Abstract
Detecting dispersal pathways is important both for understanding species range expansion and for managing nuisance species. However, direct detection is difficult. Here, we propose detecting these crucial pathways using a virtual ecology approach, simulating species dynamics using models, and virtual observations. As a case study, we developed a dispersal model based on cellular automata for the pest insect Stenotus rubrovittatus and simulated its expansion. We tested models for species expansion based on four landscape parameters as candidate pathways; these are river density, road density, area of paddy fields, and area of abandoned farmland, and validated their accuracy. We found that both road density and abandoned area models had prediction accuracy. The simulation requires simple data only to have predictive power, allowing for fast modeling and swift establishment of management plans.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dispersal is a fundamental biological process that operates at multiple temporal and spatial scales (Nathan 2001; Wilson et al. 2009). Dispersal determines the scale at which species interact with their environment, respond to perturbations, and evolve (Kinlan et al. 2003). The dispersal pathway is a crucial factor for species expansion and population dynamics (Wilson et al. 2009; Osawa and Ito 2015). For example, dispersal through multiple pathways can stabilize populations (Dey and Joshi 2006; Campbell Grant et al. 2010). Landscape corridors used by birds strongly influence seed dispersal and plant distribution ranges (Levey 2005). Understanding mechanisms of dispersal is important for ecology, especially when focusing on management of nuisance species such as invasive species, because it is one of the most important factors for successful invasion (Wilson et al. 2009; Pyšek et al. 2011; Donaldson et al. 2014). Despite their importance, however, dispersal pathways are rarely observed directly because of the difficulty involved (Nathan 2001; Campbell Grant et al. 2010).
In dealing with processes that are difficult to observe in the real world, such as dispersal, a theoretical approach is useful (Logan et al. 2003; Skarpaas et al. 2005). A theoretical model has shown that a propensity for limited dispersal through multiple pathways is especially critical for population stability and metapopulation persistence (Hill et al. 2002). The use of the virtual ecology approach—which simulates species dynamics using models—along with “virtual observation” of those species dynamics (Zurell et al. 2010; Pagel and Schurr 2012) is effective in dealing with dispersal pathways for predicting species expansion (Skarpaas et al. 2005; Cabral and Schurr 2010; Osawa and Ito 2015; Osawa et al. 2016). Using this approach, a hypothetical pathway simulating dispersal can be evaluated using actual distribution records, even if the records are limited (Osawa and Ito 2015; Osawa et al. 2016). In the present study, we propose a virtual ecology approach for detecting the crucial dispersal pathway of a pest insect.
Governmental agencies in Japan collect records of major pest insects in several regions to forecast their occurrence (Ministry of Agriculture, Forestry and Fisheries 2017; Occurrence Prediction Project). However, these types of records are usually used only to forecast the dynamics of the pest in the year and around the location that they are collected. In this study, we used such occurrence records as time-series distribution records for evaluating the results of virtual ecology simulations. As a case study, we selected the grain-feeding mirid bug Stenotus rubrovittatus (Heteroptera: Miridae) in Japan. Although S. rubrovittatus is considered to be a native species in Japan and the surrounding region (Kobayashi et al. 2011), the economic damage that it causes to crops has increased in recent years (0.3% of rice yield was degraded in 1990, 5.3% was degraded in 2012 at Akita Prefecture) as its distribution range has expanded (the range was limited in 2004 but dominated in 2010 at Akita Prefecture) (Tabuchi et al. 2017). There are some theories for the recent range expansion, such as changes in agricultural land use (Ito 2004) or climate factors (Kiritani 2006), but there are no clear plans or management strategy (Watanabe and Higuchi 2006; Higuchi 2010; Ohtomo 2013). In this study, we focus on the dispersal pathways that might have contributed to the recent expansion of S. rubrovittatus. If we can determine the crucial pathway, we should be able to predict future expansion of the species and contribute to establishing effective management plans.
In this study, we modified the virtual ecology model for predicting species dispersal previously established by Osawa and Ito (2015). This earlier model focused only on the physical form of the dispersal pathway, namely stream flow. The model was thus difficult to apply to species that can disperse unrestrained by stream flow. To apply the model to our target species, which has flying capability (i.e., unrestrained by stream flow), we incorporated the idea of “matrix resistance” in which there is substantial variability in the landscape matrix for dispersal (Ricketts 2001; Kuroe et al. 2011). In the modified model, a virtual population can expand into a neighboring landscape matrix according to the resistance of the landscape. Thus, the spatial locations of matrix types can determine dispersal pathways of the target species. Using the new model, we simulated the dispersal process of the mirid bug using some candidate pathways and validated the results with real time-series distribution records.
Materials and methods
Study area and analysis unit
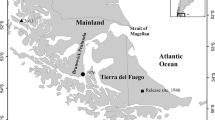
This study was conducted in Akita Prefecture, Japan (39°43′N, 140°6′E, 11637.52 km2; Fig. 1), with a mean annual precipitation of 1686.2 mm including heavy snow and a mean annual temperature of 11.7 °C (Japan Meteorological Agency 2017). This area is in the Tohoku region of Japan on the Sea of Japan side (Fig. 1) and is dominated by rice-producing farms practicing paddy agriculture (Akita Prefecture 2016).
We used a grid size of approximately 5 km, hereafter “5-km mesh” (Fig. 1). The 5-km mesh grid system is a standard Japanese unit used for several types of statistical analyses at this scale of geographic resolution, mainly of wildlife records (Biodiversity Center of Japan 2017). We used this as our analysis unit because a similar virtual ecology model at this scale performed well (Osawa and Ito 2015; Osawa et al. 2016). We used the mesh ID of the 5-km mesh as the cell ID, which was derived from the Japan Integrated Biodiversity Information System (J-IBIS 2017) available at the Biodiversity Center of Japan.
Study species
We used the mirid bug S. rubrovittatus as a case study. Grain-feeding bugs cause major damages worldwide in rice (Oryza sativa L.) and other economically important grains (Nagasawa et al. 2012). In Japan, the discoloration of rice grains caused by mirid bugs is a serious economic problem for rice cultivation (Kiritani 2006; Shintani 2009; Takada et al. 2012; Yoshioka et al. 2014) because the contamination of damaged rice with discolored grains results in a lower grade under Japanese rice quality regulations and thus a lower market price. Since the 1990s, mirid bugs including S. rubrovittatus have been recognized as major rice pests in the Tohoku and Hokuriku regions and have caused economic damage to rice farmers (Watanabe and Higuchi 2006; Tabuchi et al. 2015). During the past decade, the distribution range of S. rubrovittatus in this area expanded drastically, causing serious agricultural damage (Ohtomo 2013; Tabuchi et al. 2015).
Insect data collection
Time-series distribution records of S. rubrovittatus were obtained from local plant-protection offices within the study area. We requested the records for S. rubrovittatus occurrence as collected by sweep-netting conducted outside of paddy-field areas. These data were mainly collected originally for occurrence prediction. Summarized data have been posted on the website of the Tohoku Agricultural Research Center (Tohoku Agricultural Research Center, NARO 2015). We focused on the data collected at field boundaries and farm roads to avoid any effect of pesticide application.
The sweep-netting for this species was conducted at 120 sites with some differences in sampling effort. That is not a problem for this study because we merged and established the data on occurrence for 5-km mesh units for analysis. The sweep-netting was conducted at all 120 sites three times per year from 2003 to 2008, in mid and late July and in mid September, which corresponds to the seasonal occurrence of the species in the adjacent Iwate Prefecture (Iimura et al. 2004). Netting was also conducted three times per year at 100 sites in 2009 and 2010. In 2011 and 2012, sweep-netting was conducted at these three times and in early August and mid or late August (five times in total per year) at 100 sites. All distribution records were georeferenced using GIS software (ArcGIS 10.1, ESRI, Redland, CA) and maps from the Geospatial Information Authority of Japan (GSI 2016). Occurrence records were merged within each 5-km mesh square to establish the distribution records for each year from 2003 to 2012. The occurrence records for all years were merged to show the overall occurrence. A total of 21 cells in the study area contained sites with sweep-net data (Fig. 1).
Virtual ecology model
We used a simple cellular automata (CA) model that can predict the expansion of a target species based on candidate pathways. CA models apply flexible mathematical tools to approximate spatiotemporal dynamics and are, therefore, used for a wide range of ecological problems (Rácz and Bulla 2003; Dragićević 2010; Koike and Iwasaki 2011). The digital space in the CA model consists of a rectangular grid of square cells representing the target area (Rácz and Bulla 2003). The grid is set to the same size as the unit of predicted range expansion. The model yields a theoretical number of invasions, i.e., virtual invasion values in each cell, which serves as an invasion probability. Each cell has two parameters: cell ID and expansion path vector (Fig. 2a). The cell ID indicates the spatial location of the cell within the area of analysis. The expansion path involves four variables representing the four directional vectors into adjacent cells (Fig. 2b). Each variable is a probability value, i.e., 0–1, that indicates the probability of successful invasion. Thus, high resistance value has low probability of successful invasion. This is the matrix resistance value in this study.
Basic structure of the modified cellular automata (CA) model. a Two values are associated with each cell: 1) the cell ID “x”, a unique ID for each cell, and 2) the expansion path ex indicating four directional vectors into adjacent cells (described below). b Values e1, e2, e3, and e4 indicate the probability of dispersion using the path to the top, left, bottom, and right cells, respectively. If the dispersion path value is 1, the insect species in this cell can expand to that adjacent cell. c Example of values for e1, e2, e3, and e4 for cell “x” as probability values between 0 and 1
The analysis unit of the target insect species was the local population; that is, one cell can contain one population. Thus, the model simulates the capability for population expansion within the target area. The virtual population can expand according to the expansion path probability values (described below in detail).
Expansion path between cells
In this CA model, the expansion path is defined as the probability of expansion into four adjacent cells: those on the top, left, bottom, and right (Fig. 2b). The expansion path is defined as a vector with four probability values:
where e1, e2, e3, and e4 indicate the probability of expansion success to the top, left, bottom, and right cells, respectively (Fig. 2b). If all expansion path values are 0, the insect species in this cell cannot expand to any other cells. If all expansion path values are 1, the species in this cell can expand to all adjacent cells.
We used four candidate variables to represent potential pathways (river density, road density, area of paddy fields, and area of abandoned farmland) for setting the expansion path values. We selected these four variables because of the feeding habits of S. rubrovittatus; it feeds on inflorescences of various species of Poaceae and Cyperaceae (Hayashi and Nakazawa 1988; Kashin et al. 2009; Nagasawa and Higuchi 2012; Nagasawa et al. 2012). In general, in Japan there are grasslands with species of both Poaceae and Cyperaceae along rivers, roads, in paddy furrows, and on abandoned farmland (Hayashi 2009). The river line data were derived from the National Land Numerical Information Download Service of Japan (Ministry of Land, Infrastructure, Transport and Tourism, Japan 2017). The river lines were derived from GIS line data on national primary rivers and developed in 2007 (Ministry of Land, Infrastructure, Transport and Tourism, Japan). Although data for smaller rivers such as secondary rivers, small streams, and ditches are not available from this database, these rivers do not have large riparian areas with grassland supporting Poaceae or Cyperaceae. The road data were also derived from the National Land Numerical Information Download Service of Japan developed in 2010. The data were derived from total road length per 1-km square grid unit, and do not include other road characteristics such as width. Data for both paddy-field area and abandoned farmland area were derived from a data paper published in 2015 (Osawa et al. 2015). These areas did not drastically change from 2003 to 2012.
The expansion path was defined on the basis of the amount of a candidate pathway per unit area. The probability values for the dispersion paths e1, e2, e3, and e4 were estimated as the amount of a candidate pathway in a destination cell divided by the maximum amount of that pathway per cell in Akita Prefecture. Thus, these were relative values throughout the study area. These values reflect the type of landscape matrix preferred by the insects because if the matrix resistance is high, it is difficult for the insect to expand into that cell and vice versa. In addition, these values reflect the spatial position of the landscape matrix in the study area. If the landscape matrix shows a sequence of cells with low resistance, i.e., high probability of successful invasion, that is, there is an existing pathway, then the insect population can expand according to that pathway.
Expansion simulation
Using this CA field, we calculated the potential expansion range for all cells as the theoretical number of cells that could be expanded into using each candidate pathway. Thus, we had four CA simulation fields that were run independently. We predicted and compiled the expansion ranges from all cells; that is, we started the simulation from one cell, repeated that simulation for all cells, and summed all results. The results of one simulation from one cell show the expanded range from that starting cell. Summed cells with high values indicate that virtual insects could invade from several other cells. We used these invasion numbers (hereafter virtual invasion values) as indices of expansion success. The model was run for 999 iterations. If one virtual population failed to invade, it could try again during other iterations. Simulation models were implemented using the C language and compiled by MinGW ver. 0.6.2-beta-20131004-1 (MinGW.org Project 2017).
Model validation
Validation using the current distribution range
We validated the results of the CA model using merged occurrence records of S. rubrovittatus from 2003 to 2012. We compared average virtual invasion values between grid cells containing occurrence records and those without. We also compared average virtual invasion values between grid cells containing occurrence records and all cells in the study area. If the model prediction accurately represents S. rubrovittatus dispersion in the real world, the virtual invasion values should be higher in cells containing occurrence records than in those without, and higher than the average for all analyzed grid cells. We, therefore, used the Mann–Whitney U test to compare model predictions with field data.
Validation using time-series data
For the candidate pathways that showed predictive accuracy in the above analysis, we also validated the results of the CA model using time-series occurrence records of S. rubrovittatus from 2003 to 2012. For this analysis, we attached rank values that reflected the year of invasion for occurrence cells; that is, 2003, the first year of monitoring, was given a rank of 1, 2004 was 2, up to 2012 as 10. The small value indicates that the cell was preferred by S. rubrovittatus, thus, we assumed that the year of first invasion would reflect the preference of S. rubrovittatus. If a pathway was not effective for expansion, there should be a highly positive relationship between the year of first invasion and the virtual invasion values because species would use that pathway in later stages of an expansion. All statistical analyses were performed using R software version 3.2.1 (R Development Core Team 2015). A schematic of the whole procedure is provided in Fig. S1.
Results
Stenotus rubrovittatus distribution records
We acquired the distribution records for S. rubrovittatus from 2003 to 2012 in Akita Prefecture. A total of 21 cells within the 5-km mesh had sampling data for this time period. There was a steady increase in the number of cells where the target species was present (Fig. 3). After 2005, the number of cells with positive occurrence increased drastically until 19 of the 21 cells were occurrence cells. Because only two cells in the mesh showed no occurrence, we could not statistically compare our average virtual invasion values between cells with and without occurrence.
Relationships among candidate pathways
Our CA field contained 920 cells in total. The correlations (Pearson’s correlation coefficient) among the four candidate pathways are shown in Table 1. Three combinations had relatively high correlation coefficients and the other three combinations had relatively low correlation coefficients. The coefficient of correlation between river length and paddy area was 0.707, between river length and abandoned area was 0.635, and between paddy field and abandoned area was 0.619. These results suggest that simulations using these pathways might be expected to show similar results. On the other hand, the coefficient of correlation between river length and road length was 0.326, between road length and paddy area was 0.214, and between road length and abandoned area was 0.309. These results suggest that simulations using these pathways might be expected to show different results.
Accuracy of the cellular automata model simulations
We mapped the merged occurrence records and results of simulations (virtual invasion values) onto the study area (Fig. 4) and analyzed the models for accuracy (Table 2). All four models showed obviously higher virtual invasion values in cells with occurrence records (n = 19) than in those without (n = 2) (Table 2). A statistical comparison is not possible because there were only two cells with no occurrences. Thus, at some level our simulation has predictive accuracy. On the other hand, the virtual invasion values predicted by the river model (i.e., using river length per grid cell) for cells with occurrence records (n = 19, 39.47 ± 23.58; mean ± S.D.) did not significantly differ from that for all analyzed cells in the mesh, that is, all except those over the ocean (n = 920, 40.74 ± 25.18) (Mann–Whitney U test, U = 7281, P > 0.05). The virtual invasion values from the road model (n = 19, 14.21 ± 6.399), paddy area model (n = 19, 28.21 ± 17.27), and abandoned area model (n = 19, 15.68 ± 10.69) for cells with records of occurrence were significantly higher than those for all analyzed cells in the mesh (n = 920, 4.95 ± 6.37, 6.47 ± 12.06, 6.09 ± 9.11) (Mann–Whitney U test, U = 2055.5, 1570.5, 3061.5, P < 0.001 for all). All models except the river model accurately predicted the current distribution range. Thus, our virtual ecology models using road density, paddy area, and abandoned area are realistic. Among these three pathways, all correlation coefficients excluding that between paddy and abandoned area were relatively low: 0.214 for road–paddy, 0.309 for road–abandoned, and 0.619 for paddy–abandoned (Table 1).
We used time-series occurrence records to validate the results of these three models. Among these, the time rank of first invasion and virtual invasion values from the paddy model were significantly positively correlated even though the correlation coefficient was low (r = 0.1; Table 3). This suggests that paddy area does not reflect a preferred pathway for S. rubrovittatus. In addition, there was a high correlation between pathway values for paddy area and abandoned area (Table 1).
Discussion
We developed a dispersal simulation model based on CA with matrix resistance theory to find the crucial dispersal pathway of S. rubrovittatus from four candidate pathways. The simulation predictions from road density, paddy area, and abandoned area models had prediction accuracy for the real distribution range. Among these, paddy area as a pathway was relatively less preferred by the mirid bug. These results suggested that both road and abandoned area were crucial dispersal pathway of S. rubrovittatus. Our results provide a variety of information about the expansion mechanism of S. rubrovittatus. Thus, the virtual ecology approach is useful for detecting the expansion pathway of a species.
Estimation of efficient dispersal pathway
Among the four candidate pathway models, the road density, paddy area, and abandoned area models accurately reflected the real distribution range. The most plausible explanation for this is the existence of grassland in these three habitats, which acts as an emergence and feeding site for the mirid bug. Stenotus rubrovittatus is a euryphagous species that uses more than ten species in the Poaceae or Cyperaceae (Nagasawa and Higuchi 2012; Nagasawa et al. 2012), and it can survive in several types of grassland, such as those on slopes along roadsides and in paddy furrows and fallow soil (Yasuda et al. 2011). The extent of these areas could reflect habitat area and habitat continuity. In contrast, although there might be grasslands in riparian areas, the river model did not predict the actual distribution range of S. rubrovittatus. One possible explanation for this is anthropogenic effects. Many riverbanks in Japan have been recently concreted, the result being a lack of natural riparian zones (Osawa et al. 2011). In addition, many river banks are frequently mowed as part of river management by local governments. In Akita Prefecture, many rivers are concreted and managed. Therefore, in this study, river density did not reflect the amount of grassland. These anthropogenic effects, however, could differ by region, depending on policies and management strategies. This particular result should, therefore, be carefully interpreted and the river model should be considered in other regions.
Pathway preference by mirid bugs
Our time-series validation suggests that paddy area might not be preferred by S. rubrovittatus as an expansion pathway. Previous studies have shown that rice (O. sativa) was neither preferred for oviposition by of S. rubrovittatus nor improved its growth performance compared to other wild poaceous species (Nagasawa and Higuchi 2012; Nagasawa et al. 2012). Also, Yasuda (2012) showed that the number of S. rubrovittatus captured by sweep-netting was not affected by the extent of paddy-field area around the census area (Yasuda 2012). Nagasawa et al. (2012) indicated that S. rubrovittatus depended on a weed that had invaded a paddy field, not the rice itself. Therefore, S. rubrovittatus might not prefer a paddy field alone, either as a food resource or as an expansion pathway even there are furrows.
Nevertheless, our simulation using paddy area as a pathway was able to predict the current distribution range of S. rubrovittatus to some degree. One possible explanation is that paddy area as a pathway reflected other pathways, specifically abandoned area. In fact, the correlation between paddy area and abandoned area, which showed relatively high prediction accuracy, was high (0.619, Table 1). Thus, paddy area might act as a proxy for abandoned farmland area in our study area to some degree. This is a weak point of our model because there is some correlation between all pathway proxies, including river length and paddy area (Table 1). Although river length, which was highly correlated with other pathways, did not have high prediction accuracy in our case study, that might not always be true. To apply our approach to other areas or other species, it will be necessary to understand this problem to avoid overlooking possible pathways.
Application to a management plan
Pest management resources are generally limited in terms of both time and space, and thus it is necessary to have a spatially explicit plan for intensive management in high priority areas (Osawa and Ito 2015; Osawa et al. 2016; Tabuchi et al. 2017). Spatially explicit management plans should be developed at multiple spatial scales (Hiebert 1997; Shea et al. 2002; Foxcroft et al. 2009). Management plans at large spatial scales are suitable for guiding management, for monitoring activities, and for risk and priority assessment, whereas those at small spatial scales are preferred for management interventions (Foxcroft et al. 2009). For mirid bugs, there are already established intervention methods at small spatial scales, such as the timing of pesticide application (Kashin 2009; Yokota et al. 2009) or of mowing food plants (Yokota and Suzuki 2008), and these have been systematized to some degree (Ohtomo 2013). Also, a technique has been proposed based on local land-use maps for predicting potential damage to rice in areas in which the mirid bug has already become established (Tabuchi et al. 2017). Together with our approach for regional spatial scales, these existing techniques for small spatial scales will allow managers to establish a spatially explicit management plan with multiple spatial scales. Thus, managers can use our approach to identify priority areas with high potential for a species invasion. Subsequently, managers can implement an intervention plan for the priority areas using the small-scale methods.
One of the advantages of our method is that it is practicable without detailed information on the target species such as distribution records or monitoring data. In this study, we used time-series distribution records of S. rubrovittatus only for validation of the simulation results; thus, the simulation itself can be applied to other regions even there are no such data. Our approach could be applied as the first step in establishing a management plan. If a target area has not yet been invaded, managers should implement monitoring efforts in areas with high virtual invasion values to prevent invasion (Osawa and Ito 2015).
Conclusions
In this study, we used a virtual ecology approach to find crucial expansion pathways of S. rubrovittatus. We identified several effective pathways, and our overall results are consistent with ecological characteristics. We also demonstrated that the more important pathways can be screened in a simple manner using time-series information that is often included in pest dynamics data obtained by local governments. Thus, virtual ecology is one effective approach for testing possible expansion pathways. The expansion of all species is constrained by their pathways; thus, our approach may be applicable to other species, especially those with ecological characteristics similar to those of the mirid bug. Using a pathway simulation model, we should be able to find the important pathways of a target species, and that is useful information for managing nuisance species such as pests, weeds, and invasive species. Establishing a virtual ecology model is, therefore, an appealing approach with multiple applications in ecology.
References
Akita Prefecture, Japan. 2016. Retrieved 20 October, 2017, from http://www.pref.akita.lg.jp/pages/archive/16019 (in Japanese).
Biodiversity Center of Japan. 2017. Retrieved 19 May, 2017, from http://www.env.go.jp/press/press.php?serial=5533
Cabral, J.S., and F.M. Schurr. 2010. Estimating demographic models for the range dynamics of plant species. Global Ecology and Biogeography 19: 85–97. https://doi.org/10.1111/j.1466-8238.2009.00492.x.
Campbell Grant, E.H., J.D. Nichols, W.H. Lowe, and W.F. Fagan. 2010. Use of multiple dispersal pathways facilitates amphibian persistence in stream networks. Proceedings of the National academy of Sciences of the United States of America 107: 6936–6940. https://doi.org/10.1073/pnas.1000266107.
Dey, S., and A. Joshi. 2006. Stability via asynchrony in drosophila Metapopulations with low migration rates. Science 312: 434–436. https://doi.org/10.1126/science.1125317.
Donaldson, J.E., C. Hui, D.M. Richardson, M.P. Robertson, B.L. Webber, and J.R.U. Wilson. 2014. Invasion trajectory of alien trees: The role of introduction pathway and planting history. Global Change Biology 20: 1527–1537. https://doi.org/10.1111/gcb.12486.
Dragićević, S. 2010. Modeling the dynamics of complex spatial systems using GIS, cellular automata and fuzzy sets applied to invasive plant species propagation. Geography Compass 4: 599–615.
Foxcroft, L., D. Richardson, and M. Rouget. 2009. Patterns of alien plant distribution at multiple spatial scales in a large national park: Implications for ecology, management and monitoring. Diversity and Distributions 15: 367–378.
GSI map. 2016. Geospatial Information Authority of Japan. Retrieved 19 May, 2017, from https://maps.gsi.go.jp.
Hayashi, Y. 2009. Wilid flower of Japan. Tokyo: Yama to Keikokusha. (in Japanese).
Hayashi, H., K. Nakazawa. 1988. Studies on the bionomics and control of the sorghum plant bug, Stenotus rubrovittatus Matsumura (Hemiptera: Miridae) 1. Habitat and seasonal prevalence in Hiroshima Prefecture. Bulletin Hiroshima Prefectural Agricultural Experiment Station 51: 45–53. (in Japanese).
Hiebert, R. 1997. Prioritizing invasive plants and planning for management. Assessment and Management of Plant Invasions.
Higuchi, H. 2010. Ecology and management of rice bugs causing pecky rice. Japanese Journal of Applied Entomology and Zoology 54: 171–188. https://doi.org/10.1303/jjaez.2010.171. (in Japanese).
Hill, M.F., A. Hastings, and L.W. Botsford. 2002. The effects of small dispersal rates on extinction times in structured metapopulation models. American Naturalist 160: 389–402. https://doi.org/10.1086/341526.
Iimura, S., Y. Saitoh, and J. Gotoh. 2004. Seasonal occurrence of sorghum plant bug, Stenotus rubrovittatus Matsumura (Hemiptera: Miridae), in Iwate Prefecture. Annual Report of The Society of Plant Protection of North Japan 55: 117–121. (in Japanese).
Integrated Biodiversity Information System (J-IBIS) available at the Biodiversity Center of Japan. Retrieved 19 May, 2017, from http://gis.biodic.go.jp/webgis/sc-023.html.
Ito, K. 2004. A possible cause of recent outbreaks of rice-ear bugs-changes in the use of paddy field Kiyomitsu. Annual Report of the Society of Plant Protection of North Japan 55: 134–139. (in Japanese).
Japan Meteorological Agency. 2017. Retrieved 25 October, 2017, from http://www.data.jma.go.jp/obd/stats/etrn/view/nml_sfc_ym.php?prec_no=32&block_no=47582&year=&month=&day=&view=p1.
Kashin, J. 2009. Optimum time for chemical control of sorghum plant bug Stenotus rubrovittatus (Hemiptera: Miridae) in Scirpus juncoides Roxb. var. ohwianus-infested rice fields. Annual Report of The Society of Plant Protection of North Japan 60: 159–162. http://doi.org/10.11455/kitanihon.2009.60_159. (in Japanese).
Kashin, J., N. Hatanaka, T. Ono, J. Oyama, and T. Kidokoro. 2009. Effect of Scirpus juncoides Roxb. var. ohwianus on occurrence of sorghum plant bug, Stenotus rubrovittatus (Matsumura) (Hemiptera: Miridae) and pecky rice. Japanese Journal of Applied Entomology and Zoology 53: 7–12. (in Japanese).
Kinlan, B.P., S.D. Gaines, B.P. Kinlan, and S.D. Gaines. 2003. Propagule dispersal in marine and terrestrial environments: A community perspective. Ecology 84: 2007–2020.
Kiritani, K. 2006. Predicting impacts of global warming on population dynamics and distribution of arthropods in Japan. Population Ecology 48: 5–12. https://doi.org/10.1007/s10144-005-0225-0.
Kobayashi, T., T. Sakurai, M. Sakakibara, and T. Watanabe. 2011. Multiple origins of outbreak populations of a native insect pest in an agro-ecosystem. Bulletin of Entomological Research 101: 313–324.
Koike, F., and K. Iwasaki. 2011. A simple range expansion model of multiple pathways: the case of nonindigenous green crab Carcinus aestuarii in Japanese waters. Biological Invasions 13: 459–470.
Kuroe, M., N. Yamaguchi, T. Kadoya, and T. Miyashita. 2011. Matrix heterogeneity affects population size of the harvest mice: Bayesian estimation of matrix resistance and model validation. Oikos 120: 271–279. https://doi.org/10.1111/j.1600-0706.2010.18697.x.
Levey, D.J. 2005. Effects of landscape corridors on seed dispersal by birds. Science 309: 146–148. https://doi.org/10.1126/science.1111479.
Logan, J.A., J. Régnière, and J.A. Powell. 2003. Assessing the impacts of global warming on forest pest dynamics. Frontiers in Ecology and the Environment 1: 130–137. https://doi.org/10.1890/1540-9295(2003)001[0130:ATIOGW]2.0.CO;2.
MinGW.org Project. 2017. Retrieved 19 May, 2017, from http://mingw.org.
Ministry of Agriculture, Forestry and Fisheries. 2017. Occurrence Prediction Project. Retrieved May 19, 2017, from http://www.maff.go.jp/j/syouan/syokubo/gaicyu/.
Ministry of Land, Infrastructure, Transport and Tourism, Japan. 2017. Retrieved 19 May, 2017, from http://nlftp.mlit.go.jp/ksj-e/index.html.
Nagasawa, A., and H. Higuchi. 2012. Suitability of poaceous plants for nymphal growth of the pecky rice bugs Trigonotylus caelestialium and Stenotus rubrovittatus (Hemiptera: Miridae) in Niigata, Japan. Applied Entomology and Zoology 47: 421–427. https://doi.org/10.1007/s13355-012-0135-5.
Nagasawa, A., A. Takahashi, and H. Higuchi. 2012. Host plant use for oviposition by Trigonotylus caelestialium (Hemiptera: Miridae) and Stenotus rubrovittatus (Hemiptera: Miridae). Applied Entomology and Zoology 47: 331–339. https://doi.org/10.1007/s13355-012-0123-9.
Nathan, R. 2001. The challenges of studying dispersal. Trends in Ecology & Evolution 16: 481–483. https://doi.org/10.1016/S0169-5347(01)02272-8.
Ohtomo, R. 2013. Occurrence and control of Stenotus rubrovittatus (Hemiptera: Miridae) in Touhoku area in Japan. Japanese Journal of Applied Entomology and Zoology 57: 137–149. https://doi.org/10.1303/jjaez.2013.137. (in Japanese).
Osawa, T., and K. Ito. 2015. A rapid method for constructing precaution maps based on a simple virtual ecology model: A case study on the range expansion of the invasive aquatic species Limnoperna fortunei. Population Ecology 57: 529–538. https://doi.org/10.1007/s10144-015-0493-2.
Osawa, T., H. Mitsuhashi, H. Niwa, and A. Ushimaru. 2011. The role of river confluences and meanderings in preserving local hot spots for threatened plant species in riparian ecosystems. Aquatic Conservation: Marine and Freshwater Ecosystems 21: 358–363. https://doi.org/10.1002/aqc.1194.
Osawa, T., T. Kadoya, and K. Kohyama. 2015. 5- and 10-km mesh datasets of agricultural land use based on governmental statistics for 1970–2005. Ecological Research 30: 757. https://doi.org/10.1007/s11284-015-1290-2.
Osawa, T., S. Okawa, S. Kurokawa, and S. Ando. 2016. Generating an agricultural risk map based on limited ecological information: A case study using Sicyos angulatus. Ambio 45: 895–903. https://doi.org/10.1007/s13280-016-0782-9.
Pagel, J., and F.M. Schurr. 2012. Forecasting species ranges by statistical estimation of ecological niches and spatial population dynamics. Global Ecology and Biogeography 21: 293–304. https://doi.org/10.1111/j.1466-8238.2011.00663.x.
Pyšek, P., V. Jarošík, and J. Pergl. 2011. Alien plants introduced by different pathways differ in invasion success: Unintentional introductions as a threat to natural areas. PLoS ONE. https://doi.org/10.1371/journal.pone.0024890.
R Development Core Team, Vienna, Austria. 2015. Retrieved 25 October, 2017, from https://cran.r-project.org/.
Rácz, É. V. P., and M. Bulla. 2003. Cellular automata models of environmental processes. In Proceedings of the international conference in memoriam John von Neumann, 109–119.
Ricketts, T.H. 2001. The matrix matters: Effective isolation in fragmented landscapes. The American Naturalist 158: 87–99. https://doi.org/10.1086/320863.
Shea, K., H. Possingham, and W. Murdoch. 2002. Active adaptive management in insect pest and weed control: Intervention with a plan for learning. Ecological 12:927–936
Shintani, Y. 2009. Effect of seasonal variation in host-plant quality on the rice leaf bug, Trigonotylus caelestialium. Entomologia Experimentalis et Applicata 133: 128–135. https://doi.org/10.1111/j.1570-7458.2009.00915.x.
Skarpaas, O., K. Shea, and J.M. Bullock. 2005. Optimizing dispersal study design by Monte Carlo simulation. Journal of Applied Ecology 42: 731–739. https://doi.org/10.1111/j.1365-2664.2005.01056.x.
Tabuchi, K., T. Ichita, R. Ohtomo, J. Kashin, T. Takagi, T. Niiyama, Y. Takahashi, J. Nagamine, et al. 2015. Rice bugs in the Tohoku Region: Their occurrence and damage from 2003 to 2013. Bulletin of the National Agricultural Research Center for Tohoku Region 117: 63–115.
Tabuchi, K., T. Murakami, S. Okudera, S. Furihata, M. Sakakibara, A. Takahashi, and T. Yasuda. 2017. Predicting potential rice damage by insect pests using land use data: A 3-year study for area-wide pest management. Agriculture, Ecosystems and Environment 249: 4–11. https://doi.org/10.1016/j.agee.2017.08.009.
Takada, M. B., A. Yoshioka, S. Takagi, S. Iwabuchi, and I. Washitani. 2012. Multiple spatial scale factors affecting mirid bug abundance and damage level in organic rice paddies. Biological Control 60: 169–174. https://doi.org/10.1016/j.biocontrol.2011.11.011.
Tohoku Agricultural Research Center, NARO. 2015. Retrieved 19 May, 2017, from http://www.naro.affrc.go.jp/publicity_report/publication/files/tohoku117-v2.xlsx.
Watanabe, T., and H. Higuchi. 2006. Recent occurrence and problem of rice bugs. Plant Protection 60: 201–203. (in Japanese).
Wilson, J.R.U., E.E. Dormontt, P.J. Prentis, A.J. Lowe, and D.M. Richardson. 2009. Something in the way you move: Dispersal pathways affect invasion success. Trends in Ecology & Evolution 24: 136–144. https://doi.org/10.1016/j.tree.2008.10.007.
Yasuda, M. 2012. Does surrounding landscape composition affect agricultural pest abundances? Plant Protection 66: 366. (in Japanese).
Yasuda, M., T. Mitsunaga, A. Takeda, K. Tabuchi, K. Oku, T. Yasuda, and T. Watanabe. 2011. Comparison of the effects of landscape composition on two mirid species in Japanese rice paddies. Applied Entomology and Zoology 46: 519–525. https://doi.org/10.1007/s13355-011-0071-9.
Yokota, H., and T. Suzuki. 2008. Most effective timing of mowing on the population density of the nymphs of the overwintering generation of Stenotus rubrovittatus in the footpath between paddy fields. Annual Report of the Society of Plant Protection of North Japan 59: 116–119. (in Japanese).
Yokota, H., M. Terata, K. Chiba, and T. Suzuki. 2009. Timing of control of sorghum plant bug Stenotus rubrovittatus by regional cooperative control. Annual Report of the Society of Plant Protection of North Japan 60: 155–158. http://doi.org/10.11455/kitanihon.2009.60_155. (in Japanese).
Yoshioka, A., M.B. Takada, and I. Washitani. 2014. Landscape effects of a non-native grass facilitate source populations of a native generalist bug, Stenotus rubrovittatus, in a heterogeneous agricultural landscape. Journal of Insect Science 14: 1–14. https://doi.org/10.1093/jis/14.1.110.
Zurell, D., U. Berger, J.S. Cabral, F. Jeltsch, C.N. Meynard, T. Münkemüller, N. Nehrbass, J. Pagel, et al. 2010. The virtual ecologist approach: Simulating data and observers. Oikos 119: 622–635. https://doi.org/10.1111/j.1600-0706.2009.18284.x.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Osawa, T., Yamasaki, K., Tabuchi, K. et al. Detecting crucial dispersal pathways using a virtual ecology approach: A case study of the mirid bug Stenotus rubrovittatus. Ambio 47, 806–815 (2018). https://doi.org/10.1007/s13280-018-1026-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13280-018-1026-y