Abstract
Purpose
Despite vast improvements that have been made in the treatment of children with acute lymphoblastic leukemia (ALL), the majority of infant ALL patients (~80 %, < 1 year of age) that carry a chromosomal translocation involving the mixed lineage leukemia (MLL) gene shows a poor response to chemotherapeutic drugs, especially glucocorticoids (GCs), which are essential components of all current treatment regimens. Although addressed in several studies, the mechanism(s) underlying this phenomenon have remained largely unknown. A major drawback of most previous studies is their primary focus on individual genes, thereby neglecting the putative significance of inter-gene correlations. Here, we aimed at studying GC resistance in MLL-rearranged infant ALL patients by inferring an associated module of genes using co-expression network analysis. The implications of newly identified candidate genes with associations to other well-known relevant genes from the same module, or with associations to known transcription factor or microRNA interactions, were substantiated using literature data.
Methods
A weighted gene co-expression network was constructed to identify gene modules associated with GC resistance in MLL-rearranged infant ALL patients. Significant gene ontology (GO) terms and signaling pathways enriched in relevant modules were used to provide guidance towards which module(s) consisted of promising candidates suitable for further analysis.
Results
Through gene co-expression network analysis a novel set of genes (module) related to GC-resistance was identified. The presence in this module of the S100 and ANXA genes, both well-known biomarkers for GC resistance in MLL-rearranged infant ALL, supports its validity. Subsequent gene set net correlation analyses of the novel module provided further support for its validity by showing that the S100 and ANXA genes act as ‘hub’ genes with potentially major regulatory roles in GC sensitivity, but having lost this role in the GC resistant phenotype. The detected module implicates new genes as being candidates for further analysis through associations with known GC resistance-related genes.
Conclusions
From our data we conclude that available systems biology approaches can be employed to detect new candidate genes that may provide further insights into drug resistance of MLL-rearranged infant ALL cases. Such approaches complement conventional gene-wise approaches by taking putative functional interactions between genes into account.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
During the last four decades major improvements in the treatment of childhood acute lymphoblastic leukemia (ALL) have been made, resulting in current disease-free survival rates of approximately 85 % [1]. Unfortunately, the benefits of these improvements are not distributed equally. A majority (~80 %) of infant (< 1 year of age) ALL patients carry a chromosomal translocation involving the mixed lineage leukemia (MLL) gene, and only ~40 % of these patients have a chance of long-term survival, depending on the treatment protocol used [2, 3]. Experimental studies have shown that cellular resistance to chemotherapeutic drugs is the main obstacle for achieving a successful treatment outcome. In particular, resistance to prednisolone, an essential glucocorticoid (GC) drug administered in most currently used ALL treatment regimes, limits the chances of a successful treatment outcome [4]. A poor response to prednisolone has been observed in 30 % of MLL-rearranged infant ALL patients, while this rate drops to <10 % in non-infant (> 1 year of age) pediatric ALL patients [5]. An in-depth understanding of the mechanisms that underlie prednisolone resistance in childhood ALL is required to develop mitigation measures for this phenomenon, to achieve a full treatment potential, and to increase a successful outcome rate.
Previous studies have indicated that a high expression of the myeloid cell leukemia 1 (MCL1) gene, a member of the anti-apoptotic BCL-2 family, is a common factor involved in resistance to prednisolone in non-infant and infant MLL-rearranged ALL [6, 7]. It has been found that the MCL1 activity in pediatric ALL can be controlled by rapamycin and that the sensitivity to GCs can be restored by this drug [8]. Rapamycin has, however, been shown to be ineffective in the treatment of MLL-rearranged infant ALL, which indicates that additional factors must be responsible for GC resistance in MLL-rearranged infant ALL patients. It has also been shown that pan-BCL-2 inhibitors such as gossypol and AT-101 can induce prednisolone sensitivity in MLL-rearranged infant ALL patients by increasing the levels of pro-apoptotic proteins such as BIM, BID, BAD and NOXA, rather than by decreasing the levels of pro-survival proteins such as BCL-2, BCL-X or MCL1 [9]. In addition, it was found that free calcium in the cytosol may be associated with GC-induced lymphocytolysis [10] and that high levels of S100 calcium-binding protein family members such as S100A8 and S100A9 may be correlated with prednisolone resistance in MLL-rearranged infant ALL patients [11]. Interestingly, it was found that the expression levels of S100A8 and S100A9 in primary leukemic cells are up-regulated in infant ALL compared to non-infant ALL patients [12]. This relationship was subsequently found to be true for more members of the S100 family of proteins, including S100A4 and S100A8 and S100A12 [11]. Recently, a high level of another calcium-binding protein, annexin A2 (ANXA2), has been found to be associated with GC resistance in MLL-rearranged infant ALL cases as well [13]. The underlying mechanism of this latter effect includes Src kinase-induced phosphorylation of ANXA2, which requires the adapter protein p11 that is encoded by the S100A10 gene. Knockdown of either the ANXA2 or S100A10 genes was found to lead to inhibition of this process and, concomitantly, to an increased sensitization to prednisolone [13]. A similar sensitization to prednisolone of MLL-rearranged ALL cells could be elicited by LY294002, an inhibitor of phosphatidylinositol 3-kinase (PI3K) [14]. This latter sensitization was found to be accompanied by down-regulation of several genes, including S100A2.
Several studies on GC resistance in MLL-rearranged infant ALL cases have revealed potential targets that may inhibit this phenomenon, but its underlying mechanism(s) are still largely unknown. One major drawback of most of the previous studies may be the ignorance of putative relevant inter-gene correlations. Although the identification of differentially expressed genes may provide clues for subsequent functional analyses, gene co-expression patterns may provide leads that may have been overlooked by previous one-dimensional differential gene expression approaches. Relevant gene sets can either be deduced from existing well-characterized gene sets or be inferred from putative functional interactions between genes. Multiple studies have shown that gene networks can be used to identify new biomarkers related to complex diseases such as cancer [15–23]. Co-expression networks are the most commonly used gene networks that model correlations between gene expression profiles. In these networks, highly co-expressed (correlated) genes are grouped into co-expression modules (gene sets) for further analysis using, for example, enrichment for gene ontology (GO) or multivariate statistical methods. The subsequent identification of key regulators (‘hubs’) in these modules may provide useful information, as they may be related to significant biological functions.
Here, we set out to examine GC resistance in MLL-rearranged infant ALL patients and show that co-expression network analysis allows the detection of gene modules that are associated with GC resistance. We found that one module encompassed multiple genes, including several S100 and ANXA genes, that have previously been associated with GC resistance in MLL-rearranged infant ALL patients. We also set out to examine relationships among consensus modules (i.e., modules that are conserved in both GC sensitive and GC resistant co-expression networks), by constructing eigengene networks for each group separately and by performing differential eigengene network analyses across groups. No significant rewiring among consensus modules was observed between eigengene networks, and inter-modular relationships were found to be preserved between the GC sensitive and GC resistant groups.
2 Materials and methods
2.1 Datasets and preprocessing
The microarray datasets used were retrieved from the NCBI gene expression omnibus with GEO Series accession numbers GSE32962 [11] and GSE19143 [7]. Dataset GSE32962 consists of gene expression profiles of 43 infants (< 1 year of age) with MLL-rearranged ALL. The infants could be classified into two groups based on prednisolone sensitivity status, i.e., 19 prednisolone-sensitive and 24 prednisolone-resistant. Dataset GSE19143 consists of gene expression profiles of both infant (< 1 year of age) and non-infant (> 1 year of age) ALL cases that could again be classified into prednisolone-sensitive and prednisolone-resistant groups. Only the infant ALL samples of this latter dataset, including 13 sensitive and 12 resistant samples, were used in this study. A full description of the microarray datasets used in this study is provided in Table S1 (Supplementary Materials). The downloading of raw CEL files and the conversion of probe-level data into expression abundances were carried out using the GEOquery [24] and Affymetrix [25] R software packages. Expression data were normalized using the Variance Stabilizing Normalization (VSN) method, as implemented in the R VSN package [26]. Probe identifiers were mapped to gene identifiers using an annotation file released by Affymetrix for each platform. After these preprocessing steps, we obtained two expression matrices in which the rows represent genes and the columns represent samples.
2.2 Construction of co-expression networks
A weighted gene co-expression network was constructed to reveal correlations between gene expression profiles, as implemented in package WGCNA [27]. We calculated pairwise gene correlations using the biweight midcorrelation (bicor) measure [28] based on its robustness to outliers in comparison to the Pearson correlation coefficient. The two common types of co-expression networks were designated as signed network and unsigned network. While the former uses the correlation coefficient value, the latter uses the absolute value and hence, information regarding gene activation and repression is ignored. In this study, we constructed signed hybrid networks by removing all negative correlation values and preserving only links associated with positive correlations. Next, the non-negative similarity matrix of the signed hybrid network was converted to a weighted adjacency matrix using a soft-thresholding approach.
To compute the adjacency matrix as described above, we used the adjacency function of package WGCNA with the following parameters (corFnc = “bicor”, type = “signed hybrid”, corOptions = “maxPOutliers = 0.1”). The power parameter β , which satisfies the scale-free topology property of the co-expression network, was estimated for the input of the adjacency function by the pickSoftThreshold function of package WGCNA with the parameters stated above.
2.3 Module identification
Genes were grouped into proximity-based modules using a generalized version of the topological overlap measure (TOM) for weighted networks [29], as implemented in the TOMsimilarity function of package WGCNA. Every element of the topological overlap matrix represents the similarity of two genes in terms of shared neighbors in the co-expression network. The dissimilarity matrix 1-TOM was used as an input parameter to the average hierarchical clustering algorithm for module identification. For average hierarchical clustering we used the flashClust function with parameter method = “average”, and modules were identified from the resulting dendrogram using function cutreeDynamic with parameters deepSplit = 2 and minClusterSize = 30, and other parameters set as default.
After the detection of gene modules, the gene expression profiles of each module were summarized by the module eigengenes [27]. A module eigengene (ME) is defined for a given module as the first principle component of the expression data of that module. We computed the module eigengenes for all modules using the function moduleEigengenes from package WGCNA. To prioritize genes within modules, we calculated the module eigengene-based connectivity and intra-modular connectivity measures [27]. In the module eigengene-based connectivity, the similarity between the expression profile of gene i and the eigengene of a module q is calculated as
where x i refers to the expression profile of gene i and ME q to the expression profile of the eigengene of module q. The membership value of a gene to a given module was calculated using the ME-based connectivity, and the statistical significance of this value was provided by a simple correlation test p-value. The second connectivity measure is the intra-modular connectivity, which was calculated for the genes of each module using the intramodularConnectivity function. Genes with a strong connectivity to other genes in their respective modules, called ‘hub’ genes, were identified as key regulators in a given module.
2.4 Differential co-expression analysis
Two different statistical tests on the resulting modules were performed to identify modules that are differentially co-expressed between two groups of GC sensitive and GC resistant samples. The first test entails gene set co-expression analysis (GSCA) [30], by which the Euclidean distance between the pairwise correlation vectors of two groups is calculated to determine the extent of differential co-expression for a given gene set. Pairwise correlations within a gene set, separately measured for each group of samples, form two distinct correlation vectors. A permutation test was used to assess the significance of the difference. The second test entails gene set net correlations analysis (GSNCA) [31] by which multivariate changes in the co-expression structures for the identified gene modules between two conditions are calculated. GSNCA assigns a normalized version of the eigenvector of the correlation matrix corresponding to the largest eigenvalue as a weight vector for the genes in the gene set under each condition. The difference between the weight vectors of two conditions was quantified using a L1-norm distance measure, and to assess the significance of the difference a p-value was calculated using a permutation test. The GSNCA method is part of the Bioconductor GSAR package. In this study, we considered genes with \( {k}_{ME}^q>0.5 \) as gene sets related to the modules, and applied the two methods described above to all gene sets.
2.5 Consensus network analysis
To study relationships between the modules of co-expression networks across two conditions we used package WGCNA for consensus network analysis [32]. In doing so, TOM matrices for the GC sensitive (TOM sensitive ) and GC resistant (TOM resistant ) samples were constructed separately. To construct a consensus network among both conditions, both TOM sensitive and TOM resistant were combined into a new consensus_TOM matrix in the following manner:
In the consensus network analysis, the dissimilarity measure 1-Consensus_TOM was used to identify consensus modules. After merging close modules, an eigengene network was constructed for each group of samples by measuring correlations among module eigengenes in each group. In the eigengene network, each node is an eigengene which relates to a consensus module and the correlation between each pair of eigengenes is represented by a weighted link. To examine whether there was any rewiring in the co-expression networks at the module level, the links of the eigengene networks were compared between two groups using the preservation measure implemented in the plotEigengeneNetworks function of the WGCNA package. Through this function correlations between the eigengenes of each pair of modules between the sensitive and the resistant eigengene networks are compared. A strong preservation of each link across the two phenotypes is indicated by a high preservation measure, while a low value indicates a change in the relationship across the two phenotypes. Furthermore, to assess the amount of similarity between eigengene networks, the density measure (D) was calculated by comparing correlations of all pairs of eigengenes across the two phenotypes. A high D value depicts a strong preservation between two networks.
2.6 Transcription factor and miRNA target data analysis for module validation
Glucocorticoids are used as chemotherapeutic agents to induce apoptosis through two mechanisms, i.e., (1) activating the transcription of death-inducing genes or (2) inhibiting the transcription of survival genes through the repression of transcription factor activities [33]. We assessed genes in GC-associated modules in the context of these two mechanisms using transcription factor (TF)-target interaction data from the Transcriptional Regulatory Relationships Unraveled by Sentence-based Test mining (TRRUST) database [34]. TRRUST is a literature-curated database with data deduced from millions of Medline abstracts. Its current version includes 8015 interactions between 748 TFs and 1975 non-TF targets, and provides information about their modes of regulation, i.e., activation or repression.
MicroRNAs (miRNAs) act as post-translational regulators of gene expression. They can inhibit translation by binding to the corresponding target mRNAs. Previously, silencing of miRNAs via methylation in MLL-rearranged infant ALL cases has been studied, and 11 miRNAs were found to be down-regulated as a consequence of hypermethylation of their promoters in t(4;11)-positive (MLL-rearranged) infant ALL samples compared to normal bone marrow samples [35]. The down-regulation of at least some of these miRNAs may contribute to over-expression of their corresponding target genes. We examined such gene expression patterns in a GC-associated module using the miRTarBase database [36]. The miRTarBase database consists of experimentally validated miRNA-target interactions and contains 324,219 interactions between 2619 miRNAs and 12,738 targets. Only miRNA-target interactions validated by strong experimental evidence, i.e., reporter or Western blot assays, were considered in this study.
3 Results
We present modules that are revealed by weighted gene co-expression network analysis and highlight one module that is highly associated with GC (prednisolone) resistance in MLL-rearranged infant ALL patients. The robustness of the modules is underscored by our finding that the inter-module relationships exhibit no significant changes between two conditions (GC sensitive versus GC resistant), and that the modules are preserved in two independent infant ALL datasets. We corroborate the relevance of the highlighted module to prednisolone resistance using differential gene expression analysis and, in addition, assessing its co-expression structure using two co-expression analysis methods. Finally, we perform literature-based functional enrichment analyses for the genes in the highlighted module. The accumulated results establish a single module associated with prednisolone resistance, including multiple genes already known to be associated with prednisolone resistance.
3.1 Network construction reveals 14 co-expression modules
Independent of the prednisolone resistance status, the preprocessed gene expression profiles of MLL-rearranged infant ALL samples (i.e., GSE32962 dataset) were used to construct a weighted gene co-expression network. To this end, we chose the soft-thresholding approach with a soft power parameter β = 8 to obtain a scale-free topology fit index > 0.8 (Fig. S1, Supplementary Materials). Genes with a stable expression across two phenotypes were discarded, since they offer no or little distinction. From the ~19,000 genes in the dataset, the 5000 most varying genes were selected for network construction. The expression of these genes varied from that of S100A8 (average expression = 11.86, variance = 5.26) to that of CAD (average expression = 7.93, variance = 0.07). Next, the co-expression network was clustered into gene modules, after which a unique color was assigned to each module as shown in the upper color band of the unmerged modules in Fig. S2 (Supplementary Materials). Since some modules were not sufficiently distinct and their assignments were suboptimal, we merged close modules and subsequently adjusted the module assignments using the moduleMergeUsingKME function of package WGCNA [27]. These merged modules are indicated by the lower color bands in Fig. S2 (see Supplementary Materials). After merging, the number of modules decreased from 23 to 14, with module sizes ranging from 52 genes (light-yellow module) to 639 genes (turquoise module). We grouped 2343 genes that could not be assigned to any of the 14 modules (background genes) into one module (grey). This module was ignored in the following analyses.
3.2 Two co-expression modules are related to prednisolone resistance
Correlations between the expression levels of module eigengenes and prednisolone resistance status were calculated to identify the most significant associations. We found that 2 out of the 14 modules were significantly (FDR corrected p-value < 0.01) associated with prednisolone resistance in the MLL-rearranged infant ALL samples. One of these modules (green-yellow) contains 127 genes and a second module (brown) contains 204 genes that were positively correlated with prednisolone resistance (correlations of 0.56 and 0.51, respectively). The genes of these modules were found to be over-expressed in the prednisolone resistant ALL samples compared to the prednisolone sensitive ALL samples. The associations of all modules with prednisolone resistance are depicted in Table S2 (Supplementary Materials).
Genes in module q were found to be associated with its representative eigengene with different module membership (MM) levels, quantified by \( {k}_{ME}^q \). To identify genes associated with prednisolone resistance, we defined the gene significance (GS) measure as a Pearson correlation of gene expression with prednisolone resistance status. By doing so, we examined the correlation between MM and GS values for the genes of each module. Among the 14 modules, we observed a significant positive Pearson correlation between the MM and the GS values in the green-yellow module (correlation = 0.67, FDR corrected p-value < 0.001) and in the brown module (correlation = 0.49, FDR corrected p-value < 0.001). In Fig. S3 (Supplementary Materials, panels a and b) scatterplots are shown between the MM and GS values for both the green-yellow and the brown modules. Genes significantly associated with the prednisolone resistance status were often the most significant members of the modules associated with the prednisolone resistance status.
3.3 Prednisolone resistance module validation by differential gene expression
To further validate the modules relevant to prednisolone resistance, we set out to compare the module-centric results from the previous section with those obtained by conventional differential gene expression analysis. To this end we calculated the log2 expression fold change (\( { \log}_2^{FC} \)) using package Limma [37]. The fold change for a gene refers to the average expression ratio in the resistant samples compared to the sensitive samples. In both the green-yellow and brown modules, a significant correlation (FDR corrected p-value < 0.001) was observed between the module eigengene-based connectivity \( \left({k}_{ME}^q\right) \) and the \( { \log}_2^{FC} \). Scatterplots of \( {k}_{ME}^q \)versus \( { \log}_2^{FC} \) for the green-yellow and brown modules are depicted in Fig. S4 (Supplementary Materials). This figure shows that significantly over-expressed genes in the resistant samples have the highest level of membership to the green-yellow module. Most of the genes with \( { \log}_2^{FC} \)> 1 (i.e., 15 out of 20 genes) are located in the green-yellow module with \( {k}_{ME}^{green- yellow}>0.5 \).
In order to additionally verify the association between the resulting modules and the prednisolone resistance status using conventional differential gene expression analysis, we compared modules by their average absolute moderated t-statistic obtained from package Limma [37] as an alternative module significance measure. In accordance with the above results, we found that the green-yellow module has the highest module significance compared to the other modules (Fig. 1).
3.4 Module preservation using an independent data set
Next, a second dataset (i.e., the infant samples of the GSE19143 dataset) was used to test whether the modules resulting from the first dataset (GSE32962) can be replicated, and whether the co-expression module definition is robust across both datasets. To this end, we first applied the method reported in [22] to assess the comparability among the datasets. We found that the correlation of gene expression levels was positive and high (cor = 0.84, p-value < 1e-200) and that also the gene connectivity was high between the datasets (cor = 0.58, p-value < 1e-200) (Fig. S5, Supplementary Materials). This result indicates that the datasets are suitable for module preservation analysis. We selected 3858 genes of the second dataset that were also present among the 5000 most varying genes of the first dataset for repeating the analysis. After construction of the co-expression network using the selected genes of the second dataset, genes were assigned to modules based on the module assignment scheme in the first dataset. To assess the module preservation quantitatively, the Z-statistic was calculated for each module separately using the modulePreservation function of package WGCNA [27]. For each module, a moderate preservation in the second dataset can be concluded when 2 ≤ Z-statistic ≤ 10, while a Z-statistic > 10 implies a strong preservation [38]. Figure S6 (Supplementary Materials) shows that 5 modules had 5 ≤ Z-statistic ≤ 10 and that 9 modules had a Z-statistic > 10, indicating that some modules are moderately preserved between the two datasets while others are strongly preserved.
3.5 Differential co-expression analysis reveals hub genes
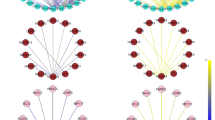
In agreement with our above findings using the module-centric approach, we found that also gene set co-expression analysis (GSCA) and gene set net correlation analysis (GSNCA) identified the green-yellow gene set as exhibiting both a differential pairwise correlation as well as a differential correlation structure between the GC resistant and GC sensitive samples. Out of the 14 modules, only the green-yellow module was identified by both GSCA and GSNCA at a significance level of 0.001. The most highly correlated genes in both the GC sensitive and GC resistant samples, identified using the minimum spanning tree-2 (MST2) structure implemented in the plotMST2.pathway function of package GSAR, are depicted in Fig. 2. The MST2 of a correlation network is formed by combining the first and second MSTs and highlights the minimal set of essential links (highest correlations) among genes in the co-expression network (see [31] for more details). The MST is defined as the acyclic tree with the shortest links (highest correlations) that connect all genes in the co-expression network. In the MST2 structure, the degree of the highly connected genes is relatively high, and they are placed in central positions. In contrast, low degree genes are placed in non-central positions in the MST2 structure [31]. Node colors indicate the value of the weight factor (w) assigned to each gene to reflect its average correlation with all other genes in the gene set. The MST2 plot for the GC sensitive samples shows that several S100 genes (S100P, S100A8, S100A9 and S100A12) are relatively highly correlated among themselves, as well as with many other genes in this gene set (i.e., hub genes). This pattern is lost in the GC resistant samples in which their w-values are reduced (especially for S100A12). This pattern likely indicates a regulatory role for the S100 genes in the GC sensitive phenotype that is lost in the GC resistant phenotype. Although the four S100 genes lost their high w-values in the GC resistant phenotype, they remained close to each other in the MST2 structure in the GC resistant phenotype, indicating that the correlations among the genes themselves were not entirely lost but rather reduced. We found, however, that their correlations with other genes in the gene set were diminished. In addition, we found that the MST2 in the GC sensitive phenotype revealed an overall better clustering, i.e., functional link between genes, than that in the GC resistant phenotype. These observations are in agreement with previous findings regarding the S100 gene family [11].
3.6 Functional enrichment analysis reveals enrichment for GC-related GO terms
The Database for Annotation Visualization and Integrated Discovery (DAVID) tool [39] was used to perform a functional enrichment analysis of the genes within the green-yellow module with a \( {k}_{ME}^{green- yellow}>0.5 \). The significantly enriched gene ontology (GO) terms for the genes within the green-yellow module are listed in Table 1. This functional enrichment analysis revealed that the module is significantly enriched for several GO terms, including defense response, inflammatory response, response to stress, response to wounding and response to stimulus (FDR < 1e-5). Other highly enriched GO terms include immune system process, immune response, response to another organism, regulation of cytokine production, regulation of immune system process, regulation of tumor necrosis factor production and phagocytosis (Table 1). In addition, we found that signaling in the immune system and hemostasis were among the most important REACTOME pathways highly enriched in the green-yellow module (p-value < 0.001). Some of the KEGG pathways in the module are also of particular interest, including the hematopoietic cell lineage (p-value < 0.001) and the leukocyte trans-endothelial migration (p-value = 0.003) pathways, as well as the inflammation mediated by chemokine and cytokine signaling pathway (p-value < 0.001) from the Panther pathways (Table S3, Supplementary Materials). Prednisolone is known to have immunosuppressive and anti-inflammatory effects, and the enrichment for several immune and inflammatory response gene sets and related GO terms in the green-yellow module further underscores its relevance to prednisolone resistance in MLL-rearranged infant ALL patients.
3.7 Consensus network analysis reveals preserved inter-modular relationships
Following the construction of the consensus network, as described in the Materials and methods section, consensus modules were detected in both the GC sensitive and GC resistant samples. After merging modules with eigengenes correlations > 0.8 (ignoring the grey module), the number of consensus modules decreased from 20 to 11. Next, the eigengene network was defined across the consensus modules for each group separately, and the links of eigengene networks were compared to identify which inter-modular relationships were rewired between the two groups. Through preservation heatmap analysis, we found that the correlations between all consensus eigengenes were strongly preserved across the two groups (Fig. 3), and that the overall preservation was 0.89 as measured by the density of preservation measure (i.e., D measure). MEs refer to the consensus module eigengenes. ME0, which represents the grey module, is ignored (see Materials and methods). Figure 3 shows that the relationships between pairs of the modules across the GC sensitive and GC resistant MLL-rearranged infant ALL patients are well-preserved.
Consensus network analysis. The dendrograms show the eigengene clustering trees in the GC sensitive and GC resistant groups (upper panel). In addition, heatmaps of the eigengene correlations in both groups are shown, in which a positive correlation is indicated by a red color and a negative correlation by a blue color. The preservations of eigengene networks between the two groups are shown in a preservation heatmap and a barplot, respectively. The preservation heatmap indicates which inter-modular relationships are highly preserved and the barplot depicts the mean preservation of correlations for each of the consensus eigengenes (overall D measure = 0.89)
4 Discussion
Through weighted gene co-expression network analysis, we identified one (the green-yellow) module as being most significantly associated with GC (prednisolone) resistance in MLL-rearranged infant ALL patients. The module was found to be preserved across two independent datasets, and conventional differential gene expression analysis revealed that the significantly over-expressed genes in the GC resistant phenotype exhibited the highest level of module membership. Functional enrichment analysis revealed GO terms and other gene sets that are biologically relevant to GC resistance. Together, these results suggest that new candidate genes from the module may be added to the list of currently known GC resistance biomarkers and, thus, that these genes warrant further analysis. In Fig. 4 (generated using the Cytoscape software platform [40]) a correlation network is depicted with nodes representing genes from the green-yellow module with the largest module membership (\( {k}_{ME}^{green- yellow}>0.5 \)) and links representing correlations > 0.7. More details regarding the genes depicted in Fig. 4 are listed in Table S4 (Supplementary Materials). The correlation network in Fig. 4 is configured such that genes with the lowest intra-modular connectivity are placed at the fringes, whereas the genes with highest connectivity (i.e., the hub genes) are placed at the core. Most of the differentially expressed genes between the GC resistant and GC sensitive samples (i.e., genes with \( { \log}_2^{FC}>1 \)) are located in the green-yellow module, and are presented by hexagons in Fig. 4.
Correlation network of the green-yellow module, in which nodes represent genes with the highest module membership and links represent significant correlations. Genes with different levels of intra-modular connectivities are placed in different concentric circles, such that genes forming the inner circles have a higher connectivity than genes forming the outer circles. Hexagons represent the differentially expressed genes of the module. Genes with a known association to GC resistance are indicated in green. Novel GC-related calcium binding genes are indicated in red. New possibly relevant genes (see text) are indicated in purple. The remaining genes of the module are indicated in blue
The relevance of the genes composing the green-yellow module in terms of GC response in MLL-rearranged ALL is emphasized by the notion that several of these genes have previously been associated with chemo-resistance in leukemia, such as S100A8, S100A9, S100A10, S100A11, S100A12 and S100P. Spijkers-Hagelstein et al. [11] studied the mRNA expression levels of the entire S100 protein family in a relatively large cohort of MLL-rearranged infant ALL patient samples and found that high levels of S100A8, S100A9, S100A10, S100A11 and S100P expression were significantly associated with prednisolone resistance. They also found that the S100A8 and S100A9 proteins, which form hetero-tetramers, function together to induce glucocorticoid resistance in MLL-rearranged ALL cells. Moreover, S100A8 and S100A11 were also found to be involved in the development of resistance to chemotherapeutic drugs other than glucocorticoids [41, 42]. A recent study has shown that a high level of ANXA2 may mediate glucocorticoid resistance in MLL-rearranged ALL cells through the activation of Src kinases, a process requiring the adaptor protein p11 that is encoded by the S100A10 gene. The authors found that knockdown of either ANXA2 or S100A10 expression in GC resistant MLL-rearranged ALL cells hampered this process, leading to increased sensitization to prednisolone [13]. The presence of the ANXA1, ANXA3 and ANXA2P2 (an ANXA2 pseudogene) genes in the green-yellow module may suggest a role in the GC response of MLL-rearranged ALL cases for all Annexin-coding genes. Four other genes present in the green-yellow module (VCAN, ITGAM, FCN1 and MPO) encode proteins that exhibit calcium binding properties, similar to the ANXA and S100 genes. The observed recurrent association of calcium binding proteins with GC resistance suggests that these proteins are of particular relevance for the acquisition of GC resistance in MLL-rearranged ALL patients.
Two other interesting genes within the green-yellow module were LGALS3 (encoding galectin-3) and ITGB2 (encoding CD18). As it is becoming increasingly evident that the bone marrow microenvironment plays an important role in the occurrence of leukemia relapses and the acquisition of drug resistance, changes in leukemic gene expression patterns mediated by bone marrow stromal cells are of particular relevance. Interestingly, it has been found that the expression of both LGALS3 and ITGB2 can be up-regulated by bone marrow stromal cells, thereby mediating drug resistance in acute [43] and chronic [44] leukemias, respectively.
Recently, Spijkers-Hagelstein and colleagues reported gene expression profiles of primary MLL-rearranged infant ALL samples either sensitive or resistant to prednisolone, including an expression signature of 66 genes significantly associated with prednisolone resistance in this type of leukemia [13]. In a subsequent study, the same group used this expression signature to perform connectivity map analyses to identify possible modulators of this specific set of genes, and found the PI3K inhibitor LY294002 could markedly sensitize otherwise resistant MLL-rearranged ALL cells to prednisolone [14]. Interestingly, they found that LY294002-induced prednisolone sensitization was accompanied by down-regulation of the PSTPIP2, RNASE2 and TLR2 genes [14], which are all present in the green-yellow module. Like RNASE2, LYZ encodes a protein associated with myeloid differentiation. As MLL-rearranged ALL is often typified by co-expression of specific myeloid cell surface markers, abundant expression of both LYZ and RNASE2 may suggest that myeloid characteristics specifically occur in GC resistant MLL-rearranged ALL cells. The latter may particularly be true, since acute myeloid leukemia (AML) cells typically do not respond to GCs.
Two transcription factors (TFs), CEBPB and CEBPD, are included in the green-yellow module and both of them were found to be over-expressed in the GC resistant ALL samples compared to the GC sensitive samples. TF-target interaction analysis of the TRRUST database indicated that CEBPB may activate target genes such as ADM, CDKN1A, GOT1, INSR, IL1B, IL6 and PTGS2. Some of these genes respond to GC stimulation whereas some, like CDKN1A, IL1B and IL6, act as positive regulators of cell proliferation and negative regulators of apoptosis, suggesting that CEBPB may play a role in GC resistance in infant ALL via the activation of these target genes. Three TFs were identified as the most likely targets of GC-induced gene repression, i.e., AP-1, NF-κB and c-Myc [33]. Linking genes from the green-yellow module to these three TFs lends further biological support to the association between this module and GC resistance. AP-1 induces the transcription of growth factors, cytokines and survival genes resulting in enhanced cellular growth and proliferation. After binding of GC to the GC receptor (GR), the activated GR binds to AP-1 and blocks its trans-activating activity, thereby suppressing the transcription of survival genes and inducing apoptosis [33]. AP-1 is a dimer composed of two oncogenic proteins, JUN and FOS. According to the TRRUST database, several genes from the green-yellow module, including LGALS3, RNASE2, ITGAM, CD14, CYBB, CCR1, NCF2, S100A10, TLR2, CD1D, ITGB2, S100A9, CSF3R, CD33, PTAFR, MPO and BCL2A1, act downstream of JUN and other genes such as CD1D, S100A9, CD33, CSF1R and ARG1 which, in turn, can be activated by FOS. Similarly, NF-κB is a heterodimeric TF that activates survival genes coding for cytokines, cytokine receptors, chemotactic proteins and cell adhesion molecules [45, 46]. Repression of the transcriptional activity of these survival factors leads to apoptosis. NFKB1, a subunit of NF-κB, can upregulate several genes present in the green-yellow module, including CCL5, BCL2A1, MPO, NCF2, CEBPB, PTAFR, CD33, CYBB, S100A9, CD1D, TLR2, RETN, CD14 and CSF1R. Most of these genes are normally upregulated by RELA, another subunit of NF-κB. Also, c-Myc is involved in cell cycle regulation, proliferation and survival. GCs can repress c-Myc expression, leading to cell cycle arrest and apoptosis. Concordantly, previous studies have revealed correlations between c-Myc suppression and GC-induced apoptosis in human leukemic cells [47]. Interestingly, several genes from the green-yellow module, such as VCAN, TLR2, RNASE2, ITGAM, LGALS3, ITGB2, PTAFR, NCF2, BCL2A1 and ARG1, may act as downstream targets of c-Myc based on the TF-target interactions listed in the TRRUST database. We found that at least some of the genes from the green-yellow module may be linked to these three TFs and, as such, they may disrupt their role in GC-induced apoptosis. These observations underscore the association between the green-yellow module and GC resistance in general. Further experimental work is, however, needed to establish which of the candidates discussed here is predominantly contributing to GC resistance in ALL.
MLL-rearrangements are more frequent in infant ALL patients than in non-infant ALL patients [2, 3] and correlate well with GC resistance. In addition to genetic factors, epigenetic modifications in MLL-rearranged infant ALL samples have also been shown to influence the clinical outcome [48]. In particular, hypermethylated CpG islands within gene promoter regions [49] can be used to distinguish different subgroups of MLL-rearranged infant ALLs, and they correlate well with down-regulation of its corresponding genes [48]. Aberrant DNA methylation is more pronounced in infant ALL patients with t(4;11) or t(11;19)-associated MLL translocations than in those with t(9;11)-associated MLL translocations or wild type MLL genes. The data reported in [48] show that some TF-encoding genes, such as DLX4, NR2F2, NR3C1, NOTCH3, TBPL1, FOXA1 and OTX2, may have hypermethylated gene promoters in MLL-rearranged infant ALL samples. We retrieved the downstream targets of these TFs from the TRRUST database [34] and found that some of them are up-regulated in the green-yellow module. We hypothesize that experimentally-validated [48] TF promoter hypermethylation patterns in MLL-rearranged infant ALL samples may explain the up-regulation of at least some of their downstream target genes. The NR3C1 gene, for example, encodes GRs that bind to glucocorticoid response elements (GREs) in the promoters of GC responsive genes and, by doing so, activate their transcription [33]. GRs are essential in inducing AP-1 and NF-κB mediated apoptosis, and the repression of NR3C1 in MLL-rearranged infant ALL cells may well affect their GC-induced mode of action.
Previously, the effect of methylation on the expression of miRNAs has been studied in MLL-rearranged infant ALL cases [35]. Eleven miRNAs were found to be down-regulated as a consequence of promoter hypermethylation in t(4;11)-positive infant ALL cells compared to normal bone marrow cells. miRNAs act as post-translational regulators of gene expression, repressing gene translation by binding to the corresponding target mRNAs [50, 51]. Here, we retrieved miRNA-target interaction data from the miRTarBase database [36] and found that miRNA down-regulation in MLL-rearranged infant ALL cases may contribute to over-expression of at least some target genes present in the green-yellow module. miRNAs such as miR-432, miR-503 and miR-148a, which are down-regulated more than 100-fold in t(4;11)-positive infant ALL cases [35] can, for instance, lead to over-expression of VCAN, TLR2, CYBB, PTAFR, CSF1R, MPO, BCL2A1, CD1D, CEBPD, CCR1, CD33, S100A10, S100A9, CSF3R and ARG1. Interestingly, we found that most of these genes, as well as other genes in the green-yellow module such as ITGB2, LGALS3, IL6R, CEBPB, RNASE2, ITGAM, NCF2, CD14 and RETN, may act as targets of one or more of the differentially expressed miRNAs in MLL-rearranged infant ALL samples. Some TFs that play key roles in cell survival and proliferation such as NFKB1, RELA, JUN, FOS and c-MYc (discussed earlier) may also be activated via the down-regulation of specific miRNAs such as miR-432, miR-152, miR-10a, miR-200a and miR-200b in the MLL-rearranged infant ALL cases.
Evidently, the presented green-yellow module does encompass multiple genes already proven to be associated with GC resistance and, therefore, it is likely to contain more genes that, although to date unacknowledged, do mediate GC responses in MLL-rearranged ALL patients. Although this implication by association requires further experimental validation, our findings show that systems biology-based computational approaches may complement conventional gene-wise methods and, as such, may provide new insights into drug resistance in human malignancies such as ALL.
References
C.-H. Pui, M. V. Relling, J. R. Downing, Acute lymphoblastic leukemia. New Engl J Med 350, 1535–1548 (2004)
M. Greaves, Infant leukaemia biology, aetiology and treatment. Leukemia 10, 372–377 (1996)
R. Pieters, M. Schrappe, P. De Lorenzo, I. Hann, G. De Rossi, M. Felice, L. Hovi, T. LeBlanc, T. Szczepanski, A. Ferster, A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): an observational study and a multicentre randomised trial. Lancet 370, 240–250 (2007)
R. Pieters, M. Den Boer, M. Durian, G. Janka, K. Schmiegelow, G. Kaspers, E. Van Wering, A. Veerman, Relation between age, immunophenotype and in vitro drug resistance in 395 children with acute lymphoblastic leukemia-implications for treatment of infants. Leukemia 12, 1344–1348 (1998)
H. Riehm, A. Reiter, M. Schrappe, F. Berthold, R. Dopfer, V. Gerein, R. Ludwig, J. Ritter, B. Stollmann, G. Henze, Corticosteroid-dependent reduction of leukocyte count in blood as a prognostic factor in acute lymphoblastic leukemia in childhood (therapy study ALL-BFM 83). Klinische Padiatrie 199, 151–160 (1986)
A. Holleman, M. H. Cheok, M. L. den Boer, W. Yang, A. J. Veerman, K. M. Kazemier, D. Pei, C. Cheng, C.-H. Pui, M. V. Relling, Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. New Engl J Med 351, 533–542 (2004)
R. W. Stam, M. L. Den Boer, P. Schneider, J. de Boer, J. Hagelstein, M. G. Valsecchi, P. de Lorenzo, S. E. Sallan, H. J. Brady, S. A. Armstrong, Association of high-level MCL-1 expression with in vitro and in vivo prednisone resistance in MLL-rearranged infant acute lymphoblastic leukemia. Blood 115, 1018–1025 (2010)
G. Wei, D. Twomey, J. Lamb, K. Schlis, J. Agarwal, R. W. Stam, J. T. Opferman, S. E. Sallan, M. L. den Boer, R. Pieters, Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell 10, 331–342 (2006)
J. A. Spijkers-Hagelstein, P. Schneider, S. M. Pinhanços, P. G. Castro, R. Pieters, R. W. Stam, Glucocorticoid sensitisation in mixed lineage leukaemia-rearranged acute lymphoblastic leukaemia by the pan-BCL-2 family inhibitors gossypol and AT-101. Eur J Cancer 50, 1665–1674 (2014)
N. Kaiser, I. S. Edelman, Calcium dependence of glucocorticoid-induced lymphocytolysis. Proc Natl Acad Sci USA 74, 638–642 (1977)
J. A. Spijkers-Hagelstein, P. Schneider, E. Hulleman, J. de Boer, O. Williams, R. Pieters, R. W. Stam, Elevated S100A8/S100A9 expression causes glucocorticoid resistance in MLL-rearranged infant acute lymphoblastic leukemia. Leukemia 26, 1255–1265 (2012)
S. Qazi, F. M. Uckun, Gene expression profiles of infant acute lymphoblastic leukaemia and its prognostically distinct subsets. Br J Haematol 149, 865–873 (2010)
J. A. Spijkers-Hagelstein, S. M. Pinhancos, P. Schneider, R. Pieters, R. W. Stam, Src kinase-induced phosphorylation of annexin A2 mediates glucocorticoid resistance in MLL-rearranged infant acute lymphoblastic leukemia. Leukemia 27, 1063–1071 (2013)
J. Spijkers-Hagelstein, S. Pinhanços, P. Schneider, R. Pieters, R. Stam, Chemical genomic screening identifies LY294002 as a modulator of glucocorticoid resistance in MLL-rearranged infant ALL. Leukemia 28, 761–769 (2014)
X. Wang, J. Wen, R. Li, G. Qiu, L. Zhou, X. Wen, Gene expression profiling analysis of castration-resistant prostate cancer. Med Sci Monitor 21, 205–212 (2014)
J. Y. Chen, Z. Yan, C. Shen, D. P. Fitzpatrick, M. Wang, A systems biology approach to the study of cisplatin drug resistance in ovarian cancers. J Bioinf Comput Biol 5, 383–405 (2007)
B. C. Browne, F. Hochgräfe, J. Wu, E. K. Millar, J. Barraclough, A. Stone, R. A. McCloy, C. S. Lee, C. Roberts, N. A. Ali, Global characterization of signalling networks associated with tamoxifen resistance in breast cancer. FEBS J 280, 5237–5257 (2013)
J. Helleman, M. Smid, M. P. Jansen, M. E. van der Burg, E. M. Berns, Pathway analysis of gene lists associated with platinum-based chemotherapy resistance in ovarian cancer: the big picture. Gynecol Oncol 117, 170–176 (2010)
W. L. Allen, L. Stevenson, V. M. Coyle, P. V. Jithesh, I. Proutski, G. Carson, M. A. Gordon, H.-J. D. Lenz, S. Van Schaeybroeck, D. B. Longley, A systems biology approach identifies SART1 as a novel determinant of both 5-fluorouracil and SN38 drug resistance in colorectal cancer. Mol Cancer Ther 11, 119–131 (2012)
S. Nam, H. R. Chang, H. R. Jung, Y. Gim, N. Y. Kim, R. Grailhe, H. R. Seo, H. S. Park, C. Balch, J. Lee, A pathway-based approach for identifying biomarkers of tumor progression to trastuzumab-resistant breast cancer. Cancer Lett 356, 880–890 (2015)
C. Clarke, S. F. Madden, P. Doolan, S. T. Aherne, H. Joyce, L. O’Driscoll, W. M. Gallagher, B. T. Hennessy, M. Moriarty, J. Crown, Correlating transcriptional networks to breast cancer survival: a large-scale coexpression analysis. Carcinogenesis 34, 2300–2308 (2013)
Giulietti M, Occhipinti G, Principato G, Piva F. Weighted gene co-expression network analysis reveals key genes involved in pancreatic ductal adenocarcinoma development. Cell. Oncol., 1–10 (2016)
W. Liu, L. Li, W. Li, Gene co-expression analysis identifies common modules related to prognosis and drug resistance in cancer cell lines. Int J Cancer 135, 2795–2803 (2014)
S. Davis, P. S. Meltzer, GEOquery: a bridge between the Gene expression omnibus (GEO) and BioConductor. Bioinformatics 23, 1846–1847 (2007)
L. Gautier, L. Cope, B. M. Bolstad, R. A. Irizarry, Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315 (2004)
W. Huber, A. Von Heydebreck, H. Sültmann, A. Poustka, M. Vingron, Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18, S96–S104 (2002)
P. Langfelder, S. Horvath, WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008)
L. Song, P. Langfelder, S. Horvath, Comparison of co-expression measures: mutual information, correlation, and model based indices. BMC Bioinformatics 13, 328 (2012)
A. M. Yip, S. Horvath, Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinformatics 8, 22 (2007)
Y. Choi, C. Kendziorski, Statistical methods for gene set co-expression analysis. Bioinformatics 25, 2780–2786 (2009)
Y. Rahmatallah, F. Emmert-Streib, G. Glazko, Gene sets net correlations analysis (GSNCA): a multivariate differential coexpression test for gene sets. Bioinformatics 30, 360–368 (2014)
P. Langfelder, S. Horvath, Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol 1, 54 (2007)
S. Greenstein, K. Ghias, N. L. Krett, S. T. Rosen, Mechanisms of glucocorticoid-mediated apoptosis in hematological malignancies. Clin Cancer Res 8, 1681–1694 (2002)
H. Han, H. Shim, D. Shin, J. E. Shim, Y. Ko, J. Shin, H. Kim, A. Cho, E. Kim, T. Lee, TRRUST: a reference database of human transcriptional regulatory interactions. Sci Rep UK 5 (2015)
D. Stumpel, D. Schotte, E. Lange-Turenhout, P. Schneider, L. Seslija, R. De Menezes, V. Marquez, R. Pieters, M. Den Boer, R. Stam, Hypermethylation of specific microRNA genes in MLL-rearranged infant acute lymphoblastic leukemia: major matters at a micro scale. Leukemia 25, 429–439 (2011)
C.-H. Chou, N.-W. Chang, S. Shrestha, S.-D. Hsu, Y.-L. Lin, W.-H. Lee, C.-D. Yang, H.-C. Hong, T.-Y. Wei, S.-J. Tu, miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucl Acids Res 44, D239–D247 (2016)
Smyth GK. Limma: linear models for microarray data. Bioinformatics and computational biology solutions using R and Bioconductor: Springer, 397–420 (2005)
P. Langfelder, R. Luo, M. C. Oldham, S. Horvath, Is my network module preserved and reproducible. PLoS Comput Biol 7, e1001057 (2011)
G. Dennis Jr., B. T. Sherman, D. A. Hosack, J. Yang, W. Gao, H. C. Lane, R. A. Lempicki, DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4, P3 (2003)
M. E. Smoot, K. Ono, J. Ruscheinski, P.-L. Wang, T. Ideker, Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 (2011)
M. Yang, P. Zeng, R. Kang, Y. Yu, L. Yang, D. Tang, L. Cao, S100A8 contributes to drug resistance by promoting autophagy in leukemia cells. PLoS One 9, e97242 (2014)
J. Szczepanek, M. Pogorzala, M. Jarzab, M. Oczko-Wojciechowska, M. Kowalska, A. Tretyn, M. Wysocki, B. Jarzab, J. Styczynski, Expression profiles of signal transduction genes in ex vivo drug-resistant pediatric acute lymphoblastic leukemia. Anticancer Res 32, 503–506 (2012)
K. Hu, Y. Gu, L. Lou, L. Liu, Y. Hu, B. Wang, Y. Luo, J. Shi, X. Yu, H. Huang, Galectin-3 mediates bone marrow microenvironment-induced drug resistance in acute leukemia cells via Wnt/β-catenin signaling pathway. J Hematol Oncol 8, 1 (2015)
M. Plander, P. Ugocsai, S. Seegers, E. Orsó, A. Reichle, G. Schmitz, F. Hofstädter, G. Brockhoff, Chronic lymphocytic leukemia cells induce anti-apoptotic effects of bone marrow stroma. Ann Hematol 90, 1381–1390 (2011)
K. De Bosscher, W. Vanden Berghe, L. Vermeulen, S. Plaisance, E. Boone, G. Haegeman, Glucocorticoids repress NF-kB-driven genes by disturbing the interaction of p 65 with the basal transcription machinery, irrespective of coactivator levels in the cell. Proc Natl Acad Sci U S A 97, 3919–3924 (2000)
K. Vazquez-Santillan, J. Melendez-Zajgla, L. Jimenez-Hernandez, G. Martínez-Ruiz, V. Maldonado, NF-κB signaling in cancer stem cells: a promising therapeutic target? Cell Oncol 38, 327–339 (2015)
R. Thulasi, D. Harbour, E. Thompson, Suppression of c-myc is a critical step in glucocorticoid-induced human leukemic cell lysis. J Biol Chem 268, 18306–18312 (1993)
D. J. Stumpel, P. Schneider, E. H. van Roon, J. M. Boer, P. de Lorenzo, M. G. Valsecchi, R. X. de Menezes, R. Pieters, R. W. Stam, Specific promoter methylation identifies different subgroups of MLL-rearranged infant acute lymphoblastic leukemia, influences clinical outcome, and provides therapeutic options. Blood 114, 5490–5498 (2009)
A. Ferraro, Altered primary chromatin structures and their implications in cancer development. Cell Oncol 39, 1–16 (2016)
V. Taucher, H. Mangge, J. Haybaeck, Non-coding RNAs in pancreatic cancer: challenges and opportunities for clinical application. Cell Oncol 39, 1–24 (2016)
M. Vitiello, A. Tuccoli, L. Poliseno, Long non-coding RNAs in cancer: implications for personalized therapy. Cell Oncol 38, 17–28 (2015)
Acknowledgments
The authors would like to thank the Center of High Performance Computing (CHPC), School of Mathematics, Statistics and Computer Science, College of Science, University of Tehran for providing their cluster to meet our computational needs. We would like to thank the anonymous reviewer/editor for scientific and English editing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights and informed consent
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Mousavian, Z., Nowzari-Dalini, A., Stam, R.W. et al. Network-based expression analysis reveals key genes related to glucocorticoid resistance in infant acute lymphoblastic leukemia. Cell Oncol. 40, 33–45 (2017). https://doi.org/10.1007/s13402-016-0303-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-016-0303-7