Abstract
The neurotransmitter acetylcholine (ACh) plays a ubiquitous role in cognitive functions including learning and memory with widespread innervation in the cortex, subcortical structures, and the cerebellum. Cholinergic receptors, transporters, or enzymes associated with many neurodegenerative diseases, including Alzheimer’s disease (AD) and Parkinson’s disease (PD), are potential imaging targets. In the present study, we have developed 2D-quantitative structure–activity relationship (2D-QSAR) models for 19 positron emission tomography (PET) imaging agents targeted against presynaptic vesicular acetylcholine transporter (VAChT). VAChT assists in the transport of ACh into the presynaptic storage vesicles, and it becomes one of the main targets for the diagnosis of various neurodegenerative diseases. In our work, we aimed to understand the important structural features of the PET imaging agents required for their binding with VAChT. This was done by feature selection using a Genetic Algorithm followed by the Best Subset Selection method and developing a Partial Least Squares- based 2D-QSAR model using the best feature combination. The developed QSAR model showed significant statistical performance and reliability. Using the features selected in the 2D-QSAR analysis, we have also performed similarity-based chemical read-across predictions and obtained encouraging external validation statistics. Further, we have also performed molecular docking analysis to understand the molecular interactions occurring between the PET imaging agents and the VAChT receptor. The molecular docking results were correlated with the QSAR features for a better understanding of the molecular interactions. This research serves to fulfill the experimental data gap, highlighting the applicability of computational methods in the PET imaging agents’ binding affinity prediction.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
According to World Health Organisation (WHO), currently, more than 55 million people live with dementia worldwide, and there are more than 10 million cases new cases every year. Dementia is characterized by the loss or decline in memory or other cognitive impairment commonly observed in neurodegenerative disorders like Alzheimer’s disease (AD), Parkinson’s disease (PD), schizophrenia, and Down’s syndrome. The severity of dementia-associated cognitive dysfunction is connected with the loss of cholinergic synaptic elements in the cortex and subcortical regions of the brain (Bohnen and Albin 2011; Hampel et al. 2018). Cholinergic neurons are accountable for synaptic transmission as well as neuronal modulation in various regions of the central and peripheral nervous systems. Cholinergic neurotransmission controls cognitive functions including learning and memory. Acetylcholine (ACh) is one of the main neurotransmitters secreted by cholinergic neurons to perform a plethora of physiological functions (Prado et al. 2013). ACh is produced at the nerve terminals from acetyl coenzyme A (acetyl CoA) and choline by Choline acetyltransferase (ChAT) enzyme. The neurotransmitters are then transported and stored in synaptic vesicles by transporters called vesicular ACh transporters (VAChTs), before being released in the synaptic cleft (Amenta and Tayebati 2008). Neurodegenerative diseases have common events of cholinergic impairment. Thus, radiolabeling of these vesicular transporters would provide a presynaptic marker of cholinergic innervation. The depletion in ChAT and AChE levels, occurring in several neurodegenerative diseases, are potential measuring targets for these imaging agents (Bergmann et al. 1978; Mountjoy 1986; Mountjoy et al. 1984) (Fig. 1). Imaging cholinergic neurotransmission in vivo with positron emission tomography (PET) provides noteworthy information about disease progression.
PET imaging of vesicular acetylcholine transporter to study its role in presynaptic cholinergic innervations. VAChT helps in the transport of Acetylcholine (ACh), the essential neurotransmitter regulating AD and PD, through the synaptic vesicles. PET imaging technology helps in the diagnosis of the increase or decrease in the number of VAChT receptors. Mechanism of VAChT: In the cytoplasm of nerve endings, ACh is synthesized by the enzyme ChAT, and then it is loaded into synaptic vesicles by VAChT. Upon any nerve impulse, vesicles fuse to the plasma membrane and release the neurotransmitter ACh. (AD: Alzheimer’s disease; PD: Parkinson’s disease; CoA: Coenzyme A; ChAT: Choline acetyltransferase)
Radio imaging of presynaptic VAChT was first done using 18F-fluoroethoxybenzovesamicol (18F-FEOBV), a PET ligand, which was later successfully rendered into clinical application. Vesamicol (2-(4-phenylpiperidino)cyclohexanol) was reported to bind to VAChT and is considered to be a useful lead for developing new PET imaging agents for mapping cholinergic signaling in vivo (Giboureau et al. 2012). Kitamura et al. (2016) found that o-methyl-trans-decalinvesamicol (OMDV) demonstrated a high binding affinity and selectivity for VAChT and can be used in the early diagnosis of Alzheimer’s disease (AD). [11C]OMDV was synthesized and investigated as a new PET ligand for VAChT imaging through in vivo evaluation. Kilbourn et al. (2009) used (2R,3R)-5-[18F]fluoroethoxybenzovesamicol in micro PET imaging to determine the regional brain pharmacokinetics of rat and rhesus monkey brains. Horsager et al. (2022) evaluated human in vivo VAChT distribution in 13 peripheral organs using a 70 min dynamic [18F]fluoroethoxybenzovesamicol ([18F]FEOBV) PET/CT protocol. Tu et al. (2009) synthesised nine fluorine-containing VAChT inhibitors and screened them as potential PET tracers for imaging the VAChT. Earlier, imaging of AD was possible through the amyloid beta detection; however, these methods were not useful to evaluate the therapeutic efficacy of the AD treatment. The dysfunction of presynaptic cholinergic neurons is associated with loss of choline acetyl transferase (ChAT) (enzyme synthesizing ACh) and the vesicular acetylcholine transporter (VAChT) (Reinikainen et al. 1990). Thus, these internal molecules function as novel cranial molecular targets for developing new imaging agents for their detection.
Driven by the continuous search for new entities with improved properties and considerably lower toxicities, theoretical approaches are of high priority within the chemical and pharmaceutical industries. This provides a logical design of chemicals or pharmaceuticals with reduced time and cost. Quantitative structure–activity relationship (QSAR) has gained immense importance in the pharmaceutical industries as an effective tool for the predictions when experimental data is limited (Gramatica 2020). QSAR has enormous applications in medicinal chemistry, drug designing, and toxicity prediction. Another chemometric approach, similarity-based quantitative read-across (Chatterjee et al. 2022), can also be used for data gap filling. This method uses a weighted average approach to quantitatively predict similar query compounds. Read-across approach, due to its transparency, has a strong potential for providing confident predictions.
In the present research, we have strived to develop a two-dimensional QSAR model with 19 PET imaging agents acting against vesicular acetyl choline transporter. The selection of a small dataset was due to the non-availability of a larger number of experimental data. Here, QSAR modeling plays a pivotal role for providing promising predictions when data is scarce. To revalidate our predictions, we have performed leave-one-out and leave-many-out cross-validation tests. We have also performed read-across based predictions to analyze the predictive ability of the features obtained from QSAR analysis. Besides these, we also have performed molecular docking analysis to corroborate its results with QSAR analysis. Further, we have used two external datasets of PET imaging agents for their VAChT binding predictions (vide infra) using our developed 2D-QSAR model.
Materials and methods
In the present study, 2D-quantitative structure–activity relationship (2D-QSAR) models were developed for 19 positron emission tomography (PET) imaging agents targeted against presynaptic vesicular acetylcholine transporter (VAChT). VAChT assists in the transport of ACh into the presynaptic storage vesicles, and it is one of the main targets for the diagnosis of various neurodegenerative diseases.
The dataset
According to the OECD principle, dataset selection with a defined endpoint is the first essential step while developing a QSAR model. For our present work, the binding affinity (Ki) values of 19 PET imaging agents acting against vesicular acetylcholine transporter were procured from different previously published literature (Kovac et al. 2010; Tu et al. 2015, 2009). The VAChT binding affinity of the dataset compounds was assayed by the same experimental protocol of the competitive displacement of 5 nM [3H]vesamicol on homogenates of PC12 cells (Zea-Ponce et al. 2005). The binding affinity data which was expressed as Ki were converted to its negative logarithmic form (pKi). The structures obtained from different sources were then represented in MarvinSketch version 15.12.7.0 software with proper explicit hydrogen addition and aromatization. The 19 PET imaging agents used for the present study is given in Table 1. The dataset compounds obtained from three sources had some common compounds. The compound IDs in Table 1 are given in such a way so that the common compounds are not repeated.
Molecular descriptors
The molecular descriptor is a fundamental component of QSAR and other in-silico models since it formally represents a molecule's structure numerically. Descriptors provide a mathematically meaningful relationship between the molecular structure and biological activities, physicochemical and toxicological properties of chemicals (Mauri et al. 2017). Descriptors can be classified into different categories depending on the process of calculation or scheme of experimental determination or concept of the origin. For ease of interpretation, the present work involved the use of eight main types of two-dimensional (2D) descriptors, viz., E-state indices, extended topochemical atom (ETA), connectivity, constitutional, functional, 2D atom pairs, ring, atom-centered fragments and molecular property descriptors. The descriptors were calculated using alvaDesc descriptor calculator (Alvascience, alvaDesc version 2.0.6, 2021, https://www.alvascience.com). With the intention to minimize the redundant and incompetent data, inter-correlated descriptors (correlation greater than 0.95) were removed from the original descriptor pool. This resulted in a final pool of 188 descriptors which was used as input variables for QSAR modeling.
Feature selection and model development
In general, a QSAR model development involves a training set and a test for model development and validation purposes respectively. However, owing to the small number of compounds in our dataset, we did not apply the general method of data division (Király et al. 2022; Kovács et al. 2021; Rácz et al. 2021). It is natural that all the descriptors calculated through AlvaDesc will not be able describe the binding properties of the PET imaging agents. Therefore, to further reduce the data pool, we have applied the Genetic Algorithm (Sukumar et al. 2014) feature selection method to choose essential features required for binding. Further, we have executed the Best Subset Selection (available from http://dtclab.webs.com/software-tools) on the reduced pool of 12 descriptors obtained from the GA. Finally, the acquired pool of descriptors was applied to develop the final model using the partial least squares (PLS) regression (Wold et al. 2001). PLS converts the original descriptors into the new latent variable space thus lowering the dimensionality and obviating the inter-correlation among the original descriptors.
Machine learning-based read across predictions
In the current work, we have employed a machine learning-based Read-across prediction tool which relies on similarity approaches. The predictions were made using the tool Quantitative Read Across v4.0 developed by Chatterjee et al. (2022) available from https://sites.google.com/jadavpuruniversity.in/dtc-lab-software/home. The main similarity approaches involved in this tool are Euclidean distance-based similarity, Gaussian kernel function, and Laplacian kernel function-based similarity estimation. Please note that read-across does not develop any statistical model like QSAR and make predictions only based on the similarity values. Thus, this approach may be good when a limited number of source compounds is available (Banerjee and Roy 2022). For read-across predictions, we have divided the dataset into training and test sets. The prediction scheme starts with the initial optimization of hyperparameters (sigma and gamma values; distance and similarity thresholds) which requires division of the training into sub-training and sub-test sets into different combinations. This step is followed by the selection of the best setting of hyperparameters which is then applied to the original training and test sets.
Molecular docking
In this study, molecular docking was performed using the most and least active compounds from the initial dataset to identify the interaction pattern with the target. Owing to the non-availability of any protein structure for VAChT in the protein data bank, we have retrieved the predicted protein structure from the AlphaFold Protein Structure Database (available from https://alphafold.ebi.ac.uk/entry/Q16572) with the UniProt: Q16572, Source organism: Homo sapiens (Human), and AlphaFold id: AF-Q16572-F1-model_v2. AlphaFold is an artificial intelligence (AI) system established by DeepMind that predicts a protein’s three-dimensional (3D) structure from its amino acid sequence (Jumper et al. 2021; Varadi et al. 2022). We have then validated the reliability of the predicted structure using the Ramachandran plot server embedded in Biovia Discovery Studio 4.1 which represents the good quality of the model (see Fig. 2). In this study, multiple active sites at the surface of the protein were predicted using the Biovia discovery studio 4.1 client platform from the “define and edit binding site” using the module “generate active site from receptor cavities”, and the ligand was docked into each site to identify the favorable binding site (identified most favorable active site coordinate x: 16.478, y: 6.38307, Z: -15.9527, the radius of the sphere: 26). Initially, a total of sixteen binding sites were identified where the standard compound “vesamicol” was docked. It was found that vesamicol binds at core of site 1 of the protein with a good binding energy (27.572 kcal/mol) and interactions (shown later in the “Results and discussion” section). Out of other 15 sites, molecular docking failed in five docking sites and in case of the rest ten sites vesamicol did not bind at the docking site (outside the grid). Thus, site 1 was chosen for further docking analysis. Ligand preparation was performed using selected high and low active compounds by running them through the Discovery Studio platform’s ‘small-molecule module’, where several ligand conformers were formed. Each of these generated conformers was subsequently employed in the CDOCKER module for molecular docking using a CHARMm-based molecular dynamic scheme (Wu et al. 2003). The CDOCKER interaction energy parameter (kcal/mol) was examined for all receptor-ligand complexes, and the highest-scoring (more negative; hence favorable to binding) poses with only non-covalent interactions (ionic bonds, hydrophobic interactions, hydrogen bonds, etc.) were kept for future investigation.
Ramachandran plot for Vesicular acetylcholine transporter model (UniProt: Q16572, Source organism: Homo sapiens (Human), and AlphaFold id: AF-Q16572-F1-model_v2). Ramachandran plot shows 435 residues (97.098%) reside in the most favored region, 10 (2.232%) residues reside in the preferable region and only 3 (0.670%) reside in the unfavorable region
Results and discussions
The present work demonstrates the contribution of different structural attributes of PET imaging agents required for binding to and quantifying the presence of vesicular acetylcholine transporter. The main work is focused on the development of a simple 2D-QSAR model to obtain the major structural features responsible for binding. These features were further validated using the structural similarity-based read-across approach as well as molecular docking techniques.
QSAR modeling of binding affinity of PET imaging agents towards VAChT
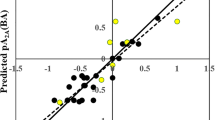
The dataset procured for this study consisted of 19 compounds. A three-descriptor partial least squares (PLS) regression model with two latent variables (LVs) was developed which could explain 71.77% of the variance. The leave-one-out cross-validated determination coefficient (i.e., \({Q}_{LOO}^{2}=0.523\)) is above the critical threshold value fulfilling the statistical reliability of the model. We have also calculated the leave-many-out squared correlation coefficient (\({Q}_{LMO(25\%)}^{2}\)), and the result obtained was above the threshold value (Roy et al. 2015). The observed versus predicted pKi (Supplementary S1) scatter plot is shown in Fig. 3. In cases, where residuals are high (Fig. 3), clearly some contributing features important for the response have remained unidentified and not included in the model. This is usual for models developed from a small data set, as due to limited variability of a particular (important) feature in the data set, the feature is not captured by the modeling algorithm. As more and more data become available, the model can be refined subsequently. However, with the available data, the presently developed model may be a good start as a tool for future predictions.
The descriptors appearing in the final PLS model are all 2D atom pair descriptors suggesting the importance of the presence of a particular atom pair in the PET tracer molecule. The 2D atom pair descriptors are mainly dependent on the topological distance between two atoms pairs. Thus, the value of the descriptors can be similar for many compounds resulting in same predicted pKi values for many dataset compounds (Fig. 3). The residual might be high in such cases, but all the compounds are inside the applicability domain of the model (vide infra). The variable importance plot (Akarachantachote et al. 2014) given in Fig. 4 shows the significance level of each descriptor toward VAChT binding affinity. The descriptor F09[N-F] was the most significant descriptor with VIP Score > 1 (VIP = 1.289) followed by F08[C-N] (VIP = 1.043) and B06[N–O] (VIP = 0.502). F09[N-F] which contributes negatively to the binding affinity, is the frequency of the N-F fragment at a topological distance 9. Compounds like 10 and 11 (Fig. 4) have nitrogen and fluorine at the topological distance 9, thereby decreasing the binding affinity towards VAChT, whereas in compounds like 21 and 23, the N-F fragment at 9 distance is absent, and the pKi values are high.
The next important 2D atom pair descriptor is F08[C-N] which denotes the frequency of C-N fragment at the topological distance 8. The positive regression coefficient indicates that with an increase in the frequency of C-N at the 8 distance, the binding affinity will increase as observed in compounds like 20 (Fig. 4), 23, and 25. These compounds have three such fragments and have high pKi values of 3.658, 3.319, and 2.700 respectively.
The least important among all the descriptors is B06[N–O] which implies the presence or absence of an N–O fragment at a topological distance 6. The negative contribution indicates that the presence of such a fragment will decrease the VAChT binding of the PET imaging agents as seen in compounds like 10 and 11 (Fig. 4). These compounds have a very low binding affinity towards (1.251 and 1.032 respectively) VAChT receptor.
The significance and validity of the developed model were further analyzed using some important PLS plots, namely, the loading plot, randomization plot, and applicability domain (AD) which are described below.
A loading plot (Fig. 5) explains the relationship between the independent variables or descriptors (X-variables) with the dependent variable or pKi values (Y-variable). The influence of the descriptors on the developed model can be recognized from the loading plot. Descriptors that are far from the plot origin (like F08[C-N] and F09[N-F]) contribute significantly more toward the binding affinity. Descriptors with different meanings appear distantly from each other in the loading plot.
Model randomization confirms that the model is not the outcome of any chance correlation (Topliss and Edwards 1979). The randomization plot determines the statistical significance and robustness of the model. Multiple models are generated during a randomization plot development by shuffling different combinations of either X-variables (X-randomization) or Y-response (Y-randomization). The Y-randomization was performed in the present study with 100 permutations for each model for random model generation. For a non-random model \({R}_{y}^{2}\) intercept should not exceed 0.3 and \({Q}_{y}^{2}\) intercept should not exceed 0.05. The randomization plot given in Fig. S1 (Supplementary File S2) shows that the developed model is non-random and robust and is suitable for prediction.
According to OECD guideline 3, a developed QSAR model should possess a defined chemical domain of applicability. AD can be interpreted as a chemical space defined by the structural information or molecular properties of the chemicals used in the model development (Gadaleta et al. 2016). Compounds present within this chemical space can only be properly predicted. In this study, the DModX (distance to model in X-space) method of AD determination (Kar et al. 2018; Vargas et al. 2018) at a 99% confidence interval (D-crit = 0.009999) was applied using SIMCA 16.0.2 software (Wu et al. 2010). DModX represents the unexplained variation (residuals), and it can be explained as the distance to the model X space corresponding to the X residuals standard deviation (Vargas et al. 2018). The DModX value of an observation i can be calculated using the formula \({S}_{i}=\sqrt{\frac{{\Sigma e}_{ik}^{2}}{(K-A)}}/\sqrt{\frac{{\Sigma e}_{k}^{2}}{(N-A-A0)((K-A)}}\), where \({e}_{ik}\) is the X-residual of the observation i and variable k, \({\Sigma e}_{k}^{2}\) is the squared sum of the residuals, N is the number of observations, \(K\) is the number of x-variables, and \(A\) is the number of latent variables, A0 is 1 if the model is centered and 0 otherwise. The DModX is asserted to be F-distributed, and thus, can be used to analyse if the observation is significantly far away from the PLS model presuming the data is normally distributed. The AD plot (Fig. 6) shows none of the compounds was an outlier. The PET compounds selected for the VAChT binding and imaging contained a basic core structure of 2-(piperidin-1-yl)cyclohexan-1-ol which is also the main core moiety of standard compound vesamicol. The QSAR model developed in the present research contains 2D-atom pair features which can predict compounds with or without the core structures as evident from the external set predictions (vide infra).
Although we developed our QSAR model from the whole set due to the limited availability of the experimental data, to further check its validity for external predictions, we have additionally split the dataset into training and test sets, and redeveloped three models with the same combination of descriptors (given in the Supplementary Section S1). The models were found to be robust and predictive.
Read-across based predictions
To explore the predictivity of the selected features used for QSAR modeling, a similarity-based read- across prediction was performed by using a group of five compounds (compound ID: 3, 11, 12, 21, and 27) as the test set (Chatterjee et al. 2022). Read-across was also previously performed on small datasets (< 20 compounds) successfully (Gajewicz et al. 2014, 2017; Gajewicz 2017a, b). In the current work, three types of similarity were measured: the Euclidean Distance-based, the Gaussian Kernel Similarity-based, and the Laplacean Kernel Similarity based predictions using Read-Across-v4.1 (https://sites.google.com/jadavpuruniversity.in/dtc-lab-software/home) tool and after hyperparameter optimization using Auto_RA_Optimizer-v1.0 tool (https://sites.google.com/jadavpuruniversity.in/dtc-lab-software/home), it was found that the external validation results obtained from quantitative Read-Across algorithm using Gaussian Kernel Similarity-based functions (\({Q}_{F1}^{2}=0.763, {Q}_{F2}^{2}=0.763, RMSE=0.414, MAE=0.331\)) was better compared to the results obtained with the other two read-across approaches (Table 2).
Molecular docking
Molecular docking must include a reasonably accurate model of energy and should be able to deal with the combinatorial complexity experienced by the molecular flexibility of the docking partners. In the present research, molecular docking studies were performed to understand the individual molecular interactions and orientation of the imaging agents occurring at the binding zone of the VAChT receptor (Fig. 7). In the present work, the protein structure for VAChT was not available in PDB, hence, we have procured the predicted protein structure from AlphaFold Protein Structure Database. The selected protein structure was further validated using the famous Ramachandran plot to improve the accuracy of prediction. From the Ramachandran plot, we found that 435 residues (97.098%) reside in the most favored region, 10 (2.232%) residues reside in the preferable region and only 3 (0.670%) reside in the unfavorable region. During docking, the physiological pH of brain was considered at which the piperidine N is protonated. The tautomers were also considered. Further, validation using the standard compound, i.e., vesamicol was performed by docking at the binding site to understand its nature of interactions. Again, both high and low-active compounds were also used for the docking study. In the case of vesamicol (compound 9), which has a moderate binding affinity (pKi = 2.261), the interaction forces include hydrogen bond interactions (both conventional and carbon-hydrogen bond interactions) and π-anion interactions. The amino acid residues engaged in vesamicol binding are Asp A:202, Asp A:483, and Ser A:480. Comparing vesamicol-VAChT binding interactions with highly active compounds like compound ID 20 (pKi = 3.658), 21 (pKi = 3.602), and 22 (pKi = 3.347), it was observed that similar interactions were also involved in their binding (Fig. 7). However, it was found that these highly active compounds were docked with higher number of interactions at their binding site with far better binding (Table 3). For compound 20, halogen (fluorine) interactions, attractive charge, π-cation, and π-alkyl interactions were active along with hydrogen bond interactions. In the case of compound 21, additional interactions include attractive charge, π-anion, and π-cation interactions. Similarly, in the case of compound 22, attractive charges, alkyl, and π-alkyl interactions were active along with conventional hydrogen bond and carbon-hydrogen bond interactions. The attractive charge interaction of Asp A:483 amino acid with the nitrogen of piperidine moiety of all three high active compounds was a noteworthy finding inferring the importance of the fragment in VAChT binding.
In the case of lower active compounds like compound 10 (pKi = 1.032) and compound 29 (pKi = 0.967) (Fig. 7), the number of molecular interactions was much less than the higher active ones (Table 3). Conventional hydrogen bond and carbon-hydrogen bond interactions were prevalent, with additional halogen and π-alkyl in the case of compound 29.
Relationship with QSAR features
From QSAR modeling, it was found that F08[C-N] is the only positively contributing descriptor. Therefore, the presence of nitrogen in the PET imaging agent is very essential for good VAChT binding. In the case of highly active compounds (compounds 20, 21, and 22) used for molecular docking, it was found that attractive charge interaction was prevalent in all three compounds which occurred between Asp A:483 amino acid with the nitrogen of piperidine moiety of the PET tracer. These two observations correlate with each other and thus it can be inferred that nitrogen (as piperidine moiety) is essential for good VAChT binding.
True external set predictions
For the analysis of the predictivity of the developed model, we have considered two PET datasets previously used by our group (De et al. 2019; De and Roy 2020) for their VAChT binding predictions (Table S1). Dataset D1 was initially used for amyloid beta imaging and dataset D2 was used for Dopamine (D2) imaging. The prediction quality was further verified using by the application of “Prediction Reliability Indicator” tool (Roy et al. 2018) available from https://dtclab.webs.com/software-tools. The prediction tool reported “Good” quality prediction for all the compounds and they were all inside the AD of the model (Supplementary Files S1 and S2). Thus, these compounds can also be considered as potential PET imaging agents for VAChT subject to experimental validation.
Conclusions
The neurotransmitter acetylcholine (ACh) plays a ubiquitous role in cognitive functions including learning and memory with widespread innervation in the cortex, subcortical structures and the cerebellum. Cholinergic receptors, transporters, or enzymes associated with many neurodegenerative diseases, including Alzheimer’s disease (AD) and Parkinson’s disease (PD), are potential imaging targets. In the present study, we have developed a 2D-QSAR model for 19 positron emission tomography (PET) imaging agents targeted against presynaptic vesicular acetylcholine transporter (VAChT). In our work, we aimed to understand the important structural features of the PET imaging agents required for their binding with VAChT. This was done by the feature selection using a Genetic Algorithm followed by the Best Subset Selection method and developing a Partial Least Squares- based 2D QSAR model using the best feature combination. The developed QSAR model showed significant statistical performance and reliability (\({R}^{2}=0.718, {Q}_{(LOO)}^{2}=0.523, {Q}_{LMO(25\%)}^{2}=0.598\)). Using the features selected in the 2D-QSAR analysis, we have also performed similarity-based chemical read-across predictions and obtained encouraging external validation statistics. From the developed QSAR model, it was found that the presence of nitrogen in the PET tracer molecule potentiates the binding affinity towards the VAChT receptor. This was further confirmed by molecular docking studies where nitrogen in the piperidine moiety produced attractive charge interaction with Asp A:483 amino acid of VAChT. In the future, this study will help in the prediction of newly developed compounds within the applicability domain of the model targeted toward VAChT.
Availability of data and material
Some of the data and materials are available in Supplementary Materials of this paper. Additional data are available from the authors on request.
Code availability
DTC Lab software tools are available from http://teqip.jdvu.ac.in/QSAR_Tools/.
References
Akarachantachote N, Saithanu K, Chadcham S, Akarachantachote N, Chadcham S, Saithanu K (2014) Cutoff threshold of variable importance in projection for variable selection. Int J Pure Appl Math 94(3):307–322
Amenta F, Tayebati SK (2008) Pathways of acetylcholine synthesis, transport and release as targets for treatment of adult-onset cognitive dysfunction. Curr Med Chem 15(5):488–498
Banerjee A, Roy K (2022) First report of q-RASAR modeling toward an approach of easy interpretability and efficient transferability. Mol Divers 26(5):2847–2862
Bergmann K, Tomlinson BE, Blessed G, Gibson PH, Perry RH (1978) Correlation of cholinergic abnormalities with senile plaques and mental test scores in senile dementia. Br Med J 2(6150):1457–1459
Bohnen NI, Albin RL (2011) The cholinergic system and Parkinson disease. Behav Brain Res 221(2):564–573
Chatterjee M, Banerjee A, De P, Gajewicz-Skretna A, Roy K (2022) A novel quantitative read-across tool designed purposefully to fill the existing gaps in nanosafety data. Environ Sci Nano 9(1):189–203
De P, Roy K (2020) QSAR modeling of PET imaging agents for the diagnosis of Parkinson’s disease targeting dopamine receptor. Theor Chem Acc 139(12):176
De P, Bhattacharyya D, Roy K (2019) Application of multilayered strategy for variable selection in QSAR modeling of PET and SPECT imaging agents as diagnostic agents for Alzheimer’s disease. Struct Chem 30(6):2429–2445
Gadaleta D, Mangiatordi GF, Catto M, Carotti A, Nicolotti O (2016) Applicability domain for QSAR models: where theory meets reality. IJQSPR 1(1):45–63
Gajewicz A (2017a) What if the number of nanotoxicity data is too small for developing predictive Nano-QSAR models? An alternative read-across based approach for filling data gaps. Nanoscale 9(24):8435–8448. https://doi.org/10.1039/C7NR02211E
Gajewicz A (2017b) Development of valuable predictive read-across models based on “real-life”(sparse) nanotoxicity data. Environ Sci Nano 4(6):1389–1403
Gajewicz A, Cronin MT, Rasulev B, Leszczynski J, Puzyn T (2014) Novel approach for efficient predictions properties of large pool of nanomaterials based on limited set of species: nano-read-across. Nanotechnology 26(1):015701. https://doi.org/10.1088/0957-4484/26/1/015701
Gajewicz A, Jagiello K, Cronin MTD, Leszczynski J, Puzyn T (2017) Addressing a bottle neck for regulation of nanomaterials: quantitative read-across (Nano-QRA) algorithm for cases when only limited data is available. Environ Sci Nano 4(2):346–358. https://doi.org/10.1039/C6EN00399K
Giboureau N, Mat Som I, Boucher-Arnold A, Guilloteau D, Kassiou M (2012) PET radioligands for the vesicular acetylcholine transporter (VAChT). Curr Top Med Chem 10(15):1569–1583
Gramatica P (2020) Principles of QSAR modeling: comments and suggestions from personal experience. IJQSPR 5(3):61–97
Hampel H, Mesulam MM, Cuello AC, Farlow MR, Giacobini E, Grossberg GT et al (2018) The cholinergic system in the pathophysiology and treatment of Alzheimer’s disease. Brain 141(7):1917–1933
Horsager J, Okkels N, Van Den Berge N, Jacobsen J, Schact A, Munk OL et al (2022) In vivo vesicular acetylcholine transporter density in human peripheral organs: an [18F]FEOBV PET/CT study. EJNMMI Res 12(1):1–11
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O et al (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596(7873):583–589
Kar S, Roy K, Leszczynski J (2018) Applicability domain: a step toward confident predictions and decidability for QSAR modeling. In: Computational toxicology: methods and protocols, pp 141–169
Kilbourn MR, Hockley B, Lee L, Sherman P, Quesada C, Frey KA et al (2009) Positron emission tomography imaging of (2R,3R)-5-[18F]fluoroethoxybenzovesamicol in rat and monkey brain: a radioligand for the vesicular acetylcholine transporter. Nucl Med Biol 36(5):489–493
Király P, Kiss R, Kovács D, Ballaj A, Tóth G (2022) The relevance of goodness-of-fit, robustness and prediction validation categories of OECD-QSAR principles with respect to sample size and model type. Mol Inform 41(11):2200072
Kitamura Y, Kozaka T, Miwa D, Uno I, Azim ul MA, Ogawa K et al (2016) Synthesis and evaluation of a new vesamicol analog o-[11C]methyl-trans-decalinvesamicol as a PET ligand for the vesicular acetylcholine transporter. Ann Nucl Med 30(2):122–129
Kovac M, Mavel S, Deuther-Conrad W, Méheux N, Glöckner J, Wenzel B et al (2010) 3D QSAR study, synthesis, and in vitro evaluation of (+)-5-FBVM as potential PET radioligand for the vesicular acetylcholine transporter (VAChT). Bioorg Med Chem 18(21):7659–7667
Kovács D, Király P, Tóth G (2021) Sample-size dependence of validation parameters in linear regression models and in QSAR. SAR QSAR Environ Res 32(4):247–268
Mauri A, Consonni V, Todeschini R (2017) Molecular descriptors. In: Handb Comput Chem. pp 2065–93
Mountjoy CQ (1986) Correlations between neuropathological and neurochemical changes. Br Med Bull 42(1):81–85
Mountjoy CQ, Rossor MN, Iversen LL, Roth M (1984) Correlation of cortical cholinergic and GABA deficits with quantitative neuropathological findings in senile dementia. Brain 107(2):507–518
Prado VF, Roy A, Kolisnyk B, Gros R, Prado MAM (2013) Regulation of cholinergic activity by the vesicular acetylcholine transporter. Biochem J 450(2):265–274
Rácz A, Bajusz D, Héberger K (2021) Effect of dataset size and train/test split ratios in QSAR/QSPR multiclass classification. Molecules 26(4):1111
Reinikainen KJ, Soininen H, Riekkinen PJ (1990) Neurotransmitter changes in alzheimer’s disease: implications to diagnostics and therapy. J Neurosci Res 27(4):576–586
Roy K, Kar S, Das RN (2015) Statistical Methods in QSAR/QSPR. A Prim. QSAR/QSPR Model. Springer, Cham pp 37–59
Roy K, Ambure P, Kar S (2018) How precise are our quantitative structure-activity relationship derived predictions for new query chemicals? ACS Omega 3(9):11392–11406
Sukumar N, Prabhu G, Saha P (2014) Applications of genetic algorithms in QSAR/QSPR modeling. In: Appl Metaheuristics Process Engg. pp 315–24
Topliss JG, Edwards RP (1979) Chance factors in studies of quantitative structure-activity relationships. J Med Chem 22(10):1238–1244
Tu Z, Efange SMN, Xu J, Li S, Jones LA, Parsons SM et al (2009) Synthesis and in vitro and in vivo evaluation of 18F-labeled positron emission tomography (PET) ligands for imaging the vesicular acetylcholine transporter. J Med Chem 52(5):1358–1369
Tu Z, Zhang X, Jin H, Yue X, Padakanti PK, Yu L et al (2015) Synthesis and biological characterization of a promising F-18 PET tracer for vesicular acetylcholine transporter. Bioorg Med Chem 23(15):4699–4709
Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G et al (2022) AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 50(D1):D439–D444
Vargas JM, Nielsen S, Cárdenas V, Gonzalez A, Aymat EY et al (2018) Process analytical technology in continuous manufacturing of a commercial pharmaceutical product. Int J Pharm 538(1–2):167–178
Wold S, Sjöström M, Eriksson L (2001) PLS-regression: a basic tool of chemometrics. Chemom Intell Lab Syst 58(2):109–130
Wu G, Robertson DH, Brooks CL, Vieth M (2003) Detailed analysis of grid-based molecular docking: a case study of CDOCKER—A CHARMm-based MD docking algorithm. J Comput Chem 24(13):1549–1562
Wu Z, Li D, Meng J, Wang J (2010) Introduction to SIMCA-P and its application. In: Handbook of Partial Least Squares: Concepts, Methods and Applications. pp 757–774
Zea-Ponce Y, Mavel S, Assaad T, Kruse SE, Parsons SM, Emond P et al (2005) Synthesis and in vitro evaluation of new benzovesamicol analogues as potential imaging probes for the vesicular acetylcholine transporter. Bioorganic Med Chem 13(3):745–753
Acknowledgements
PD thanks Indian Council of Medical Research, New Delhi, for awarding with a Senior Research Fellowship (file no. ISRM/11(61)/2019). Financial assistance for SERB, New Delhi under the MATRICS scheme (MTR/2019/000008) is thankfully acknowledged.
Funding
Funding is provided by ICMR, New Delhi (PD), SERB, New Delhi (KR).
Author information
Authors and Affiliations
Contributions
PD: computation, analysis, and initial draft. KR: conceptualization, editing, supervision, and funding.
Corresponding author
Ethics declarations
Conflict of interest
None.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
De, P., Roy, K. Computational modeling of PET imaging agents for vesicular acetylcholine transporter (VAChT) protein binding affinity: application of 2D-QSAR modeling and molecular docking techniques. In Silico Pharmacol. 11, 9 (2023). https://doi.org/10.1007/s40203-023-00146-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s40203-023-00146-4