011) Microbial community structure and diversity as indicators for evaluating soil quality. In: Lichtfouse E (ed) Biodiversity, biofuels, agroforestry and conservation agriculture. Sustainable agriculture reviews, vol 5. Springer, Netherlands, pp 317–358. https://doi.org/10.1007/978-90-481-9513-8_11
Shen CC, Xiong JB, Zhang HY, Feng YZ, Lin XG, Li XY, Liang WJ, Chu HY (2013) Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol Biochem 57:204–211. https://doi.org/10.1016/j.soilbio.2012.07.013
Sun H, Terhonen E, Koskinen K, Paulin L, Kasanen R, Asiegbu FO (2013) The impacts of treatment with biocontrol fungus (Phlebiopsis gigantea) on bacterial diversity in Norway spruce stumps. Biol Control 64(3):238–246. https://doi.org/10.1016/j.biocontrol.2012.11.015
Sun RB, Zhang XX, Guo XS, Wang DZ, Chu HY (2015) Bacterial diversity in soils subjected to long-term chemical fertilization can be more stably maintained with the addition of livestock manure than wheat straw. Soil Biol Biochem 88:9–18. https://doi.org/10.1016/j.soilbio.2015.05.007
Thorburn PJ, Meier EA, Collins K, Robertson FA (2012) Changes in soil carbon sequestration, fractionation and soil fertility in response to sugarcane residue retention are site-specific. Soil Tillage Res 120:99–111. https://doi.org/10.1016/j.still.2011.11.009
Timothy DB, Mark A, Elizabeth MH, Rahul S (2011) The emerging role for bacteria in lignin degradation and bio-product formation. Curr Opin Biotechnol 22:394–400. https://doi.org/10.1016/j.copbio.2010.10.009
Wang XB, Dai K, Zhang DC, Zhang XM, Wang Y, Zhao QS, Cai DX, Hoogmoed WB, Oenema O (2011) Dryland maize yields and water use efficiency in response to tillage/crop stubble and nutrient management practices in China. Field Crop Res 120:47–57. https://doi.org/10.1016/j.fcr.2010.08.010
Wang XH, Yang HS, Liu J, Wu JS, Chen WP, Wu J (2015a) Effects of ditch-buried straw return on soil organic carbon and rice yields in a rice-wheat rotation system. Catena 127:56–63. https://doi.org/10.1016/j.catena.2014.10.012
Wang SN, Zou HT, Zhang YL, Yu N, Zhang YL, Fan QF (2015b) Effect of straw deep returning on the soil water features and soil organic carbon components. Journal of Soils Water Conserve 29:154–158 (in Chinese, with English abstract)
Wang H, Shen M, Hui D, Chen J, Sun G, Wang X, Zhang Y (2019) Straw incorporation influences soil organic carbon sequestration, greenhouse gas emission, and crop yields in a Chinese rice (Oryza sativa L.) - wheat (Triticum aestivum L.) cropping system. Soil Tillage Res 195:104377. https://doi.org/10.1016/j.still.2019.104377
Weilandbräuer N, Fischer MA, Schramm KW, Schmitz RA (2017) Polychlorinated biphenyl (PCB)-degrading potential of microbes present in a cryoconite of Jamtalferner glacier. Front Microbiol 8:1105. https://doi.org/10.3389/fmicb.2017.01105
Xu JB, Feng YZ, Wang YM (2013) Soil microbial mechanisms of Stevia rebaudiana (Bertoni) residue returning increasing crop yield and quality. Biol Fertil Soils 49:839–846. https://doi.org/10.1007/s00374-013-0777-7
Yin HJ, Zhao WQ, Li T, Cheng XY, Liu Q (2018) Balancing straw returning and chemical fertilizers in China: role of straw nutrient resources. Renew Sust Energ Rev 81:2695–2702. https://doi.org/10.1016/j.rser.2017.06.076
Yuan H, Ge T, Zhou P, Liu S, Roberts P, Zhu H, Zou Z, Tong C, Wu J (2013) Soil microbial biomass and bacterial and fungal community structures responses to long-term fertilization in paddy soils. J Soils Sediments 13:877–886. https://doi.org/10.1007/s11368-013-0664-8
Zhao K, Xue PJ, Gu GY (2008) Study on determination of reducing sugar content using 3,5-Dinitrosalicylic acid method. Food Science 29(08):534–536 (in Chinese, with English abstract)
Zhao J, Ni T, Li Y, Xiong W, Ran W, Shen Q, Shen Q, Zhang R (2014) Responses of bacterial communities in arable soils in a rice-wheat cropping system to different fertilizer regimes and sampling times. PLOS ONE 9:eR5301. https://doi.org/10.1371/journal.pone.0085301
Zhao SC, Li KJ, Zhou W, Qiu SJ, Huang SW, He P (2016) Changes in soil microbial community, enzyme activities and organic matter fractions under long-term straw return in north-central China. Agric Ecosyst Environ 216:82–88. https://doi.org/10.1016/j.agee.2015.09.028
Zou HT, Wang SN, Yan HL, Ma YB, Fan QF, Huang Y, Zhang YL (2014) Effects of straw deep returning on soil structure moisture in semiarid region of Northeast China. Agricultural Research in the Arid Areas 32(2):52–60 (in Chinese, with English abstract)
Zou HT, Ye XH, Li JQ, Lu J, Fan QF, Yu N, Zhang YL, Dang XL, Yulong Z (2016) Effects of straw return in deep soils with urea addition on the soil organic carbon fractions in a semi-arid temperate cornfield. PLoS One 11(4):e0153214. https://doi.org/10.1371/journal.pone.0153214
Acknowledgements
We thank Professor Youzhi Feng, of the Institute of Soil Science, Chinese Academy of Sciences, for advice and help.
Funding
This experiment was funded by the National Key Technology R&D Program (2015BAD23B0203), Special Fund for Agro-scientific Research in the Public Interest (201303125), Liaoning agricultural science and technology innovation talents training program (Grant No. 2015051), and team project of science and technology innovation in Liaoning Province (2015213001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 151 kb)
Rights and permissions
About this article
Cite this article
Li, J., Ye, X., Zhang, Y. et al. Maize Straw Deep-Burying Promotes Soil Bacteria Community Abundance and Improves Soil Fertility. J Soil Sci Plant Nutr 21, 1397–1407 (2021). https://doi.org/10.1007/s42729-021-00448-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42729-021-00448-6












