Abstract
Biological control agents are preferred over chemicals for managing plant diseases, with Trichoderma species being particularly effective against soil-borne pathogens. This study examines the use of a highly antagonistic strain, Trichoderma asperellum A10, and a virulent strain, Sclerotium rolfsii Sr38, identified and confirmed through ITS, β-tubulin (T. asperellum), TEF 1α, and RPB2 (S. rolfsii) sequences. In vitro and in planta experiments compared the antagonistic potential of A10 with other antagonistic fungi and fungicides against S. rolfsii. A10 achieved 94.66% inhibition of S. rolfsii in dual culture assays. In greenhouse trials with tomato variety Pusa Ruby, A10 showed significant pre- and post-inoculation effectiveness, with disease inhibition of 86.17 and 80.60%, respectively, outperforming T. harzianum, Propiconazole, and Carbendazim. Additionally, microbial priming with A10 was explored to enhance plant defense responses. Pre-treatment of tomato plants with T. asperellum A10 led to significant upregulation of several defense-related genes, including PR1, PR2, PR3, PR5, PR12, thioredoxin peroxidase, catalase, polyphenol oxidase, phenylalanine ammonia lyase, isochorismate synthase, laccase, prosystemin, multicystatin, WRKY31, MYC2, lipoxygenase A, lipoxygenase C, proteinase inhibitor I, proteinase inhibitor II, and ethylene response 1 associated with various signaling pathways such as salicylic acid (SA)-mediated and jasmonic acid/ethylene (JA/ET)-mediated responses. This upregulation was particularly evident at 48 h post-inoculation in A10-primed plants challenged with S. rolfsii, inducing resistance against collar rot disease. This study underscores the effectiveness of T. asperellum A10 in controlling collar rot and highlights its potential for inducing resistance in plants through microbial priming, providing valuable insights into sustainable disease management strategies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tomato (Lycopersicum esculentum Miller), one of the most important vegetable crops, is attacked by a large number of pathogens. Sclerotium rolfsii is one of the important fungal pathogen that causes collar rot disease resulting in substantial yield losses of around 30% in India (Mahato et al. 2017). Like many other soil-borne fungal diseases, S. rolfsii continues to be a difficult pathogen to control due to its wide host range of infecting more than 600 plant species and the production of abundant persistent sclerotia. Biological control of the disease using the beneficial microbes is an environmentally sound, effective and economically useful by enhancing the productivity in a sustainable manner (Abdelrahman et al. 2016). Biological control agents (BCAs) are the natural antagonists that can compete with the plant pathogens either direct through mycoparasitism, production of enzymes, and secondary metabolites (Hjeljord et al. 1998; Kubicek and Pentilla 1998) or by inducing resistance and plant defense reactions (Paparizas 1985; Howell 1998, 2003) through microbial priming could results into pre-activation of immune response against the challenging pathogens (Walters et al. 2013; Singh et al. 2020).
Trichoderma is a group of fungi that is well known for their biocontrol potential against wide range of plant pathogens, especially soil-borne pathogens. Trichoderma species helps to promote the plant growth and induces systemic resistance against subsequent pathogen attack (Al-Ani et al. 2020). Trichoderma interactions with plants may lead to the expression of defense-related genes, which in turn stimulate the plant’s immune systems in response to pathogen challenges and impact growth and development (Galletti et al. 2020; Pimentel et al. 2020). Trichoderma asperellum is one of the species in the Hypocreaceae family, which is having dual activity as it possesses both biocontrol and plant growth promoting properties. Through the enhanced expression of JA (Jasmonic Acid)/ET (Ethylene) and SA (Salicylic Acid) inducing genes, the colonization of the Arabidopsis root by T. asperellum SKT-1 and its culture filtrate results in an Induced Systemic Resistance (ISR) against Pseudomonas syringae pv. tomato DC3000 (Yoshioka et al. 2011). T. asperellum also triggers the ISR by the JA/ET signaling pathway, much like Plant Growth-Promoting Rhizobacteria (PGPR) (Shoresh et al. 2005). Six defense-related genes (CaPDF1.2, SOD, APx, GPx, PR-2, and PR-5) were expressed in pepper, when seeds were primed with T. asperellum, T. harzianum, and in combination with treatment of T. asperellum + T. harzianum against Colletotrichum truncatum (Yadav et al. 2023).
In many studies, Trichoderma spp. mediated priming against phytopathogens and its effect on key defense have been demonstrated (Boccardo et al. 2019). However, studies on T. asperellum mediated defense programming against the collar rot of tomato are limited. Therefore, the present study was undertaken to evaluate the expression profile changes of key defense genes in tomato, pre-inoculated with T. asperellum, following the infection of the S. rolfsii pathogen to gain a better insights into possible relationship in disease resistance.
Materials and methods
Fungus culture
Following our previous research, we selected a highly aggressive strain of S. rolfsii, known as Sr38, and a particularly promising antagonistic strain of T. asperellum, referred to as A10, for a subsequent investigation. The aim of this study was to assess the defense-boosting effects of T. asperellum on tomato plants through priming when facing the challenge of S. rolfsii.
Morpho-molecular identification and phylogenetic analysis
The identification of S. rolfsii strain Sr38 involved an examination of its morphological traits, which included observing mycelial growth patterns and the production of sclerotia on PDA medium. For T. asperellum strain A10, its morphological characteristics were determined by studying mycelial growth on PDA and conducting microscopic analysis. This microscopic assessment encompassed aspects such as conidiophores branching, the shape and size of phialides, and the characteristics of conidia using a compound microscope.
To confirm the identity of S. rolfsii Sr38 and T. asperellum strain A10, we conducted molecular identification procedures. The DNA from both strains was extracted using the Cetyl Trimethyl Ammonium Bromide (CTAB) method, following the protocol outlined by Cullings (1992). Approximately 500 mg of fungal mycelia were harvested from a 7-day-old culture on PDB and subsequently ground to a fine powder using liquid nitrogen and a pestle and mortar. We added 700 µL of CTAB buffer and incubated the mixture at 60 °C for 1 h in a water bath. After incubation, the sample was centrifuged at 10,000 rpm for 10 min at 24 °C. We collected the supernatant (500 µL) and added an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1), followed by another round of centrifugation at 10,000 rpm for 20 min at 24 °C. Next, we transferred 500 µL of the supernatant to a new tube and added 300 µL of isopropanol along with 50 µL of sodium acetate. This mixture was left to incubate overnight at 4 °C. The following day, we centrifuged the sample at 10,000 rpm for 10 min at 4 °C, carefully removing the supernatant without disturbing the pellet at the bottom of the tube. The pellet was then washed with 75% alcohol, and the alcohol was carefully discarded. Subsequently, the pellet was left to dry and was re-suspended in 50 µL of TE buffer.
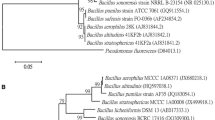
For PCR amplification, we targeted the Translation Elongation Factor 1α (TEF 1α) and RNA polymerase II (RPB2) genes to identify S. rolfsii, while for T. asperellum, we focused on the Internal Transcribed Spacer (ITS) and Beta-tubulin (β-tub) regions. The PCR mixture was prepared with a total volume of 25 µL, which included 12.5 µL of DreamTaq Green PCR master mix (Thermo Scientific, India) containing 0.25 mM of each dNTP, 2 mM of MgCl2, and Taq DNA polymerase, along with 9.5 µL of nuclease-free water. Additionally, 1 µL of each forward and reverse primer at a concentration of 10 Pmol/µL, and 1 µL of fungal DNA at a concentration of 100 ng/µL were added. The PCR amplification was conducted in thermal cycler (Bio-Rad) with specific protocols for each target region. For the ITS region, the PCR process began with an initial denaturation step at 94 °C for 3 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 57 °C for 1 min, and primer extension at 72 °C for 2 min. A final extension step was carried out at 72 °C for 5 min. In the case of the β-tubulin region, the PCR procedure commenced with an initial denaturation at 94 °C for 3 min. This was followed by 30 cycles consisting of denaturation at 94 °C for 30 s, annealing at 61 °C for 40 s, and primer extension at 72 °C for 30 s. The process concluded with a final extension step at 72 °C for 7 min. For the TEF 1α region, the PCR started with an initial denaturation at 94 °C for 2 min. Subsequently, 34 cycles of denaturation at 94 °C for 1 min, annealing at 53 °C for 1 min, and primer extension at 72 °C for 1 min were carried out. The procedure concluded with a final extension at 72 °C for 5 min. Finally, for the RPB2 region, the PCR initiated with an initial denaturation at 94 °C for 2 min, followed by 34 cycles of denaturation at 94 °C for 1 min, annealing at 62 °C for 1 min, and primer extension at 72 °C for 1 min. The process ended with a final extension step at 72 °C for 5 min. To visualize the PCR results, electrophoresis was performed using a 1.2% agarose gel containing 0.5 mg/L of ethidium bromide in 1 × TAE buffer. Electrophoresis was carried out for 30 min at 100 V and 400 mA. A 1 kb DNA ladder (Thermo Fisher Scientific, Waltham, MA, USA) was utilized to estimate the sizes of the PCR products. Subsequently, the samples underwent sequencing utilizing Sanger sequencing technique through outsourcing (Anuvanshiki (OPC) Pvt. Ltd.), and the species identification was performed using the NCBI Nucleotide BLAST. For the purpose of conducting phylogenetic analysis, we utilized the combined partial TEF 1α and RPB2 sequences for S. rolfsii and the ITS and β-tubulin sequences for T. asperellum and constructed a phylogenetic tree. The maximum-likelihood method was employed for this analysis, utilizing MEGA version 11 as the computational tool. In our analysis, along with T. asperellum strain A10, we incorporated other Trichoderma asperellum isolates and other Trichoderma species, namely T. harzianum, T. hamatum, T. virens, T. longibrachiatum, and Chaetomium globosum CBS 160.62 serving as a reference or outgroup. For S. rolfsii, our analysis involved S. rolfsii Sr38 with various S. rolfsii isolates and other Sclerotium species, including S. coffeicola, S. hydrophilum, S. tuliparum, S. rhizoides, S. perniciosum, and Sclerotinia sclerotiorum 2C, serving as the outgroup. To evaluate the stability and reliability of the branches in our constructed phylogenetic tree, we conducted a bootstrap analysis comprising 1000 replications.
Biocontrol assay
In vitro evaluation
To assess the in vitro antagonistic activity of various fungal strains, including T. asperellum strain A10, T. harzianum (ITCC-8105), T. longibrachiatum (ITCC-7794), T. virens (ITCC-7359), Chaetomium globosum (ITCC-6210), and Aspergillus niger (ITCC-7797), we conducted a dual culture assay against S. rolfsii strain Sr38. In this assay, a 5 mm disc of antagonistic fungi and S. rolfsii were placed opposite each other on individual PDA plates, positioned 2 cm from the plate’s edge. The efficacy of Carbendazim 50% WP and Propiconazole 25% EC fungicides was evaluated against S rolfsii strain Sr38. Both fungicides were tested at a concentration of 200 ppm. To prepare the fungicide-amended media, 20 mg of each fungicide was thoroughly mixed with autoclaved and cooled Potato Dextrose Agar (PDA) medium in conical flasks to achieve the desired concentration. The amended PDA medium was then poured into sterile Petri plates. After solidification of the medium, the plates were inoculated aseptically with mycelial discs of S. rolfsii Sr38. The cultures were then incubated for 7 days at 28 °C. Control plates containing only the pathogen (S. rolfsii) were maintained, each with three replicates. To quantify the inhibition percentage, we employed the formula: Inhibition (%) = 100 [(C−T)/C], where C represents the pathogen’s growth in the control plate, and T represents the pathogen’s growth in the treatment plate (Garcia 1991). The treatment values were subjected to comparison using Duncan’s multiple-range test.
In-planta evaluation
An in-planta study was conducted to assess the antagonistic efficacy of various agents, including T. asperellum strain A10, T. harzianum (ITCC-8105), T. longibrachiatum (ITCC-7794), T. virens (ITCC-7359), Chaetomium globosum (ITCC-6210), Aspergillus niger (ITCC-7797), Carbendazim (2 g/liter), and Propiconazole (2 ml/liter), against S. rolfsii strain Sr38. The effectiveness of all tested treatments against collar rot was evaluated using 1-month-old Pusa Ruby variety tomato plants. This experiment was carried out at the Center for Protected Cultivation Technology (CPCT) at IARI, New Delhi.
To multiply the test pathogen S. rolfsii, sorghum grains were used. Initially, 100 g of sorghum grains were soaked in water overnight and then placed in 500 mL Erlenmeyer flasks. The flasks were sealed with non-absorbent cotton and autoclaved at 15 psi for 30 min at 121 °C. After cooling to room temperature, the flasks were inoculated with mycelial discs from a 7-day-old culture of S. rolfsii under sterile conditions. The flasks were subsequently incubated at 28 °C for 7 days to allow complete mycelial growth coverage of the sorghum grains. To prepare the infected soil for the experiment, 10 g of S. rolfsii inoculum were mixed per kilogram of soil.
To prepare spore suspensions of all antagonistic fungi, a 7-day-old pure culture was used. Five milliliters of sterile distilled water were added to each culture plate, and spores were gently scraped off using a sterile spatula. The spore suspensions were then filtered, and their concentrations were adjusted to 108 spores/ml. These suspensions were further multiplied by inoculating 10 mL of spore suspension into 250 g of sterilized Farm Yard Manure (FYM) and incubating at 28 °C for 7 days.
The evaluation was carried out using both pre- and post-inoculation approaches. In the post-inoculation approach, 100 g of FYM enriched with antagonistic fungi were applied per pot after 5 days of transplanting the tomato plants into the S. rolfsii-infected soil. In the pre-inoculation approach, the tomato seedlings were dipped in the spore suspension of antagonistic fungi for 60 min before being transplanted into soil that had already been enriched with antagonistic fungi-enriched FYM and infected with S. rolfsii.
The experiment was designed using a Completely Randomized Design (CRD) and was conducted under greenhouse conditions. Each treatment was replicated three times, and the experiment was performed on three separate occasions to ensure robustness and reliability of the results. To compare the effects, four different treatments were employed: a control group treated with distilled water, a pathogen-treated group, a biocontrol-treated group, and a group treated with both the pathogen and the biocontrol agent. The Disease Severity Index (DSI) was recorded from pathogen-treated group and both the pathogen and the biocontrol agent treated group after 10 days of treatment, based on a scale provided by Le et al. (2012), where: 0 = no disease symptoms; 1 = disease symptoms without visible fungal outgrowth; 2 = disease symptoms with visible fungal growth; 3 = partial wilting of the plant; and 4 = complete wilting and plant death. The DSI was calculated for both pre- and post-inoculation conditions using the following formula:
Experimental design, biological treatments, and sample collection
The experimental setup involved using tomato plants of the Pusa Ruby variety, which were subjected to four distinct treatments: a control group treated with distilled water, a biocontrol treatment using T. asperellum strain A10, a pathogen treatment involving inoculation with S. rolfsii strain Sr38, and a biocontrol treatment followed by counter-inoculation with the pathogen. Each treatment was replicated 15 times. In the biocontrol treatment, a double dose was administered, first by dipping the seedlings in a spore suspension of T. asperellum strain A10 for 60 min, and second by applying T. asperellum strain A10-enriched Farm Yard Manure (FYM) at a rate of 50 g per pot into soil that had been previously amended with the pathogen to induce disease. To induce pathogen infection, the pathogen inoculum was applied at a rate of 10 g per kilogram of soil. Samples were collected at three different time points (12 h post-inoculation, 24 h post-inoculation, and 48 h post-inoculation) from all four treatments. These samples were immediately immersed in liquid nitrogen and stored at −80 °C for subsequent analysis.
RNA extraction and cDNA synthesis
Total RNA extraction from all the samples was performed using the Qiagen RNeasy Plant Mini Kit following the manufacturer’s instructions. The concentration and quality of the RNA samples were determined by measuring the optical density (OD) at 260/230 and 260/280 nm using a Nanodrop spectrophotometer. To verify the integrity of the extracted RNA, the presence of 28S and 18S bands was assessed using a 1.0% agarose gel. Subsequently, cDNA synthesis was carried out according to the manufacturer’s protocol using the NeoScript 1st Strand cDNA Synthesis Kit (Genes2Me). The concentration of the cDNA samples was quantified using a Nanodrop spectrophotometer (Thermo Fisher Scientific).
Defense gene expression analysis
To examine the expression patterns of defense-related genes in tomato plants across various treatments, real-time PCR was employed. The qPCR reactions were conducted in 96-well plates, each with a reaction volume of 20 µL per well. This volume comprised 10 µL of TB Green Premix Ex-taqII (SYBR green, TaKaRa), 7 µL of RNase-free ddH2O, 1 µL each of forward and reverse primers, and 1 µL of the cDNA template. The quantitative analysis was carried out using the CFX96 Touch Real-time PCR detection system (Bio-Rad, USA). The analysis focused on the expression profiles of several defense genes, including pathogenesis-related proteins (PR1, PR2, PR3, PR5, PR12), Thioredoxin peroxidase (TPX), Catalase, Polyphenol oxidase (PPO), Isochorismate synthase (associated with the shikimate pathway for salicylic acid synthesis), Lipoxygenase A and C (involved in jasmonic acid synthesis), Proteinase inhibitor I and II (inducible by jasmonic acid), Phenylalanine ammonia Lyase (PAL), Laccase, Prosystemin, Multicystatin, MYC2, WRKY31, and the Ethylene signaling pathway gene ETR1 (which codes for the ethylene receptor). These genes were assessed across all four treatments and at three different time points. The primer sequences for these genes were provided (Table 1). As an internal reference gene, Actin was utilized. Relative transcript quantification was carried out using the comparative 2−ΔΔCT method (Livak and Schmittgen 2001), with each treatment having three biological replicates.
Statistical analysis
The data analysis was conducted using the statistical software WASP 2.0 (Web Agri Stat Package), which can be accessed at https://ccari.icar.gov.in/waspnew.html (as of July 20, 2024). One-way analysis of variance (ANOVA) was employed to examine potential variations in parameter values among the treatments. Each experiment included three replications. Duncan’s multiple-range test was utilized to evaluate differences between treatments at a significance level of 5%. For the determination of relative gene expression, the output from the qPCR runs was analyzed using the CFX ManagerTM software, which involved assessing cycle threshold (Ct) values.
Results
Morpho-molecular identification and phylogenetic analysis
The morphological features of S. rolfsii strain Sr38 was identified by fast growing fluffy mycelial growth and abundant sclerotial production on the Potato Dextrose Agar (PDA) medium. The T. asperellum strain A10 grew faster on the PDA and the mycelial colonies were initially white on 3 days after incubation (DAI) and turned greenish after sporulation on 7 DAI. Microscopic examination revealed branching, typically paired conidiophores, ampuliform phialides, and light green conidia that ranged in shape from slightly oval to globose (Fig. 1).
Morphological characteristics of S. rolfsii and T. asperellum. a Colony growth of S. rolfsii on potato dextrose agar. b Colony growth of T. asperellum on potato dextrose agar. c Microscopic images showing conidiophores structures and branching pattern of T. asperellum (magnification at 40×). d Phialides arrangement of T. asperellum with conidia (magnification at 40×). e T. asperellum conidia (magnification at 100×)
We confirmed the amplified products generated by the TEF 1α and RPB2 gene primers for S. rolfsii, as well as the ITS and β-tubulin gene primers for T. asperellum, through exact band in agarose gel electrophoresis. Following confirmation, these products underwent sequencing for analysis. We confirmed the species identities of the strains using NCBI BLAST searches. Subsequently, all the obtained sequences were submitted to GenBank, where they were assigned unique accession numbers: ITS (OR137590) and β-tubulin (OR193710) for T. asperellum, and TEF 1α (OR335644) and RPB2 (OR227561) for S. rolfsii. We confirmed the taxonomic identities of the strains using phylogenetic analysis. The results revealed that our S. rolfsii strain Sr38 shares a close genetic relationship with other S. rolfsii isolates, distinguishing it from other Sclerotium species and the outgroup (Fig. 2). Similarly, our T. asperellum strain A10 exhibited a close genetic affinity with other T. asperellum isolates, setting it apart from other Trichoderma species and the outgroup (Fig. 3).
Maximum-likelihood phylogenetic tree based on concatenated translation elongation factor 1α (TEF 1α) and RNA Polymerase 2 (RPB2) sequences of the Sclerotium (= Athelia) rolfsii strain Sr38 and its related species and reference S. rolfsii isolates retrieved from the NCBI database. The tree was rooted using Sclerotinia sclerotiorum as an outgroup
Maximum-likelihood phylogenetic tree based on concatenated Internal Transcribed Spacer (ITS) and β-tubulin sequences of the Trichoderma asperellum strain A10 and its related species and reference T. asperellum isolates retrieved from the NCBI database. The tree was rooted using Chaetomium globosum as an outgroup
Antagonistic efficacy
In vitro evaluation
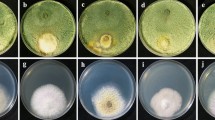
In the dual culture assay, all the antagonistic fungi significantly inhibited the mycelial growth of S. rolfsii, with inhibition percentages ranging from 20.87% to 94.66% (p < 0.05). Notably, T. asperellum strain A10 exhibited the highest inhibition rate of 94.66% among all the tested antagonists after 7 days of inoculation. T. harzianum also showed considerable effectiveness with an 82.64% inhibition rate. In comparison, T. longibrachiatum and T. virens had moderate inhibition rates of 51.38% and 52.35%, respectively. Chaetomium globosum and Aspergillus niger AN-11 were less effective, with inhibition rates of 20.87% and 30.93%, respectively. In addition to the antagonistic fungi, the fungicidal efficacy of Carbendazim exhibited a significant inhibition rate of 63.75%, indicating its effectiveness in reducing the mycelial growth of S. rolfsii. Propiconazole demonstrated an even higher inhibition rate of 71.89%, showing its superior efficacy compared to Carbendazim. These fungicides, although not as effective as the top-performing antagonistic fungus T. asperellum strain A10, still provided substantial inhibition of S. rolfsii mycelial growth. This highlights the efficacy of T. asperellum strain A10 as the most potent antagonist in suppressing S. rolfsii (Table 2, Fig. 4).
In-planta evaluation
The results indicate that all treatments were effective in reducing disease severity, with varying degrees of efficacy. The T. asperellum strain A10 consistently demonstrated superior performance, achieving the highest disease inhibition rates of 86.17% and 80.60% in the pre- and post-inoculation treatments, respectively. This was significantly better compared to the other treatments, with T. harzianum showing moderate efficacy with 77.80% and 72.22% reduction in disease severity. In contrast, T. longibrachiatum and T. virens exhibited lower disease inhibition, with reductions of 58.33% and 50.00%, and 36.11% and 33.33%, respectively. Chaetomium globosum, while effective, resulted in the least reduction in disease severity, with only 11.11% and 8.33% reduction in the pre- and post-inoculation treatments. Similarly, Aspergillus niger AN-11 showed moderate inhibition, with 22.22% and 16.67% reductions. The chemical controls, Carbendazim and Propiconazole, displayed intermediate efficacy, with reductions of 55.55% and 47.22%, and 61.11% and 55.56%, respectively, by comparing the positive control showing 100% DSI (Table 3, Fig. 5).
In-planta antagonistic evaluation of biocontrol agents and fungicides against S. rolfsii. a Trichoderma virens. b Trichoderma longibrachiatum. c Trichoderma asperellum A10. d Trichoderma harzianum. e Aspergillus niger. f Chaetomium globosum. g Carbendazim. h Propiconazole; C Control, B Biocontrol, P Pathogen, B + P Biocontrol + Pathogen, Ch Chemical, Ch + P Chemical + Pathogen
Expression profile of defense-related genes
Significant changes in the expression of defense genes in tomato plants were observed in response to T. asperellum strain A10 when exposed to S. rolfsii. The analysis included the gene expression patterns for all four treatments (Control, Biocontrol-treated, Pathogen-treated, and combined Biocontrol and Pathogen-treated) at three different time points (12 h post-inoculation, 24 h post-inoculation, and 48 h post-inoculation). It was evident that all the tested genes exhibited significant expression across all three time points.
Expression profile of pathogenesis-related genes
The pathogenesis-related protein (PR1) gene expression significantly different at 12, 24, and 48 h post-inoculation (hpi) across four treatments. At 12 hpi, the Biocontrol treatment exhibited the highest expression level at 2.332-fold, compared to the Biocontrol + Pathogen treatment (1.242-fold), the Pathogen treatment (1.061-fold), and the Control treatment (0.592-fold). By 24 hpi, the Biocontrol + Pathogen treatment showed a notable increase in PR1 expression to 6.537-fold, surpassing the Biocontrol treatment (4.485-fold), the Pathogen treatment (2.244-fold), and the Control treatment (0.893-fold). At 48 hpi, the Biocontrol + Pathogen treatment continued to exhibit the highest expression at 14.211-fold, significantly higher than the Biocontrol treatment (8.555-fold), the Pathogen treatment (5.306-fold), and the Control treatment (1.022-fold). Overall, the Biocontrol + Pathogen treatment demonstrated the most significant increase in PR1 expression at all time points (Fig. 6a, Online Resource 5).
Gene expression analysis of the defense-related genes encoding a PR-1, b PR-2, c PR-3, d PR-5, and e PR-12 in tomato var. pusa ruby grown in soil existing different treatments viz., Control, Biocontrol (T. asperellum strain A10 treated), Pathogen (S. rolfsii strain Sr38 treated), and Biocontrol + Pathogen (both). Actin was used as the reference gene. Each data point represents the average of three independent biological replicates. Error bars indicate standard deviation (SD)
The expression of PR2 (β-1, 3 glucanase) under different treatments showed significant differences at varied time points. At 12 hpi, the Control treatment exhibited the lowest expression level at 1.013-fold, while the Biocontrol + Pathogen treatment had the highest at 2.674-fold, indicating a significant early induction of PR2 in the presence of both biocontrol agents and the pathogen. By 24 hpi, the Biocontrol + Pathogen treatment continued to show the highest expression at 5.380-fold, followed by the Biocontrol treatment at 2.687-fold. The Pathogen treatment showed a moderate increase to 2.012-fold, while the Control remained relatively unchanged at 1.022-fold. At 48 hpi, the Biocontrol + Pathogen treatment maintained the highest expression level at 11.932-fold, significantly surpassing the Biocontrol treatment (11.153-fold) and the Pathogen treatment (9.256-fold). The Control treatment showed minimal change with an expression level of 1.005-fold. The Biocontrol + Pathogen treatment consistently demonstrated the highest PR2 expression levels at every time point, making it the most significant treatment overall (Fig. 6b, Online Resource 1).
The PR3 (Chitinase) expression demonstrated notable variability depending on the treatment and time points. At 12 hpi, the Biocontrol + Pathogen treatment exhibited the highest expression level at 7.596-fold, markedly higher than the Biocontrol treatment (2.510-fold), the Pathogen treatment (1.789-fold), and the Control treatment (1.072-fold). By 24 hpi, the Biocontrol + Pathogen treatment continued to show the highest expression at 13.664-fold, surpassing the Pathogen treatment (1.496-fold), the Biocontrol treatment (1.489-fold), and the Control treatment (0.861-fold). At 48 hpi, the Biocontrol + Pathogen treatment maintained the highest expression level at 26.457-fold, significantly higher than the Pathogen treatment (18.269-fold), the Biocontrol treatment (3.598-fold), and the Control treatment (1.022-fold). Among all treatments, Biocontrol + Pathogen showed the most pronounced effect; with PR3 expression reaching the highest levels at all measured time points (Fig. 6c, Online Resource 1).
The PR5 gene expression showed significant differences under different treatments at varied time points, starting at 12 hpi. At this time, the Biocontrol treatment had the highest expression level at 1.234-fold, compared to the Control treatment (1.142-fold), the Pathogen treatment (1.089-fold), and the Biocontrol + Pathogen treatment (0.613-fold). By 24 hpi, the Biocontrol treatment continued to exhibit the highest expression at 2.060-fold, followed by the Biocontrol + Pathogen treatment (1.480-fold), the Control treatment (1.055-fold), and the Pathogen treatment (0.602-fold). At 48 hpi, the Biocontrol + Pathogen treatment showed a marked increase in PR5 expression to 4.706-fold, significantly higher than the Pathogen treatment (1.959-fold), the Biocontrol treatment (1.469-fold), and the Control treatment (1.171-fold). Overall, the Biocontrol + Pathogen treatment demonstrated the most significant increase in PR5 expression at 48 hpi (Fig. 6d, Online Resource 6).
There were marked differences in the expression of PR12 under different treatment conditions and at different time points. At 12 hpi, the Biocontrol treatment exhibited the highest expression level at 2.611-fold, compared to the Pathogen treatment (1.676-fold), the Control treatment (1.609-fold), and the Biocontrol + Pathogen treatment (1.089-fold). By 24 hpi, the Pathogen treatment showed a notable increase to 2.544-fold, surpassing the Biocontrol treatment (2.052-fold), and the Biocontrol + Pathogen treatment (1.584-fold), and the Control treatment (1.103-fold). At 48 hpi, the Biocontrol + Pathogen treatment demonstrated the highest expression at 3.584-fold, significantly higher than the Pathogen treatment (1.593-fold), the Biocontrol treatment (1.026-fold), and the Control treatment (1.054-fold). Among all treatments, Biocontrol + Pathogen showed the most pronounced effect, with PR12 expression reaching the highest levels at all measured time points (Fig. 6e, Online Resource 6).
Expression profile of TPX, CAT, ICS, PPO, and PAL genes
The expression levels of the thioredoxin peroxidase (Tpx) gene varied considerably across different treatments at different time points. In the control group, expression remained relatively stable with fold changes of 0.90 at 12 hpi, 1.08 at 24 hpi, and 1.04 at 48 hpi. The biocontrol treatment showed a moderate increase, with fold changes of 1.02 at 12 hpi, 2.28 at 24 hpi, and 1.37 at 48 hpi. The pathogen treatment led to a marked increase in expression, starting at 0.49-fold at 12 hpi, rising significantly to 3.64-fold at 24 hpi and reaching 1.55-fold at 48 hpi. The most pronounced effect was observed in the combined biocontrol and pathogen treatment, where Tpx expression was downregulated to 0.28-fold at 12 hpi, followed by a substantial increase to 3.54-fold at 24 hpi, and a striking 10.80-fold at 48 hpi. This indicates that the combined treatment of biocontrol and pathogen had the most significant impact on upregulating Tpx gene expression, especially notable at 48 hpi (Fig. 7a, Online Resource 2).
Gene expression analysis of the defense-related genes encoding a Thioredoxin peroxidase, b Catalase, c Isochorismate synthase, d Polyphenol oxidase, and e Phenyl alanine ammonia lyase in tomato var. pusa ruby grown in soil existing different treatments viz., Control, Biocontrol (T. asperellum strain A10 treated), Pathogen (S. rolfsii strain Sr38 treated), and Biocontrol + Pathogen (both). Actin was used as the reference gene. Each data point represents the average of three independent biological replicates. Error bars indicate Standard deviation (SD)
Across different treatments, the expression levels of the Catalase (Cat) gene demonstrated considerable differences over time. In the control group, expression levels were relatively low and stable, with fold changes of 0.39 at 12 hpi, 1.05 at 24 hpi, and 0.80 at 48 hpi. The biocontrol treatment led to a moderate increase in gene expression, with fold changes of 0.53 at 12 hpi, 1.82 at 24 hpi, and 1.01 at 48 hpi. The pathogen treatment caused a gradual increase, showing fold changes of 0.77 at 12 hpi, 1.76 at 24 hpi, and 1.85 at 48 hpi. The most significant upregulation was observed in the combined biocontrol and pathogen treatment, with fold changes of 1.04 at 12 hpi, 2.03 at 24 hpi, and a substantial 5.50 at 48 hpi. This observation highlights that the combined biocontrol and pathogen treatment yielded the highest Cat gene expression levels, particularly at 48 hpi (Fig. 7b, Online Resource 2).
The expression levels of Isochorismate Synthase (ICS) varied significantly among treatments at different time points. In the control group, ICS expression was relatively steady, with fold changes of 0.74 at 12 hpi, 1.18 at 24 hpi, and 1.06 at 48 hpi. The biocontrol treatment resulted in a notable increase in ICS expression, with fold changes rising from 0.41 at 12 hpi to 5.64 at 24 hpi, and then slightly decreasing to 3.14 at 48 hpi. The pathogen treatment showed an initial increase to 1.42-fold at 12 hpi, with further upregulation to 2.14-fold at 24 hpi and 2.40-fold at 48 hpi. The most pronounced effect was observed in the combined biocontrol and pathogen treatment, where ICS expression surged from 1.77-fold at 12 hpi to a dramatic 14.00-fold at 24 hpi, and remained high at 9.21-fold at 48 hpi. This indicates that the highest expression of ICS occurred with the biocontrol and pathogen combination, especially notable at 24 hpi (Fig. 7c, Online Resource 4).
The expression levels of Polyphenol Oxidase (PPO) gene exhibited distinct variations across different treatments and time intervals. In the control scenario, PPO activity remained relatively stable with values of 1.14-fold at 12 hpi, 1.31-fold at 24 hpi, and 1.09-fold at 48 hpi. The biocontrol intervention led to a significant rise in PPO levels, starting from 0.37-fold at 12 hpi and escalating to 1.31-fold at 24 hpi, before reaching 1.59-fold at 48 hpi. In contrast, the pathogen treatment showed a gradual increase from 0.62-fold at 12 hpi to 0.72-fold at 24 hpi, and then a substantial spike to 2.80-fold at 48 hpi. The most dramatic increase was seen in the combined treatment of pathogen and biocontrol, where PPO activity jumped from 1.96-fold at 12 hpi to an impressive 7.40-fold at 24 hpi, and peaked at 9.86-fold at 48 hpi. This pattern suggests that the highest PPO expression was associated with the combined pathogen and biocontrol treatment, particularly remarkable at 48 hpi (Fig. 7d, Online Resource 3).
The Phenylalanine Ammonia Lyase (PAL) gene expression displayed notable differences across treatments and time points. In the control group, PAL activity remained fairly constant with values of 1.16-fold at 12 hpi, 1.25-fold at 24 hpi, and 1.10-fold at 48 hpi. The biocontrol treatment caused a substantial increase in PAL levels, starting from 2.24-fold at 12 hpi and rising to 3.51-fold at 24 hpi, before slightly decreasing to 2.80-fold at 48 hpi. For the pathogen treatment, PAL activity increased from 1.07-fold at 12 hpi to 2.87-fold at 24 hpi, and then significantly surged to 5.68-fold at 48 hpi. The most significant expression was observed in the combined biocontrol and pathogen treatment, where PAL levels dramatically rose from 0.62-fold at 12 hpi to 9.01-fold at 24 hpi, and further escalated to 10.11-fold at 48 hpi. This indicates that the highest PAL expression was achieved with the combined treatment of biocontrol and pathogen, particularly noteworthy at 24 hpi and 48 hpi (Fig. 7e, Online Resource 7).
Expression profile of LoxA, LoxC, PIN I, PIN II, and ETR genes
The Lipoxygenase A (Lox A) gene expression levels under various treatments showed significant differences at different time points. In the control group, expression remained relatively stable, with fold changes of 1.03 at 12 hpi, 0.94 at 24 hpi, and 1.00 at 48 hpi. The biocontrol treatment resulted in an initial downregulation to 0.83-fold at 12 hpi, followed by a substantial upregulation to 4.07-fold at 24 hpi, and a moderate increase to 1.40-fold at 48 hpi. Plants treated with the pathogen alone exhibited a downregulation at 12 hpi (0.45-fold), a significant upregulation to 3.25-fold at 24 hpi, and further increased to 4.75-fold at 48 hpi. The combined treatment of biocontrol and pathogen led to a downregulation at 12 hpi (0.72-fold), followed by a dramatic upregulation to 6.44-fold at 24 hpi, and a remarkable 15.89-fold increase at 48 hpi. This indicates that the most significant upregulation of gene expression occurred in the combined biocontrol and pathogen treatment, particularly evident at 48 hpi (Fig. 8a, Online Resource 10).
Gene expression analysis of the defense-related genes encoding a LoxA, b LoxC, c PIN I, d PIN II, and e ETR I in tomato var. pusa ruby grown in soil existing different treatments, viz., Control, Biocontrol (T. asperellum strain A10 treated), Pathogen (S. rolfsii strain Sr38 treated), Biocontrol + Pathogen (both). Actin was used as the reference gene. Each data point represents the average of three independent biological replicates. Error bars indicate standard deviation (SD)
The expression level of the Lipoxygenase C (Lox C) gene under various treatments showed distinct patterns over time. In the control group, expression levels decreased from 1.14-fold at 12 hpi to 0.76-fold at 24 hpi and then slightly increased to 0.85-fold at 48 hpi. The biocontrol treatment resulted in a steady upregulation of the gene, with fold changes of 1.16, 1.78, and 1.99 at 12, 24, and 48 hpi, respectively. Plants inoculated with the pathogen alone exhibited a marked increase in gene expression, with fold changes rising from 0.77 at 12 hpi to 2.55 at 24 hpi and 2.91 at 48 hpi. Notably, the combined treatment of biocontrol and pathogen led to the most significant upregulation of the gene, with fold changes of 0.74, 4.64, and 5.47 at 12, 24, and 48 hpi, respectively. The results indicate that the biocontrol plus pathogen treatment produced the highest gene expression, particularly at 24 and 48 hpi (Fig. 8b, Online Resource 4).
The expression level of Proteinase Inhibitor I (PIN I) exhibited significant differences across various treatments at different time points. In the control group, PIN I expression remained relatively stable, showing only slight upregulation with fold changes of 1.00, 1.03, and 1.02 at 12, 24, and 48 hpi, respectively. The biocontrol treatment initially resulted in a downregulation of PIN I at 12 hpi (0.37-fold) and 24 hpi (0.86-fold), followed by a substantial upregulation to 5.10-fold at 48 hpi. In contrast, plants inoculated with the pathogen alone displayed a gradual increase in PIN I expression, with fold changes of 0.81, 0.94, and 6.44 at 12, 24, and 48 hpi, respectively. Notably, the combined treatment of biocontrol and pathogen led to the highest upregulation of PIN I, with fold changes of 1.51, 3.04, and a remarkable 15.03 at 12, 24, and 48 hpi, respectively. This indicates that the most significant upregulation of PIN I occurs in the biocontrol-treated plants that were also inoculated with the pathogen, particularly evident at 48 hpi (Fig. 8c, Online Resource 3).
Response of the Proteinase Inhibitor II (PIN II) gene expression differed with each treatment across the different time points. In the control group, expression levels remained relatively consistent with a slight increase at 24 hpi, resulting in fold changes of 1.01 at 12 hpi, 2.10 at 24 hpi, and 1.01 at 48 hpi. The biocontrol treatment led to a gradual upregulation of the gene, with fold changes of 0.86 at 12 hpi, 3.69 at 24 hpi, and 2.33 at 48 hpi. The pathogen treatment caused an initial upregulation of the gene at 12 hpi (1.41-fold), followed by a stable expression at 24 hpi (2.17-fold) and a slight decrease at 48 hpi (1.39-fold). Notably, the combined treatment of biocontrol and pathogen induced the most significant gene expression changes, starting with a downregulation at 12 hpi (0.37-fold), followed by a substantial increase at 24 hpi (2.33-fold), and reaching a remarkable 7.90-fold upregulation at 48 hpi. This observation highlights that the gene expression was most elevated with the biocontrol and pathogen combination, notably at 24 and 48 hpi (Fig. 8d, Online Resource 5).
The Ethylene Response (ETR) gene showed fluctuating expression levels depending on the treatment and the time point. In the control group, gene expression remained relatively stable, with fold changes of 1.11 at 12 hpi, 1.03 at 24 hpi, and 1.06 at 48 hpi. The biocontrol treatment led to a moderate upregulation, showing fold changes of 0.90 at 12 hpi, a significant increase to 2.25 at 24 hpi, and 1.34 at 48 hpi. The pathogen treatment resulted in an initial upregulation to 1.86-fold at 12 hpi, followed by a slight decrease to 1.01-fold at 24 hpi, and an increase again to 2.17-fold at 48 hpi. The most notable changes were observed in the combined treatment of biocontrol and pathogen, which exhibited a downregulation to 0.74-fold at 12 hpi, a substantial upregulation to 2.59-fold at 24 hpi, and a remarkable 5.22-fold increase at 48 hpi. This indicates that the highest gene expression was achieved with the biocontrol and pathogen combination treatment, particularly at 48 hpi (Fig. 8e, Online Resource 9).
Expression profile of MYC2, multicystatin, laccase, WRKY-31, and prosystemin genes
The expression of MYC2 showed significant differences under different treatments at varied time points, starting at 12 hpi. At this time, the Biocontrol + Pathogen treatment exhibited the highest expression level at 1.710-fold, compared to the Pathogen treatment (1.252-fold), the Control treatment (0.633-fold), and the Biocontrol treatment (0.501-fold). By 24 hpi, the Biocontrol + Pathogen treatment continued to show the highest expression at 4.571-fold, surpassing the Biocontrol treatment (2.860-fold), the Pathogen treatment (1.682-fold), and the Control treatment (0.536-fold). At 48 hpi, the Biocontrol + Pathogen treatment maintained the highest expression level at 9.110-fold, significantly higher than the Pathogen treatment (7.031-fold), the Biocontrol treatment (3.757-fold), and the Control treatment (0.965-fold). Overall, the Biocontrol + Pathogen treatment demonstrated the most significant increase in MYC2 expression at all time points (Fig. 9a, Online Resource 10).
Gene expression analysis of the defense-related genes encoding a MYC2, b Multicystatin, c Laccase, d WRKY31, e Prosystemin in tomato var. pusa ruby grown in soil existing different treatments, viz., Control, Biocontrol (T. asperellum strain A10 treated), Pathogen (S. rolfsii strain Sr38 treated), Biocontrol + Pathogen (both). Actin was used as the reference gene. Each data point represents the average of three independent biological replicates. Error bars indicate standard deviation (SD)
Significant variations in the multicystatin gene expression were observed under different treatments, starting from 12 hpi. At this time, the Biocontrol treatment exhibited the highest expression level at 1.803-fold, followed by the Pathogen treatment (1.430-fold), the Biocontrol + Pathogen treatment (1.276-fold), and the Control treatment (1.180-fold). By 24 hpi, the Pathogen treatment had the highest expression at 5.430-fold, significantly higher than the Biocontrol + Pathogen treatment (4.427-fold), the Biocontrol treatment (1.357-fold), and the Control treatment (1.120-fold). At 48 hpi, the Biocontrol + Pathogen treatment showed a dramatic increase in multicystatin expression to 19.088-fold, surpassing both the Biocontrol treatment (4.947-fold) and the Pathogen treatment (4.037-fold), with the Control treatment remaining relatively low at 1.097-fold. The Biocontrol + Pathogen treatment showed the most significant increase in multicystatin expression overall, especially at 48 hpi (Fig. 9b, Online Resource 9).
The laccase gene expression varied significantly across different treatments and time points. At 12 hpi, the highest expression was observed in the Biocontrol treatment at 1.434-fold, while the Control, Pathogen, and Biocontrol + Pathogen treatments showed lower expressions at 1.109-fold, 0.898-fold, and 0.602-fold, respectively. By 24 hpi, the Biocontrol + Pathogen treatment showed a dramatic increase to 18.177-fold, significantly higher than the Biocontrol treatment at 4.621-fold, the Pathogen treatment at 3.609-fold, and the Control at 1.055-fold. At 48 hpi, the Biocontrol + Pathogen treatment maintained the highest expression at 15.031-fold, followed by the Biocontrol treatment at 6.056-fold, the Pathogen treatment at 4.244-fold, and the Control at 1.126-fold. Significant enhancement in laccase expression was observed in the Biocontrol + Pathogen treatment, particularly at 24 and 48 hpi (Fig. 9c, Online Resource 7).
The expression levels of WRKY31 showed notable differences across various treatments and time points. At 12 hpi, the Biocontrol treatment exhibited the highest expression at 2.210-fold, while the Pathogen, Control, and Biocontrol + Pathogen treatments had lower expressions at 1.677-fold, 1.186-fold, and 1.080-fold, respectively. By 24 hpi, WRKY31 expression peaked in the Biocontrol + Pathogen treatment, reaching 5.312-fold, significantly higher than the Pathogen treatment at 4.073-fold, the Biocontrol treatment at 2.287-fold, and the Control at 1.133-fold. At 48 hpi, the expression was most pronounced in the Biocontrol + Pathogen treatment at 19.109-fold, followed by the Biocontrol treatment at 6.708-fold, the Pathogen treatment at 6.374-fold, and the Control at 1.023-fold. The Biocontrol + Pathogen treatment consistently demonstrated the most substantial increase in WRKY31 expression, especially evident at 48 hpi (Fig. 9d, Online Resource 8).
The expression of prosystemin showed significant differences under different treatments at varied time points, starting at 12 hpi. At this time, the Biocontrol treatment had the highest expression level at 1.421-fold, compared to the Control treatment (1.030-fold), the Pathogen treatment (0.796-fold), and the Biocontrol + Pathogen treatment (0.604-fold). By 24 hpi, the Pathogen treatment exhibited the highest expression at 3.844-fold, closely followed by the Biocontrol + Pathogen treatment (3.714-fold), the Biocontrol treatment (3.189-fold), and the Control treatment (1.064-fold). At 48 hpi, the Biocontrol + Pathogen treatment showed a marked increase in prosystemin expression to 13.887-fold, significantly higher than the Biocontrol treatment (9.869-fold), the Pathogen treatment (5.815-fold), and the Control treatment (1.022-fold). The most notable increase in prosystemin expression at 48 hpi was observed with the Biocontrol + Pathogen treatment (Fig. 9e, Online Resource 8).
Discussion
In our research, we employed Trichoderma asperellum strain A10 as the antagonist against S. rolfsii strain Sr38, confirmed through a detailed analysis of both morphological and molecular characteristics. Morphologically, T. asperellum displayed rapid growth, with globose conidia, predominantly paired branches, and ampulliform phialides, aligning with the previous studies indicated that T. asperellum conidia were sub-globose to ovoid, with finely warted ornamentation, as observed through scanning electron microscopy (SEM). The conidiophores showed lateral branching, and the conidial color ranged from olive green to dark green when dries (Sriram et al. 2013). Similarly, light and SEM observations of T. asperellum Ta13 revealed regularly branched, typically paired conidiophores, straight phialides, and globose to sub-globose conidia measuring 2.91 μm × 2.37 μm, with inconspicuous ornamentation (Pandian et al. 2016). Likewise, a microscopic examinations of T. asperellum isolates confirmed the presence of branched, mostly paired conidiophores, ampulliform phialides, and slightly ovoid to globose light green conidia (Shanmugaraj et al. 2023). These observations are consistent with our findings, reinforcing the characteristic morphological traits of T. asperellum.
Relying solely on morphological characteristics for species identification can sometimes be erroneous, uncertain, or unclear due to overlapping features, which may not provide precise results. Therefore, we also conducted molecular characterization based on internal transcribed spacer (ITS) and β-tubulin region sequence analysis for more reliable identification. This method enabled us to accurately identify the T. asperellum species, demonstrating the effectiveness of molecular markers for dependable species determination. This consistent with the previous studies have confirmed that ITS and β-tubulin markers can effectively differentiate between various Trichoderma species (Thokala et al. 2021). Similarly, using a combination of ITS and β-tubulin sequences significantly distinguished all T. asperellum isolates from other species (Shanmugaraj et al. 2023). These observations are consistent with our findings, reinforcing the molecular confirmation of T. asperellum.
The morphology of the S. rolfsii strain Sr38 was characterized by its rapid mycelial growth and the production of abundant, mustard-colored sclerotia on potato dextrose agar (PDA). This study examined morphological traits such as mycelial growth rate, colony color, mycelial dispersion, appearance, and various aspects of sclerotia including color, weight, numbers, arrangement, and maturity days. These findings align with the previous research, which confirmed morphologically and categorized their S. rolfsii isolates into white, extra white, cottony white, light white, and dull white mycelium. The sclerotia color ranged from brown to dark brown, with shapes varying from spherical to oval and irregular. The sclerotial weight ranged from 3.7 to 8.6 mg, and their arrangement was observed to be scattered, peripheral, and central (Mahato and Biswas 2017). Similarly, a rapidly growing, dense mycelium with a white, woolly appearance and smooth edges was observed. Initially, the mycelium produced abundant white sclerotia on PDA medium, which turned brown after 3–4 days of vegetative growth, eventually becoming black (Sikder et al. 2024).
Identifying species based only on morphological traits can often be inaccurate, ambiguous, or confusing due to shared characteristics, leading to imprecise results. Therefore, we also conducted molecular characterization based on TEF 1α and RPB2 region sequence analysis for more reliable identification. These consistent with the previous studies showed that a collection of 238 single-sclerotial S. rolfsii isolates was obtained from diseased sugar beet plants from Chile, Italy, Portugal, and Spain were confirmed molecularly utilizing TEF 1α and RPB2 markers (Remesal et al. 2013). Similarly, all the 50 isolates of S. rolfsii from different locations of India, previously identified based on morphology, were molecularly confirmed by amplifying TEF 1α and RPB2 genomic regions (Shanmugaraj et al. 2024). These observations are consistent with our findings, reinforcing the molecular confirmation of S. rolfsii.
Furthermore, we assessed the biocontrol efficacy of the A10 strain of T. asperellum, along with other antagonistic fungi; against the collar rot pathogen S. rolfsii strain Sr38. In our in vitro antagonistic assays, T. asperellum strain A10 demonstrated significant inhibition of S. rolfsii growth, achieving a remarkable 94.66% reduction in the dual plate assay. These results are consistent with the previous studies, which found that temperature-tolerant isolates of T. asperellum reduced S. rolfsii hyphal growth by 50–68% under in vitro conditions (Poosapati et al. 2014). Moreover, T. asperellum significantly decreased the mycelial growth of Pythium aphanidermatum, Pythium debaryanum, and S. rolfsii Sr3 by 43.57%, 38.16%, and 54.87%, respectively, according to Manjunath et al. (2017). Additionally, the antagonistic effectiveness of various Trichoderma species against the collar rot pathogen was documented, showing inhibition rates ranging from 13.02% to 63.02% among different isolates, with T. asperellum achieving the highest inhibition at 63.02% (Dinesh et al. 2018). Furthermore, isolates Tricho05 and Tricho06 demonstrated significant inhibition rates of 70.37% and 63.33%, respectively, against S. rolfsii mycelial growth (Akash et al. 2022). Similarly, in dual culture assays, T. asperellum isolates significantly inhibited the mycelial growth of Athelia rolfsii, with inhibition percentages ranging from 20.87% to 94.66% (P < 0.05). The A10 isolate exhibited the highest inhibition at 94.66%, followed by A11 and A17, which showed inhibition rates of 82.64% and 81.19%, respectively (Shanmugaraj et al. 2023). Likewise, in a dual culture assay involving S. rolfsii and Trichoderma species, T. asperellum exhibited approximately 54% inhibition of the pathogenic fungi’s growth, followed by T. harzianum with a 47.5% inhibition rate (Sikder et al. 2024). These observations confirm with our findings, highlighting the in vitro antagonistic potential of T. asperellum against S. rolfsii by competition and antibiosis by lytic enzymes as per our previous study (Shanmugaraj et al. 2023).
In our in-planta study conducted on tomato plants (var. Pusa Ruby), the application of T. asperellum to the soil before disease development significantly reduced collar rot incidence. In contrast, plants treated with the pathogen alone exhibited 100% disease incidence with prominent symptoms. The application of A10 strain in infected soil resulted in lower disease incidence (13.83%) in pre-inoculation and 19.40% in post-inoculation compared to the pathogen-alone treatment. These findings are consistent with the previous research found that the application of T. asperellum reduced the severity of disease symptoms caused by S. rolfsii in onions as a result of the constitutive and induced activity of glucanase, chitinase, and peroxidase enzymes (Guzman-valle et al. 2014). Similarly, under greenhouse conditions, soil treatment with T. asperellum TR08 resulted in the lowest incidence of collar rot disease at 9.78%, indicating its potential as an effective biological control agent for collar rot in tomatoes (Islam et al. 2017). Likewise, applying a mixture of T. asperellum strains to cucumber plants significantly reduced the incidence and severities of Fusarium root and stem rot (FRSR) disease. The study found that treating cucumber plants with the individual antagonistic strains TAS23 and TAS27 of T. asperellum effectively delayed the onset of FRSR in greenhouse trials. However, the combined treatment with the strain mixture TASMix (TAS23 + TAS27) had a synergistic effect, leading to the greatest reduction (P < 0.05) in disease incidence and severity index, by 51% and 59.6%, respectively (El-komy et al. 2022). Similarly, collar rot in tomato was significantly inhibited by all the tested T. asperellum isolates ranging from a maximum of 86.17% and 80.60% disease inhibition from the A10 isolate to the minimum disease inhibition of 11.11% and 8.33% from the isolate A15 during pre- and post-inoculation treatment in the green house pot assay (Shanmugaraj et al. 2023). These confirms with our findings, highlighting the biopriming potential of T. asperellum against S. rolfsii by inducing plant defenses.
In our current research, we conducted qPCR analysis to profile the expression of defense-related genes. Notably, the expression levels of pathogenesis-related genes, such as PR1, PR2, PR3, PR5, and PR12, exhibited a significant increase over time following treatment. The highest fold expression was observed in plants treated with both the biocontrol agent and the pathogen, particularly at 48 h post-inoculation. Subsequently, the biocontrol agent alone and the pathogen-alone treatments also showed elevated gene expression compared to the control group. These findings align with the previous research that demonstrated a significant increase in PR1, PR2, and PR3 expression in the stems and roots of plants treated with T. asperellum and infected with Fusarium oxysporum f. sp. lycopersici (Fol), compared to those treated with Fol alone (El-komy et al. 2016). Furthermore, tomato plants treated with Trichoderma isolates in conjunction with Fol exhibited the highest expression levels of pathogenesis-related genes compared to the control group, which consisted of plants treated with the pathogen only (Sallam et al. 2019). Similarly, in plants primed with T. erinaceum, there was an observed induction of PR proteins such as chitinases and glucanases. Specifically, increased expression of PR-3 proteins (chitinases) PR-2 proteins (glucanases) was noted within the initial 0–24 h after bioprimed treatment with T. erinaceum, followed by challenge with Fusarium oxysporum f. sp. lycopersici (Fol) + T. erinaceum in root tissues (Aamir et al. 2019).
Likewise, in an experiment where tomato seedlings were treated with T. asperellum strain DQ-1, the PR2 gene in roots showed its highest activity at 6 h after infection, declining afterward, while in leaves, its activity peaked at 24 h and then decreased, with a secondary increase noted at 48 h. In contrast, when seedlings were first exposed to strain DQ-1 for 24 h before being sprayed with B. cinerea, PR2 gene activity increased steadily in both roots and leaves up to 24 h, reaching its peak before gradually declining (Wang et al. 2021). Similarly, in chilli pepper seeds treated with biopriming, the genes PR-2 and PR-5 showed significant expression when exposed to pathogen challenges compared to untreated seeds (control). PR-2 expression was notably higher in seeds primed with T. asperellum and T. harzianum together, followed by seeds primed with T. asperellum alone, and those inoculated with C. truncatum, all showing increased expression during pathogen challenge at 2 days post-inoculation (dpi). PR-5 expression peaked in seeds primed with both T. asperellum and T. harzianum, followed by T. asperellum alone, T. harzianum, and pathogen-challenged seeds, compared to untreated and unchallenged samples, showing a decrease by 4 dpi. These findings suggest that biopriming enhances the activation of defense genes in chilli pepper seeds during pathogen encounters (Yadav et al. 2023). These observations confirm with our findings, highlighting the expression of pathogenesis-related genes upon biopriming with Trichoderma that induce resistance.
In our current study, we assessed the expression levels of several genes, including Thioredoxin peroxidase, Catalase, Isochorismate synthase, Polyphenol oxidase, and phenylalanine ammonia lyase, and noted a significant increase in their expression over time following treatment. The most substantial fold expression was observed in plants subjected to both the biocontrol agent and the pathogen, particularly at the 48-h post-inoculation mark. These findings are consistent with prior research that demonstrated the activation of systemic acquired resistance (SAR) and Induced systemic resistance (ISR) pathways in tomato plants treated with T. asperellum strain DQ1, leading to enhanced resistance, as indicated by the elevated expression of Thioredoxin peroxidase genes (Wang et al. 2021). Similarly, plants treated with T. virens, especially the susceptible Ratna variety, showed the highest catalase expression at 1 day post-inoculation (dpi), with slightly lower levels at 2 and 4 dpi. However, when T. virens was combined with R. solani inoculation, catalase expression decreased consistently over all 4 days of observation. Plants inoculated only with R. solani showed decreased catalase expression up to 3 dpi, followed by a significant increase at 4 dpi. On average, catalase expression was lower in all treatments compared to the control plants. These results suggest that T. virens treatment alone or in combination with R. solani affects catalase expression differently in susceptible plants, highlighting potential interactions between biocontrol agents and pathogens in plant defense mechanisms (Dubey et al. 2018).
Likewise, Moran-Diez et al. (2020) also reported that plants inoculated with Pseudomonas syringae pv. tomato DC3118 and treated with Trichoderma exhibited heightened expression of SA-related defense genes, including pathogenesis-related genes, Isochorismate synthase (ICS1), and phenylalanine ammonia lyase (PAL), compared to mock-treated plants. Furthermore, in a qPCR assay, in untreated tomato plants, catalase gene expression peaked at 48 h and 96 h, with no significant changes observed by 72 h. Compared to controls, plants treated with Fusarium oxysporum fsp. lycopersici (Fol) alone showed 2.33- to 4.94-fold increases over 24 to 96 h, while Fol + T. harzianum (Th) treatments exhibited higher fold changes of 3.73- to 6.33-fold. PAL gene expression in controls varied, peaking at 24 and 72 h but declining at 48 and 96 h. In contrast, both Fol and Fol + Th treatments consistently increased PAL expression, with peaks at 72 h (4.97-fold for Fol and 6.73-fold for Fol + Th). PPO gene expression decreased non-significantly in control plants at 24, 72, and 96 h but increased at 48 h. In treated plants, PPO expression consistently rose across all time points, peaking at 48 h (Zehra et al. 2023). These studies aligned with our findings by highlighting the mechanisms of resistance in tomato plants biopriming with Trichoderma against the plant pathogens, contributing to enhanced defense responses.
We conducted an analysis of gene expression levels, which encompassed various genes, including Lipoxygenase A (LoxA), Lipoxygenase C (LoxC), Proteinase inhibitor I (PIN I), Proteinase inhibitor II (PIN II), and Ethylene response I (ETR I). Notably, we observed a significant rise in their expression levels over time following a specific treatment regimen. The most substantial increase in gene expression was observed in plants that had been exposed to both the biocontrol agent and the pathogen, particularly at the 48-h post-inoculation stage. These results are consistent with previous research and highlight that the transcriptional activation of JA-related genes, such as PIN I and PIN II, corresponds to the heightened resistance to Botrytis cinerea seen in tomato plants treated with the T. harzianum strain T22. Additionally, two other JA-responsive genes, TomLoxA and TomLoxC, often exhibited increased expression that coincided with Trichoderma-mediated tolerance to B. cinerea, as reported by Tucci et al. (2011). Furthermore, in non-challenge inoculations, tomato seedling roots were treated with a spore suspension of T. asperellum strain DQ-1 (+ T − P): The ETR1 gene expression in the roots peaked at 12 h post-inoculation (hpi), dropped to their lowest levels at 24 hpi, and then increased again. For challenge inoculations, the tomato seedling roots were treated with strain DQ-1 for 24 h and then spray-inoculated with B. cinerea. In this case, the ETR1 gene expression significantly increased in both roots and leaves from 12 hpi, reaching its highest point at 24 hpi, which is involved in the ET/JA signaling pathway in plants. This activation of ETR1 gene expression contributed to the induction of resistance against B. cinerea, as reported by Wang et al. (2021).
Similarly, in potato roots exposed to R. solani in the presence of T. harzianum, there was a notable induction of the Lox gene. At 24 h post-inoculation (hpi), the expression of the Lox gene was significantly elevated, reaching a value of 6.03 ± 1.19 (Gallou et al. 2009). Likewise, the observed increase in the expression of TomLoxA and PIN II genes in tomato roots, particularly under stress conditions with Trichoderma presence, suggests an enhanced synthesis of jasmonic acid (JA). This upregulation indicates that Trichoderma may help mitigate stress in tomatoes by boosting JA production, which is known to play a crucial role in plant stress responses and defense mechanisms (Racic et al. 2018). Furthermore, the expression of LOX1, LOX2 genes were peaked in T. longibrachiatum H9 inoculated, Botrytis cinerea challenged plants (+ T + B) than non-inoculated, challenged plants (− T + B) at 96 hpi in cucumber plants (Yuan et al. 2019).
We conducted an assessment of gene expression levels, which encompassed a variety of genes, including Laccase, Prosystemin, Multicystatin, WRKY31, and MYC2. Remarkably, we observed a substantial increase in their expression levels over time following a specific treatment protocol. The most pronounced elevation in gene expression was observed in plants that had been subjected to both the biocontrol agent and the pathogen, particularly at the 48hpi phase. These findings are consistent with prior research, noted that an increased expression of the JA-responsive genes PI II, MC, and PS, which encode proteinase inhibitor II, multicystatin, and prosystemin respectively, was observed in plants treated with T. harzianum compared to untreated plants. Interestingly, in the absence of a pathogen, T. harzianum-inoculated plants exhibited little induction of these genes. This suggests that the presence of T. harzianum primes the plants for a stronger JA-dependent defense response, which is activated upon pathogen attack, thereby enhancing resistance against B. cinerea (Martinez-Medina et al. 2013). Likewise, noted differential expression of genes encoding laccase in tomato roots induced by T. harzianum (De Palma et al. 2019). Similarly, when analyzing the time-dependent expression patterns in tomato plants under four different treatments (unprimed, Fol challenged, T. erinaceum primed, and Fol + T. erinaceum), they found that the most substantial upregulation occurred in the transcript profile of SlWRKY31 in T. erinaceum bioprimed-treated plants at 24 h post-inoculation, with a remarkable 16.51-fold increase (Aamir et al. 2019).
In summary, our study suggest that T. asperellum strain A10 exhibits promising potential as a biocontrol agent for managing collar rot disease induced by the pathogen S. rolfsii. The mechanism involved behind the defense priming in tomato against S. rolfsii by Trichoderma spp. involves the induction and accumulation of pathogenesis-related (PR) proteins (Yedidia et al. 2000), enhancement of reactive oxygen species (ROS) generation, and lignification (Patel et al. 2017; Meshram et al. 2019). PR 1, 2, 3, and 5 genes play crucial roles in plant defense, with PR 1 involved in the salicylic acid-mediated pathway (Kumari et al. 2019) and PR 2, 3, and 5 encoding enzymes that degrade fungal cell walls (Lopes et al. 2023). Catalase (CAT) regulate ROS scavenging (Dubey et al. 2018) and WRKY transcription factors involved in JA-responsive gene expression (SlWRKY31) (Lippok et al. 2007; Brotman et al. 2013; Liu et al. 2015; Li et al. 2016). Trichoderma-induced systemic resistance (TISR) involves the priming of jasmonic acid (JA)-dependent defenses, increasing the expression of PIN, TomLoxA, and LoxC genes (Tucci et al. 2011; Chehab and Braam 2012; Martinez Medina et al. 2013; Racic et al. 2018). The JA-related priming also involves genes, such as multicystatin, prosystemin, PinI, and PinII, which enhance resistance against B. cinerea (Tucci et al. 2011; Segarra et al. 2013; Brotman et al. 2013). Additional genes, such as ETR (ethylene receptor), TPx (thioredoxin peroxidase), ICS (salicylic acid biosynthesis), PPO (polyphenol oxidase), PAL (phenylalanine ammonia lyase), Laccase (lignification), and MYC2 (jasmonate signaling), further enhance the plant’s defense against biotic stresses by regulating various defensive compounds and responses (Nguyen et al. 2018; Abouzeid et al. 2019; Ali and Baek 2020; Deshpande and Mitra 2023). Collectively, these mechanisms enable tomato plants to mount an effective defense against S. rolfsii through a robust network of biochemical and molecular responses.
Conclusion
In recent years, biological control has garnered increasing attention as a promising alternative to chemical methods for managing plant pathogens. In our current investigation, T. asperellum strain A10 has emerged as a highly promising antagonist for effectively controlling S. rolfsii, as demonstrated in both laboratory and greenhouse conditions. Furthermore, our study indicates that the application of T. asperellum prior to disease establishment significantly reduces the incidence of collar rot. Our research clearly illustrates that biopriming tomato plants with T. asperellum strain A10 leads to a significant upregulation in the expression of defense-related genes encompass PR1, PR2, PR3, PR5, PR12, Thioredoxin peroxidase, Catalase, Polyphenol oxidase, Phenylalanine ammonia lyase, Isochorismate synthase, Laccase, Prosystemin, Multicystatin, WRKY31, MYC2, Lipoxygenase A, Lipoxygenase C, Proteinase Inhibitor I, Proteinase inhibitor II, and Ethylene response 1, involved in various signaling pathways like SA (salicylic acid)-mediated and JA/ET (Jasmonic acid/ethylene)-mediated. Collectively, our findings strongly suggest that T. asperellum strain A10, along with its biopriming effect, holds immense potential for effectively managing collar rot diseases by stimulating the expression of a wide array of defense-related genes. Given that the performance of biological control agents (BCAs) can vary under diverse climatic conditions, it is imperative to assess their efficacy across multiple locations and crops to validate their robustness and antagonistic capabilities.
Data availability
The sequences obtained in this study were submitted to GenBank with the following accession numbers: ITS (OR137590) and β-tubulin (OR193710) for Trichoderma asperellum, and TEF 1α (OR335644) and RPB2 (OR227561) for Sclerotium rolfsii.
References
Aamir M, Kashyap SP, Zehra A, Dubey MK, Singh VK, Ansari WA, Upadhyay RS, Singh S (2019) Trichoderma erinaceum biopriming modulates the WRKYs defense programming in tomato against the Fusarium oxysporum f.sp. lycopersici (Fol) challenged condition. Front Plant Sci 10:911. https://doi.org/10.3389/fpls.2019.00911
Abdelrahman M, Abdel-Motaal F, El-Sayes M, Jogaiah S, Shigyo M, Ito S, Tran LP (2016) Dissection of Trichoderma longibrachiatum-induced defense in onion (Allium cepa L.) against Fusarium oxysporum f.sp. cepa by target metabolite profiling. Plant Sci 246:128–138. https://doi.org/10.1016/j.plantsci.2016.02.008
Abouzeid S, Beutling U, Selmar D (2019) Stress-induced modification of indole alkaloids: phytomodificines as a new category of specialized metabolites. Phytochem 159:102–107. https://doi.org/10.1016/j.phytochem.2018.12.015
Akash AU, Ramya V, Uma Devi G, Pushpavalli SNCVL, Triveni S (2022) Antagonist activities of native rhizosphere micro-flora against groundnut stem rot pathogen, Sclerotium rolfsii Sacc. Egypt J Biol Pest Control 32(1):133. https://doi.org/10.1186/s41938-022-00631-6
Al-Ani LKT, Aguilar-Marcelino L, Fiorotti J, Sharma V, Sarker MS, Furtado EL, Wijayawardene NN, Herrera-Estrella A (2020) Biological control agents and their importance for the plant health. In: Vimal SJ, Singh JS (eds) Microbial services in restoration ecology. Elsevier, pp 13–36. https://doi.org/10.1016/B978-0-12-819978-7.00002-6
Ali MS, Baek KH (2020) Jasmonic acid signaling pathway in response to abiotic stresses in plants. Int J Mol Sci 21(2):621. https://doi.org/10.3390/ijms21020621
Boccardo NA, Segretin ME, Hernandez I, Mirkin FG, Chacon O, Lopez Y, Borras-Hidalgo O, Bravo-Almonacid FF (2019) Expression of pathogenesis-related proteins in transplastomic tobacco plants confers resistance to filamentous pathogens under field trials. Sci Rep 9(1):2791. https://doi.org/10.1038/s41598-019-39568-6
Brotman Y, Landau U, Cuadros-Inostroza A, Takayuki T, Fernie AR, Chet I, Viterbo A, Willmitzer L (2013) Trichoderma-plant root colonization: escaping early plant defense responses and activation of the antioxidant machinery for saline stress tolerance. PLoS Pathog 9(3):e1003221. https://doi.org/10.1371/journal.ppat.1003221
Chandrashekar N, Ali S, Grover A (2018) Exploring expression patterns of PR-1, PR-2, PR-3, and PR-12 like genes in Arabidopsis thaliana upon Alternaria brassicae inoculation. 3 Biotech 8:1–10. https://doi.org/10.1007/s13205-018-1259-2
Chehab EW, Braam J (2012) Jasmonates in plant defense responses. In: Witzany G, Baluska F (eds) Biocommunication of plants. Signaling and communication in plants, vol 14. Springer, Berlin, pp 67–88. https://doi.org/10.1007/978-3-642-23524-5_5
Cullings KW (1992) Design and testing of a plant-specific PCR primer for ecological and evolutionary studies. Mol Ecol 1:233–240. https://doi.org/10.1111/j.1365-294X.1992.tb00182.x
De Palma M, Salzano M, Villano C, Aversano R, Lorito M, Ruocco M, Docimo T, Piccinelli AL, Agostino N, Tucci M (2019) Transcriptome reprogramming, epigenetic modifications and alternative splicing orchestrate the tomato root response to the beneficial fungus Trichoderma harzianum. Hortic Res 6:5. https://doi.org/10.1038/s41438-018-0079-1
Deshpande S, Mitra S (2023) β-Cyclocitral-mediated metabolic changes optimize growth and defense responses in Solanum lycopersicum L. Metabolites 13(3):329. https://doi.org/10.3390/metabo13030329
Dinesh K, Sinha B, Bai AT (2018) In vitro studies on efficacy of various Trichoderma spps. against collar rot of tomato caused by Sclerotium rolfsii Sacc. in Manipur. Int J Chem Stud 6(6):1654–1656
Dubey SC, Tripathi A, Tak R (2018) Expression of defense-related genes in mung bean varieties in response to Trichoderma virens alone and in the presence of Rhizoctonia solani infection. 3 Biotech 8:1–8. https://doi.org/10.1007/s13205-018-1453-2
El-Komy MH, Saleh AA, Ibrahim YE, Hamad YK, Molan YY (2016) Trichoderma asperellum strains confer tomato protection and induce its defense-related genes against the Fusarium wilt pathogen. Trop Plant Pathol 41:277–287. https://doi.org/10.1007/s40858-016-0098-0
El-Komy MH, Al-Qahtani RM, Ibrahim YE, Almasrahi AA, Al-Saleh MA (2022) Soil application of Trichoderma asperellum strains significantly improves Fusarium root and stem rot disease management and promotes growth in cucumbers in semi-arid regions. Eur J Plant Pathol 162:637–653. https://doi.org/10.1007/s10658-021-02427-0
Galletti S, Paris R, Cianchetta S (2020) Selected isolates of Trichoderma gamsii induce different pathways of systemic resistance in maize upon Fusarium verticillioides challenge. Microbiol Res 233:126406. https://doi.org/10.1016/j.micres.2019.126406
Gallou A, Cranenbrouck S, Declerck S (2009) Trichoderma harzianum elicits defence response genes in roots of potato plantlets challenged by Rhizoctonia solani. Eur J Plant Pathol 124:219–230. https://doi.org/10.1007/s10658-008-9407-x
Garcia EF (1991) Screening of fungal antagonist to control Sclerotium cepivorum. In: Jensen DF (ed) New approaches in biological control of soil borne diseases. IOBCM/PRS Bull, Copenhagen, pp 79–81
Guzman-Valle P, Bravo-Luna L, Montes-Belmont R, Guigon-Lopez C, Sepulveda-Jimenez G (2014) Induction of resistance to Sclerotium rolfsii in different varieties of onion by inoculation with Trichoderma asperellum. Eur J Plant Pathol 138:223–229. https://doi.org/10.1007/s10658-013-0336-y
Hjeljord L, Tronsmo A, Harman GE, Kubicek CP (1998) Trichoderma and Gliocladium in biological control: an overview. In: Kubicek CP, Harman GE (eds) Trichoderma and Gliocladium, enzymes, biological control and commercial applications. Taylor and Francis, London, pp 131–151. https://doi.org/10.1201/9781482267945
Howell CR (1998) The role of antibiosis in biocontrol. In: Harman GE, Kubicek CP (eds) Trichoderma and Gliocladium, vol 2. Taylor and Francis, London, pp 173–184
Howell CR (2003) Mechanisms employed by Trichoderma species in the biological control of plant diseases: the history and evolution of current concepts. Plant Dis 87:4–10. https://doi.org/10.1094/pdis.2003.87.1.4
Islam MM, Hossain DM, Nonaka M, Harada N (2017) Biological control of tomato collar rot induced by Sclerotium rolfsii using Trichoderma species isolated in Bangladesh. Arch Phytopathol Plant Prot 50(3–4):109–116. https://doi.org/10.1080/03235408.2016.1265243
Jayanna SK, Umesha S (2017) Enhancement of the expression of defense genes in tomato against Ralstonia solanacearum by N-octanoyl-L-homoserine lactone. Afr J Microbiol Res 11(5):194–203. https://doi.org/10.5897/AJMR2016.8370
Kubicek CP, Pentilla ME (1998) Regulation of production of plant polysaccharide degrading enzymes by Trichoderma. In: Harman GE, Kubicek CP (eds) Trichoderma and Gliocladium. Enzymes biological control and commercial applications, vol 2. Taylor and Francis Ltd, London, pp 49–71
Kumari P, Kumari R, Sharma B, Prasad SK, Singh RK (2019) Abiotic stress response of field crops: recent approach. Int J Curr Microbiol App Sci 8(4):1761–1769. https://doi.org/10.20546/ijcmas.2019.804.205
Le CN, Mendes R, Kruijt M, Raaijmakers JM (2012) Genetic and phenotypic diversity of Sclerotium rolfsii in groundnut fields in central Vietnam. Plant Dis 96(3):389–397. https://doi.org/10.1094/PDIS-06-11-0468
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Lopes NDS, Santos AS, De Novais DPS, Pirovani CP, Micheli F (2023) Pathogenesis-related protein 10 in resistance to biotic stress: progress in elucidating functions, regulation and modes of action. Front Plant Sci 14:1193873. https://doi.org/10.3389/fpls.2023.1193873
Mahato A, Biswas MK (2017) Cultural, morphological and pathogenic variability of different isolates of Sclerotium rolfsii obtained from rice-tomato-rice cropping system of undulating red and lateritic zone of West Bengal, India. Int J Curr Microbiol App Sci 6(3):1843–1851. https://doi.org/10.20546/ijcmas.2017.603.210
Mahato A, Biswas MK, Patra S (2017) Prevalence of collar rot of tomato caused by Sclerotium rolfsii (Sacc.) under the red and lateritic zone of West Bengal, India. Int J Curr Microbiol Appl Sci 6(11):3231–3236. https://doi.org/10.20546/ijcmas.2017.611.378
Manjunath M, Singh A, Tripathi AN, Prasanna R, Rai AB, Singh B (2017) Bioprospecting the fungicides compatible Trichoderma asperellum isolate effective against multiple plant pathogens in vitro. J Environ Biol 38:553. https://doi.org/10.22438/jeb/38/4/mrn-339
Martinez-Medina A, Fernandez I, Sanchez-Guzman MJ, Jung SC, Pascual JA, Pozo MJ (2013) Deciphering the hormonal signalling network behind the systemic resistance induced by Trichoderma harzianum in tomato. Front Plant Sci 4:206. https://doi.org/10.3389/fpls.2013.00206
Meshram S, Patel JS, Yadav SK, Kumar G, Singh DP, Singh HB, Sarma BK (2019) Trichoderma mediate early and enhanced lignifications in chickpea during Fusarium oxysporum f.sp. ciceris infection. J Basic Microbiol 59(1):74–86. https://doi.org/10.1002/jobm.201800212
Moran-Diez ME, Tranque E, Bettiol W, Monte E, Hermosa R (2020) Differential response of tomato plants to the application of three Trichoderma species when evaluating the control of Pseudomonas syringae populations. Plants 9(5):626. https://doi.org/10.3390/plants9050626
Nguyen HC, Lin KH, Ho SL, Chiang CM, Yang CM (2018) Enhancing the abiotic stress tolerance of plants: from chemical treatment to biotechnological approaches. Physiol Plant 164(4):452–466. https://doi.org/10.1111/ppl.12812
Pandian RTP, Raja M, Kumar A, Sharma P (2016) Morphological and molecular characterization of Trichoderma asperellum strain Ta13. Indian Phytopathol 69:297–303. https://epubs.icar.org.in/index.php/IPPJ/article/view/60166. Accessed 12 April 2024
Paparizas GC (1985) Trichoderma and Gliocladium: biology, ecology and potential for biocontrol. Annu Rev Phytopathol 23:23–54. https://doi.org/10.1146/annurev.py.23.090185.000323
Patel JS, Kharwar RN, Singh HB, Upadhyay RS, Sarma BK (2017) Trichoderma asperellum (T42) and Pseudomonas fluorescens (OKC)-enhances resistance of pea against Erysiphe pisi through enhanced ROS generation and lignifications. Front Microbiol 8:306. https://doi.org/10.3389/fmicb.2017.00306
Pimentel MF, Arnao E, Warner AJ, Subedi A, Rocha LF, Srour A, Bond JP, Fakhoury AM (2020) Trichoderma isolates inhibit Fusarium virguliforme growth, reduce root rot, and induce defense-related genes on soybean seedlings. Plant Dis 104(7):1949–1959. https://doi.org/10.1094/PDIS-08-19-1676-RE
Poosapati S, Ravulapalli PD, Tippirishetty N, Vishwanathaswamy DK, Chunduri S (2014) Selection of high temperature and salinity tolerant Trichoderma isolates with antagonistic activity against Sclerotium rolfsii. Springerplus 3:1–11. https://doi.org/10.1186/2193-1801-3-641
Racic G, Vukelic I, Prokic L, Curcic N, Zoric M, Jovanovic L, Pankovic D (2018) The influence of Trichoderma brevicompactum treatment and drought on physiological parameters, abscisic acid content and signalling pathway marker gene expression in leaves and roots of tomato. Ann Appl Biol 173(3):213–221. https://doi.org/10.1111/aab.12454
Remesal E, Landa BB, Jimenez-Gasco MDM, Navas-Cortes JA (2013) Sequence variation in two protein-coding genes correlates with mycelial compatibility groupings in Sclerotium rolfsii. Phytopathol 103(5):479–487. https://doi.org/10.1094/PHYTO-07-12-0151-R
Safaie-Farahani A, Taghavi SM (2017) Transcript analysis of some defense genes of tomato in response to host and non-host bacterial pathogens. Mol Biol Res Commun 6(4):177. https://doi.org/10.22099/mbrc.2017.25600.1273
Sallam NM, Eraky AM, Sallam A (2019) Effect of Trichoderma spp. on Fusarium wilt disease of tomato. Mol Biol Rep 46:4463–4470. https://doi.org/10.1007/s11033-019-04901-9
Segarra G, Elena G, Trillas I (2013) Systemic resistance against Botrytis cinerea in Arabidopsis triggered by an olive marc compost substrate requires functional SA signalling. Physiol Mol Plant Pathol 82:46–50. https://doi.org/10.1016/j.pmpp.2013.02.002
Shanmugaraj C, Kamil D, Kundu A, Singh PK, Das A, Hussain Z, Gogoi R, Shashank PR, Gangaraj R, Chaithra M (2023) Exploring the potential biocontrol isolates of Trichoderma asperellum for management of collar rot disease in tomato. Horticulturae 9(10):1116. https://doi.org/10.3390/horticulturae9101116
Shanmugaraj C, Kamil D, Gogoi R, Das A, Kumar GS, Hussain Z, Singh PK, Iquebal MA (2024) Characterizing genetic diversity of Sclerotium rolfsii isolates by biomapping of mycelial compatibility groupings and multilocus sequence analysis. World J Microbiol Biotechnol 40(8):251. https://doi.org/10.1007/s11274-024-04049-0
Shavit R, Ofek-Lalzar M, Burdman S, Morin S (2013) Inoculation of tomato plants with rhizobacteria enhances the performance of the phloem-feeding insect Bemisia tabaci. Front Plant Sci 4:306. https://doi.org/10.3389/fpls.2013.00306
Shoresh M, Yedidia I, Chet I (2005) Involvement of jasmonic acid/ethylene signaling pathway in the systemic resistance induced in cucumber by Trichoderma asperellum T203. Phytopathol 95:76–84. https://doi.org/10.1094/PHYTO-95-0076
Sikder MM, Ahmmed MS, Akter P, Shetu FA, Akhter B, Alam NB, Bhowmik DD, Alam MN (2024) First report of Agroathelia rolfsii (Sclerotium rolfsii Sacc.) causing basal stem rot disease on sunflower in Bangladesh. Asian J Biol Sci 17(3):448–461. https://doi.org/10.3923/ajbs.2024.448.461
Singh A, Singh VK, Dwivedy AK, Deepika TS, Dwivedi A, Dubey NK (2020) Biological control of plant diseases: opportunities and limitations. In: Varma A, Tripathi S, Prasad R (eds) Plant microbiome paradigm. Springer, Cham, pp 121–146. https://doi.org/10.1007/978-3-030-50395-6_7
Sriram S, Savitha MJ, Rohini HS, Jalali SK (2013) The most widely used fungal antagonist for plant disease management in India, Trichoderma viride is Trichoderma asperellum as confirmed by oligonucleotide barcode and morphological characters. Curr Sci 104(10):1332–1340
Thokala P, Narayanasamy P, Kamil D, Choudhary SP (2021) Polyphasic taxonomy of Indian Trichoderma species. Phytotaxa 502:1–27. https://doi.org/10.11646/phytotaxa.502.1.1
Tucci M, Ruocco M, De Masi L, De Palma M, Lorito M (2011) The beneficial effect of Trichoderma spp. on tomato is modulated by the plant genotype. Mol Plant Pathol 12(4):341–354. https://doi.org/10.1111/j.1364-3703.2010.00674.x
Walters DR, Ratsep J, Havis ND (2013) Controlling crop diseases using induced resistance: challenges for the future. J Exp Bot 64(5):1263–1280. https://doi.org/10.1093/jxb/ert026
Wang R, Chen D, Khan RAA, Cui J, Hou J, Liu T (2021) A novel Trichoderma asperellum strain DQ-1 promotes tomato growth and induces resistance to gray mold caused by Botrytis cinerea. FEMS Microbiol Lett 368(20):fnab140. https://doi.org/10.1093/femsle/fnab140
Yadav M, Divyanshu K, Dubey MK, Rai A, Kumar S, Tripathi YN, Shukla V, Upadhyay RS (2023) Plant growth promotion and differential expression of defense genes in chilli pepper against Colletotrichum truncatum induced by Trichoderma asperellum and T. harzianum. BMC Microbiol 23(1):54. https://doi.org/10.1186/s12866-023-02789-x
Yedidia I, Benhamou N, Kapulnik Y, Chet I (2000) Induction and accumulation of PR proteins activity during early stages of root colonization by the mycoparasite Trichoderma harzianum strain T-203. Plant Physiol Biochem 38(11):863–873. https://doi.org/10.1016/S0981-9428(00)01198-0
Yoshioka Y, Ichikawa H, Naznin HA, Kogure A, Hyakumachi M (2011) Systemic resistance induced in Arabidopsis thaliana by Trichoderma asperellum SKT-1, a microbial pesticide of seed borne diseases of rice. Pest Manag Sci 68:60–66. https://doi.org/10.1002/ps.2220
Yuan M, Huang Y, Ge W, Jia Z, Song S, Zhang L, Huang Y (2019) Involvement of jasmonic acid, ethylene and salicylic acid signaling pathways behind the systemic resistance induced by Trichoderma longibrachiatum H9 in cucumber. BMC Genom 20:1–13. https://doi.org/10.1186/s12864-019-5513-8
Zehra A, Aamir M, Dubey MK, Ansari WA, Meena M, Swapnil P, Upadhyay RS, Ali MA, Al-Ghamdi AA, Lee J (2023) Enhanced protection of tomato against Fusarium wilt through biopriming with Trichoderma harzianum. J King Saud Univ Sci 35(2):102466. https://doi.org/10.1016/j.jksus.2022.102466
Acknowledgements
The authors are thankful to the Director, ICAR-Indian Agricultural Research Institute for providing support, encouragement, and facilities during the course of this investigation. First author is grateful to the ICAR-IARI for providing SRF fellowship toward the Ph.D. program.
Funding
The authors have not disclosed any funding. This research did not receive any specific grant from funding agencies in the public, commercial, or not for profit sectors.
Author information
Authors and Affiliations
Contributions
C.S. and D.K. were involved in the conceptualization of the project, study design, and critical inputs. C.S. contributed to the lab work, statistical analysis, and wrote the first draft. C.S., D.K., A.D., R.G., M.A.I., P.R.S., and Z.H. finalized the outline and prepared schematics. P.K.S. and Z.H. helped in carries out an in-planta experiment. C.S. and R.P. carried out Gene expression analysis. C.S., P.R.S., M.A.I., A.D., R.G., and K.N. were helped with statistical analysis and editing of the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declaring that this paper has not been submitted to any other journal for simultaneous consideration. Not been published previously and no manipulation of data has been done. There was no conflict of interest among authors, co-authors, and the institute where the work has been carried out.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shanmugaraj, C., Kamil, D., Parimalan, R. et al. Deciphering the defense response in tomato against Sclerotium rolfsii by Trichoderma asperellum strain A10 through gene expression analysis. 3 Biotech 14, 210 (2024). https://doi.org/10.1007/s13205-024-04040-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-024-04040-4