Abstract
Grey mould caused by Botrytis cinerea leads to severe economic loss on commercial tomato production. Application of beneficial microorganism offers an eco-friendly alternative for mitigation of tomato fungal disease damage, considering negative influences of fungicides. In the present study, an antagonistic Trichoderma afroharzianum isolate TM24 was evaluated for its biocontrol potential on tomato grey mould. The isolate TM24 showed obviously antagonistic effect on B. cinerea mycelium growth and production of glucanase and chitinase. Leaf spraying with spore suspension of isolate TM24 showed a biocontrol efficiency of over 54% against tomato grey mould in greenhouse pot experiment. The activities of plant defense-related enzymes including polyphenol oxidase, phenylalanine ammonialyase, superoxide dismutase, and peroxidase were all increased to varying degrees in tomato leaves after isolate TM24 treatment. Transcriptome analysis showed that, a total of 1941, 1753 and 38 differentially expressed genes (DEGs) were obtained at 24, 48 and 72 hpi, respectively, in tomato leaves pretreated with T. afroharzianum TM24, and then challenged with B. cinerea inoculation. The DEGs were mainly enriched in MAPK signaling pathway and plant hormones signal transduction pathway. Multiple genes that regulated crucial nodes of defense-related pathways, like flavonoid, phenylpropanoid, jasmonic acid and ethylene metabolisms were also identified, which may have positive correlations with the biocontrol potential of isolate TM24 in tomato plants. These promising results provided valuable information on using T. afroharzianum TM24 as a beneficial biocontrol agent in tomato grey mould management.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Grey mould caused by Botrytis cinerea Pers. ex Fr., is one of the most serious fungal diseases in commercial tomato cultivation worldwide [1]. B. cinerea is a ubiquitous pathogen that causes serious loss on greenhouse plants, and usually attacks leaves, flowers, fruits, sometimes can grow through the petiole into the stem. Fungicides play major roles in management of tomato grey mould in field or greenhouse facility [2]. Considering the emergence of fungicide-resistant pathogens and public concerns regarding negative impacts of the chemicals, biological control may be an opportune alternative in green protection of the fungal diseases including grey mould in tomato plantation [1]. During the past few decades, several beneficial microorganisms such as Bacillus sp. [3], Pseudomonas sp. [4], Trichoderma sp. [5], Streptomyces philanthi [6] and Simplicillium lamellicola [7] have been screened for biocontrol of grey mould in different plants. However, since B. cinerea is a typical necrotrophic fungus with wide-ranging hosts and great genetic diversity [8], it is imperative to explore more beneficial and effective microbial strains for nature-friendly biocontrol approach.
It is widely known that, fungal species belonging to the genus Trichoderma have good adaptability to various ecological environments due to their effective reproductive ability and diverse metabolic pattern [9]. Many Trichoderma species act as effective biocontrol agents against a range of plant pathogens including Fusarium oxysporum [10,11,12], Alternaria porri [13], Verticillium dahliae [14], Rhizoctonia solani [15], and B. cinerea [5, 16]. Besides the recognized biocontrol potential on various plant diseases, the associated action mechanisms of Trichoderma spp. were known as competing for space and nutrients, inhibiting or parasitizing pathogens through antibiotics or extracellular enzymes production, promoting seed germination and plant growth, and enhancing the plant defensive capacity [17, 18]. In recent years, some transcriptome analyses have been conducted to disclose the gene expression profiles of host plants in relation to their interaction with beneficial microorganisms [19, 20]. The inoculation of defined Trichoderma strains in host plants up-regulated a series of functional genes, which participated in plant physiological and biochemical metabolisms, as well as in plant responses to biotic and abiotic stress [21].
Recently, a Trichoderma isolate TM24 was obtained from the rhizosphere soil of a healthy tomato plant in the greenhouse in Shunyi District of Beijing suburb in China. The isolate TM24 was identified as Trichoderma afroharzianum, which had good spore production and obvious plant growth promotion ability [22]. However, the biocontrol potential of isolate TM24 against tomato grey mould, especially its influence on defense enzymes activities and gene expression levels of tomato plants are still unknown. Therefore, the present study was conducted (i) to confirm the B. cinerea inhibition and grey mould biocontrol effect of T. afroharzianum TM24 on tomato plants; (ii) to investigate the potential mechanisms that might be correlated to the biocontrol process of T. afroharzianum TM24 through defense enzymes detection and transcriptome analysis of the tomato plants.
Materials and Methods
Fungal Isolates and Culture Conditions
Trichoderma afroharzianum TM24 was obtained from the rhizosphere soil of a healthy tomato plant, grown in the greenhouse of Shunyi vegetable base in Beijing suburb of China [22]. The isolate TM24 was deposited in the biocontrol microbiology laboratory of Institute of Plant and Environment Protection (IPEP), Beijing Academy of Agriculture and Forestry Sciences (BAAFS). Conidium suspension of T. afroharzianum (106 cfu/mL) was prepared by incubating the mycelia discs in potato dextrose (PD) liquid media at 28 °C and 180 rpm for 7 days. Botrytis cinerea FH was used as the candidate pathogen in the present study, which was isolated previously from the diseased leaves of tomato plant in the greenhouse of Shunyi vegetable base of Beijing suburb and confirmed by Koch's postulates. Spore suspension of the pathogen was prepared by scraping the colony surface of B. cinerea culture with 0.01% (v/v) Tween 80, filtering through four layers of gauze, and adjusted to 106 conidia·mL−1 for inoculation in the biocontrol experiment.
Pathogen Antagonism and Hydrolytic Enzymes Production
The confrontation assay was conducted to detect the antagonism of T. afroharzianum TM24 toward common phytopathogenic fungi including Botrytis cinerea FH, Fusarium oxysporium f. sp. cucumerinum HK, Colletotrichum capsici LJTJ, Monilinia fructicola THF and F. oxysporum f. sp. niveum XK, which were isolated from tomato, cucumber, pepper, peach and watermelon, respectively, grown in the fruits and vegetables base in Beijing suburb of China by the biocontrol microbiology laboratory of IPEP, BAAFS. Mycelial discs of T. afroharzianum isolate TM24 and the fungal pathogens were placed 5 cm apart from each other on PDA plates. The fungal pathogens were inoculated 24 h prior to isolate TM24, while plates inoculated with pathogens only were used as control. After incubation for 4 days at 25 °C in darkness, the mycelial growth inhibition was calculated according to the following formula, inhibition percentage = [(R1 − R2)/R1] × 100, where R1 = radial growth of the pathogen in control plates, R2 = radial growth of the pathogen in test plates.
For measurement of the chitinase and β-1, 3-glucanase production, the isolate TM24 was grown in colloidal chitin [23] and pachyman liquid media [24], respectively. After 5 days of incubation with 150 rpm shaking at 25 °C, the culture filtrate of isolate TM24 was obtained and used as crude enzymes. Chitinase activity was determined by measuring the concentration of released N-acetyl-glucosamine (GlcNAc) after the hydrolysis of colloidal chitin in sodium acetate buffer [13]. The β-1, 3-glucanase activity was measured by the release of glucose after incubating laminarin in sodium acetate buffer with the crude enzymes [10].
Biocontrol Assay in Greenhouse
To evaluate the biocontrol effect of T. afroharzianum isolate TM24, pot experiments were carried out with the tomato plants (cv. Jingyan 16) in a greenhouse. The tomato seeds were sown with one seedling per pot and then cultivated with a 14 h day/10 h night cycle at 25 °C and 70% RH. The leaves of tomato plants were chose for pathogen inoculation since the B. cinerea strain FH used in this study was isolated originally from diseased tomato leaves. The 3-compound leaf stage seedlings were sprayed with conidium suspension of T. afroharzianum TM24 at a concentration of 106 cfu·mL−1. Sterile water was used instead of Trichoderma conidium suspension as the control. Challenge inoculation of B. cinerea was performed by spraying the pathogen spore suspension (106 cfu·mL−1) 24 h post the isolate TM24 or sterile water inoculation. In total, two different groups were established, namely T. afroharzianum TM24 inoculation treatment challenged with B. cinerea (TM24 + B.c) and sterile water control challenged with B. cinerea (Control + B.c). The experiment was arranged in a completely randomized block design with three replications of ten tomato plants per treatment and replicated two times. Disease severity (DS) was divided into six grades according to Yuan et al. [25], where grade 0 = no visible disease symptoms, grade 1 = 1–5%, grade 3 = 6–15%, grade 5 = 16–25%, grade 7 = 26–50%, grade 9 = 51–100% of the leaf surface covered with grey mould. The disease index of grey mould and biocontrol effect of T. afroharzianum TM24 on tomato plants were calculated 7 and 10 days after pathogen inoculation, according to the formulas described by Yuan et al. [25].
Detection of Plant Defense Related Enzymes Activity
Fresh tomato leaves from the two different groups described above were sampled at 24, 48 and 72 h post pathogen inoculation (hpi), homogenized with potassium phosphate buffer for detection of the defense enzymes activity. The homogenate was centrifuged at 12,000 rpm and 4 °C for 10 min, the resultant supernatant was stored at 4 °C for enzyme extraction. The polyphenol oxidase (PPO), phenylalanine ammonialyase (PAL), superoxide dismutase (SOD), and peroxidase (POD) activities in tomato leaves were measured by using the corresponding test kits as described by Jiang et al. [26] according to the instructions (Nanjing Jiancheng Biological Engineering Research Institute, Nanjing, China).
Transcriptome Analysis by RNA-Seq
Fresh tomato leaves sampled from the two different groups and three time points described above were also collected for transcriptome analysis. Thus, a total of 18 tomato leaf samples including three biological replicates were used for cDNA libraries construction and RNA sequencing. Total RNA was extracted from each sample using RNeasy Plant Mini Kit according to the manufacturer’s instructions. The quantity and purity of RNA was assessed using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The library was prepared and then sequenced using the Illumina HiSeq™ 2000 (San Diego, CA, USA) platform with pair-end method at Shanghai Majorbio Bio-pharm Technology Co. Ltd., China.
The high-quality reads were obtained after removing adaptor sequences, empty reads and low-quality reads, which were then mapped to the Solanum lycopersicum reference genome (Solanum lycopersicum, SL2.50: http://plants.ensembl.org/Solanum_lycopersicum/Info/Index) using HISAT2 version 2.1.0. The sequencing data were saved as FASTQ files, and deposited into National Center for Biotechnology Information (NCBI) (BioProject Accessions: PRJNA666988; BioSample Accessions: SAMN16339768–SAMN16339785).
GO Annotation and KEGG Analysis of the DEGs
The differentially expressed genes (DEGs) between the treatment and control groups were determined with the absolute value of Log2 fold change ≥ 1.0 and the adjusted P value < 0.05 using DESeq2 software. The Gene Ontology (GO, http://www.geneontology.org) database was adopted for function annotation of the DEGs in relation to biological processes (BP), cellular component (CC) and molecular functions (MF) terms. In addition, the DEGs which especially related to plant disease defense, such as plant hormone signal transduction and MAPK signaling pathways, were conducted by the Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/) for enrichment analysis.
Validation of DEGs by qRT-PCR
Quantitative real-time PCR (qRT-PCR) of twelve representative DEGs was carried out to validate the accuracy of RNA-Seq data. Total RNA of tomato leaf samples was extracted and purified as the steps described above. The cDNA was synthesized from purified RNA samples using the FastQuant RT Kit (Tiangen Biotech Co. Ltd, Beijing, China). Primers were designed using the primer premier 5.0 software and listed in Supplementary Table S1. The qRT-PCR amplification was performed using CFX 96 Real-Time PCR detection system (Bio-Rad, Hercules, CA, United States) under the following conditions: 95 °C for 3 min, followed by 40 cycles of 15 s at 95 °C, 30 s at 60 °C, 30 s at 72 °C, and end at 72 °C for 5 min. Each PCR reaction consisted of 1 μl of cDNA, 1 μl of each primer (10 μmol·L−1), 10 μl of 2 × TransStart Top Green qPCR SuperMix (TransGen, Beijing, China), and 7 μl of RNase-free water. Elongation factor 1-alpha (EF1α) gene (Solyc06g005060.2) was used as an internal reference to normalize the gene expression level by the 2−ΔΔCT method [27].
Statistical Analysis
Significant differences between means of different treatments were compared by analysis of variance (ANOVA) using SPSS Statistics 17.0 software (SPSS Inc., Chicago, United States). The RNA-Seq data were analyzed on the free online platform of Majorbio Cloud Platform (www.majorbio.com).
Results
Pathogen Antagonism and Hydrolytic Enzymes Production
Confrontation assay showed that T. afroharzianum isolate TM24 had broad-spectrum antagonistic effects on the test fungal pathogens. The inhibition effects of isolate TM24 on B. cinerea, Fusarium oxysporum f. sp. cucumerinum, Colletotrichum capsici, Monilinia fructicola and F. oxysporum f. sp. niveum reached 74.2%, 43.4%, 51.9%, 66.7% and 51.0%, respectively. The isolate TM24 displayed continuous overgrowth on the mycelia of pathogens and reduced the colony size of the tested pathogens obviously in later stages (Fig. 1). The detection of hydrolytic activities indicated that the isolate TM24 exhibited the highest activities of chitinase (12.67 ± 1.16 U·mL−1) and β-1, 3-glucanase (11.75 ± 1.12 U·mL−1) on the 5th day of incubation in colloidal chitin and pachyman liquid induction media, respectively.
Confrontation culture of Trichoderma afroharzianum TM24 against the plant fungal pathogens on PDA plates. a–e Colonies of Botrytis cinerea, Fusarium oxysporum f. sp. cucumerinum, Colletotrichum capsici, Monilinia fructicola and F. oxysporum f. sp. niveum incubated with T. afroharzianum TM24. f–j Colonies of B. cinerea, F. oxysporum f. sp. cucumerinum, C. capsici, M. fructicola and F. oxysporum f. sp. niveum incubated without T. afroharzianum TM24
Biocontrol Effect and Defense Enzymes Activity in Greenhouse Tomato Plants
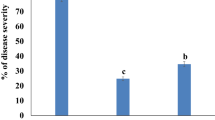
Pot experiments were conducted to verify the biocontrol effect of T. afroharzianum TM24 on tomato grey mould in the greenhouse. The results indicated that isolate TM24 could significantly reduce disease incidence and disease severity on tomato leaves, compared with the sterile water control (Supplementary Fig. S1). The disease index of tomato grey mould was also reduced by T. afroharzianum TM24 treatment, yielding a biocontrol efficiency of 54.3% after 7 days inoculation of B. cinerea. In addition, grey mould reduction effect on tomato plants triggered by T. afroharzianum TM24 was stable, which was 56.7% at the 10th day (Table 1).
T. afroharzianum TM24 had obvious influences on the change of polyphenol oxidase (PPO), phenylalanine ammonialyase (PAL), superoxide dismutase (SOD), and peroxidase (POD) activities at some period of the time under the condition of B. cinerea challenge inoculation. The PPO activity of tomato leaves treated with TM24 was higher than in the control at 24 hpi, and increased gradually to the top level as 307.90 ± 9.75 U/g at 72 hpi, while the value was sharply decreased to 181.20 ± 11.41 U/g in the control (Supplementary Fig. S2a). The PAL activity of tomato treated with TM24 was increased rapidly and got to the maximum of 204.40 ± 13.46 U/g at 48 hpi, and still maintain a relative high level at 72 hpi (Supplementary Fig. S2b). The rapid induction of SOD activity in the tomato leaves treated by T. afroharzianum TM24 was observed at 48 hpi (252.43 ± 10.11 U/g), and followed by a slight decline at 72 hpi (Supplementary Fig. S2c). The same trend was also observed in POD activity during 48–72 hpi. The POD activities of tomato leaves by T. afroharzianum treatment (455.67 ± 19.16 U/g) were significantly higher than the control at 48 hpi, while no significant difference was observed at 72 hpi (Supplementary Fig. S2d).
Transcriptome Data Analysis and Identification of the DEGs
To get insights into potential mechanisms of T. afroharzianum TM24 in biocontrol of tomato grey mould, a comparative transcriptome analysis was conducted using tomato leaves treated with T. afroharzianum TM24 or sterile water, under the condition of B. cinerea challenge inoculation. RNA-Seq of tomato leaves from two different groups at three time points (24, 48 and 72 hpi) generated a number of raw reads ranging from 43,116,197 to 46,332,128 (Table 2). The Q20 and Q30 percentage of each library were exceeded than 98% and 95%, respectively. The percentage of high-quality reads mapped onto the tomato reference genome was > 93% for all samples. These data showed that the RNA-Seq quality was applicable for further analysis.
The results indicated that T. afroharzianum TM24 can raise quick responses in tomato leaves against B. cinerea attack. It was observed that during the process of T. afroharzianum interaction with tomato plants against B. cinerea, the gene transcription levels were influenced strongly at 24 and 48 hpi, which obtained 1941 DEGs (1061 down-regulated and 880 up-regulated) and 1753 DEGs (925 down-regulated and 828 up-regulated), respectively (Supplementary Table S2).
GO Annotation and KEGG Pathway Enrichment of the DEGs
GO annotation was conducted for the global functional analysis of the DEGs, which were classified into “biological process”, “cellular component”, and “molecular function” (Supplementary Fig. S3). In comparison with the sterile water control, the most enriched terms for “biological process” were “metabolic process”, “cellular process”, and “biological regulation” in the T. afroharzianum TM24 treatment group. Moreover, the “cell part”, “membrane”, and “membrane part” were most enriched in the term “cellular component”. The “catalytic activity” term was the most frequent in the “molecular function”, followed by “binding” and “transporter activity”. The above results show that T. afroharzianum TM24 could influence the physiological and biochemical process of tomato plants during the grey mould control.
To further investigate the significant biochemical metabolic and signal transduction process, all DEGs were mapped to the terms in KEGG database and mainly classified into five pathways including metabolism, genetic information processing, environmental information processing, cellular processes, and organism system. Among which, the metabolism pathway contained the largest number of DEGs, followed by genetic information processing. In addition, T. afroharzianum TM24 treatment displayed more significant influence on the metabolism pathways of tomato leaves at 24 and 48 hpi, while the effects and the number of DEGs decreased at 72 hpi (Fig. 2a). The top 30 enriched KEGG pathways associated with DEGs under T. afroharzianum TM24 treatment at three time points were shown in Fig. 2b. Some KEGG pathways associated with plant defense functions were most enriched, such as the MAPK signaling pathway, plant hormone signaling transduction, plant-pathogen interaction, flavonoid biosynthesis and phenylpropanoid biosynthesis.
KEGG pathway classification and function enrichment of the differentially expressed genes (DEGs) in tomato leaves. a KEGG pathway classification of the DEGs at three time points. X-axis represented the number of DEGs. Y-axis represented functional classification of the pathways. b Pathway functional enrichment of the DEGs at three time points. X-axis represented enrichment factor. Y-axis represented pathway name. Control + B.c represented sterile water control challenged with B. cinerea inoculation; TM24 + B.c represented T. afroharzianum TM24 treatment challenged with B. cinerea inoculation (Color figure online)
DEGs Involved in the Plant Hormone Signal Transduction Pathway
In this study, a total of 63 DEGs were involved in the synthesis and regulation of phytohormones (Supplementary Table S3). The expression of these DEGs were analyzed and validated to be correlated with the biocontrol process of T. afroharzianum against grey mould on tomato leaves (Fig. 3a). The signal transduction pathways of auxin, cytokinin, gibberellins, abscisic acid, ethylene, brassinosteroid, jasmonic acid and salicylic acid were positively influenced by T. afroharzianum TM24 treatment under B. cinerea challenge inoculation (Fig. 3b). Five DEGs were identified in the ethylene (ET) transduction pathway, consisting of ETR, CTR1, EBF1/2, EIN3 and ERF1/2, which were all up-regulated by T. afroharzianum TM24 treatment at 24 hpi. Moreover, two of the DEGs, ETR and EBF1/2, were still up-regulated at 48 hpi (Supplementary Fig. S4). The transcription factor MYC2 in the jasmonic acid (JA) transduction pathway was up-regulated, while JAR1 and JAZ were down-regulated at 24 hpi (Supplementary Fig. S4a). In addition, a total of four up-regulated DEGs, including PYR/PYL, PP2C, SnRK2, and ABF, were found in the abscisic acid signaling pathways at 24 hpi, while the PP2C were down-regulated at 48 hpi (Supplementary Fig. S4b).
Analysis of DEGs related to plant hormone signal transduction pathway in the comparisons of T. afroharzianum TM24 pretreated or sterile water treated tomato leaves then both challenged with B. cinerea. a The heat map for expression of DEGs involved in plant hormone signaling transduction pathway. Expression level was displayed as the value of Log10 (TPM + 1). The redder color indicated higher expression; bluer color indicated lower expression. b The model of plant hormone signal transduction pathway based on KEGG pathway enrichment analysis. The yellow background in the rectangle represented the reference gene transcript; the red border in the rectangle represented the up-regulated gene transcript. CK1, CK2, and CK3 represented tomato samples pretreated with sterile water under condition of B. cinerea challenge inoculation at 24, 48, and 72 dpi, respectively. TM1, TM2, and TM3 represented tomato samples pretreated with T. afroharzianum TM24 under condition of B. cinerea challenge inoculation at 24, 48, and 72 dpi, respectively (Color figure online)
DEGs Involved in the MAPK Signaling Pathway
A lot of differentially expressed genes (DEGs) were involved in plant responses to pathogen infection, pathogen attack and phytohormones (Fig. 4a, Supplementary Table S4). The expression of such genes in the MAPK signaling pathway was also positively affected by T. afroharzianum treatment under B. cinerea challenge inoculation (Fig. 4b). Three DEGs including FLS2, MAPK3/6, and WRKY33 were identified as down-regulated in the MAPK signaling pathway at 24 hpi, which were associated with plant response to pathogen inhibition (Supplementary Fig. S5a). The WRKY 2229 was up-regulated in both of the pathogen infection and pathogen attack pathways at 48 hpi, which was related to the early defense response for pathogen (Supplementary Fig. S5b). However, the PR1 related to late defense response was down-regulated in both of the two pathways. The five DEGs related to defense response in the ethylene pathway including ETR/ERS, CTR1, EIN3/EIL, ERF1, EBF1/2 were all up-regulated at 24 hpi, among which, ETR/ERS and EBF1/2 were also up-regulated at 48 hpi (Supplementary Fig. S5b). These results indicated that the MAPK signaling pathway may be associated with the defense activity of tomato plants induced by the TM24 treatment.
Analysis of DEGs related to MAPK signaling pathway in the comparisons of T. afroharzianum TM24 pretreated or sterile water treated tomato leaves then both challenged with B. cinerea. a The heat map for expression of DEGs involved in MAPK signaling pathway. Expression level was displayed as the value of Log10 (TPM + 1). The redder color indicated higher expression; bluer color indicated lower expression. b The model of MAPK signaling pathway based on KEGG pathway enrichment analysis. The yellow background in the rectangle represented the reference gene transcript; the red border in the rectangle represented the up-regulated gene transcript. CK1, CK2, and CK3 represented tomato samples pretreated with sterile water under condition of B. cinerea challenge inoculation at 24, 48, and 72 dpi, respectively. TM1, TM2, and TM3 represented tomato samples pretreated with T. afroharzianum TM24 under condition of B. cinerea challenge inoculation at 24, 48, and 72 dpi, respectively (Color figure online)
Transcription Factors Prediction of DEGs Associated with Biocontrol Process
The expression of transcription factors (TFs) encode DEGs was significantly influenced in the tomato leaves that were pretreated with T. afroharzianum TM24 and then challenged with B. cinerea at different time points (Supplementary Fig. S6a). A total of 33 kinds of TFs were identified in T. afroharzianum TM24 treated tomato, such as WRKY, MYB, bZIP, NAC, TCP, ERF, bHLH, MIKC, which may function in plant defense activity. In addition, the types and numbers of TFs were the most in the tomato leaves at 24 hpi, which included some TFs related to plant defense activity similarly, such as MYB, WRKY, NAC, bZIP, ERF, TCP (Supplementary Fig. S6b).
Validation of DEG by qRT-PCR
To confirm the reliability of the RNA-seq data, the relative expression level of twelve genes involved in the MAPK signaling pathway (Solyc02g082930.2, Solyc01g095080.2, Solyc02g082920.2), plant hormone signal transduction (Solyc06g008580.2, Solyc12g096980.1, Solyc07g006860.2, Solyc06g053840.2), plant-pathogen interaction (Solyc02g077370.1), and four randomly selected genes (Solyc07g047850.2, Solyc03g005780.1, Solyc03g115980.1, Solyc02g089160.2) were selected for qRT-PCR analysis at three time points. As shown in Fig. 5, the patterns of gene expression detected by qRT-PCR were highly consistent with the RNA sequencing analysis, indicating high quality of the RNA sequencing datasets.
Comparison of the RNA-seq and qPCR results validated the relative expression level of the differentially expressed genes (DEGs). The graph showed the Log twofold change in expression of the twelve genes in tomato leaves. r2 indicated the degree of correlation between RNA-seq and qPCR results. Error bars represented the standard deviations of three replicates (Color figure online)
Discussion
Trichoderma species are saprophytic filamentous fungi of worldwide distribution, which are well-known biological control agents against various plant fungal diseases. Several Trichoderma spp. including T. harzianum [16, 28,29,30], T. atroviride [31], and T. viride [32] have been reported to have particularly effect in control of grey mould on many plants. In this study, we tried to provide Trichoderma afroharzianum isolate TM24 for biocontrol potential on tomato grey mould. Results indicated that the isolate TM24 displayed obvious antagonism against common plant pathogens including B. cinerea, as well as definite biocontrol effect on grey mould of tomato in pots.
Different mechanisms have been suggested as being responsible for the biocontrol potential of Trichoderma species [25, 33, 34]. The direct actions of Trichoderma against plant fungal diseases were traditionally targeted on the phytopathogens through competition, antibiosis and mycoparasitism [12, 35]. In this work, potent competition and mycoparasitism activity seem to be involved in the antagonism of T. afroharzianum TM24 against B. cinerea during confrontation culture. The isolate TM24 grew rapidly in Petri dish, invaded the colony of B. cinerea through a marked process of hyperparasitism, and finally covered the entire plates. This ability of rapid growth might give the isolate an obvious advantage in competition for nutrient, space and dominance over the target pathogen. In addition, the isolate TM24 was able to produce chitinase and β-1, 3-glucanase, which may be related to the mycoparasitism of Trichoderma by degrading cell wall of the fungal pathogens [36].
Defense activity in host plants has been reported as the indirect action way by some Trichoderma species for execution of the biocontrol function [28, 33]. Previous studies indicated that the enhanced activities of defense-related enzymes in plant tissues were positively associated with induced systemic resistance and plant disease suppression [34, 37]. Murugesan and Se [38] suggested that polyphenol oxidase (PPO), superoxide dismutase (SOD), and peroxidase (POD) activities usually acted as essential elements in oxidation of phenolic compounds and defense against phytopathogens. Our results indicated that the activities of these enzymes were significantly increased in T. afroharzianum TM24 treated tomato plants compared with the control under the condition of B. cinerea challenge inoculation. The accumulation of such defense-related enzymes appear to be associated with the enhanced defense activity in tomato plants against B. cinerea infection. In this study, the PPO activity reached to the maximum at 72 h, while the PAL, SOD, and POD activities had their maxima earlier at 48 h. The high PAL and POD activities lasted for a longer time until 72 h, however, the SOD activity was decreased at 72 h. The maximum level and duration of these enzymes activities occurred at different stages in our study, which might be explained by the different time when antioxidants were activated.
In recent years, transcriptome studies have provided abundant information for identifying the functional genes associated with metabolism regulation and signaling transduction from the molecular level [37]. Yu et al. [39] indicated that Trichoderma asperellum ACCC30536 and its eliciting plant response protein Epl1-Tas could induce the woody plant resistance to Alternaria alternata by regulating the expression of defense-related genes in the salicylic acid, jasmonic acid and auxin signal transduction pathways. Zhang et al. [20] reported that most different expression genes (DEGs), which were obtained by the biocontrol fungus Chaetomium globosum CEF-082 treatment, were enriched mainly in the same signaling pathways. In this study, the results showed that the isolate TM24 treatment has brought about the up-regulation of many genes involved in MAPK signaling pathway, plant hormone signaling transduction, and plant-pathogen interaction.
Plant hormone signaling transduction may modulate the defense network by translating Trichoderma-induced upstream signaling events into the activation of defense responses [40]. Shang et al. [41] showed that T. asperellum TC01 has the ability to induce host systematic resistance through the JA- and ET-regulated priming mechanism in tomato plants. In this study, we also analyzed the expression level of these genes in the phytohormones signal pathway, and found that some DEGs related to the pathways of jasmonic acid (JA), salicylic acid (SA), ethylene (ET) and so on were all induced in tomato plants treated with T. afroharzianum. In addition, the T. afroharzianum TM24 treatment has also induced the up-regulation of flavonoid and phenylpropanoid biosynthesis related genes. These findings suggested that the biocontrol Trichoderma may play an important role in reinforcing the plant cell wall and improving plant defense, through modulation of the plant secondary metabolism, such as synthesization of the flavonoids, lignin, and phenolic compounds [42, 43].
Transcription factors (TFs) such as WRKY, AP2-ERF, MYB, and NAC have been demonstrated to play essential roles in plants by regulating the expression of genes involved in various cellular processes [44]. Among which, WRKY factors have been verified to fulfill essential regulatory function and modulate pathogen-triggered cellular response in a number of plant species, most WRKY factors were found to participate in the SA signaling pathway [45]. Some ERF factors have been demonstrated to participate in the JA signaling pathway, which were involved in the interaction between plants and fungal pathogens [46]. In our study, we also identified 63 TFs in the biocontrol process of T. afroharzianum TM24. Two families including WRKY and ERF were both identified in our sequencing data, indicating that the two signaling pathways, SA and JA, took part in the biocontrol process of T. afroharzianum on tomato grey mould. In addition, NAC proteins obtained in this study were also reported as another kind of plant-specific TF that could regulate immune responses in plants [47]. Future study will focus on the function and mechanism of these key genes obtained by transcriptome analysis, in order to provide theoretical basis for the interactions between Trichoderma and tomato plants, as well as B. cinerea and tomato plants.
Conclusions
In this study, the biocontrol potential of Trichoderma afroharzianum TM24 on tomato grey mould was investigated through pot experiments and transcriptome analysis. T. afroharzianum TM24 displayed obvious B. cinerea inhibition and grey mould biocontrol effect on tomato. The isolate TM24 was also able to trigger significantly high levels of plant defense-related enzymes activity under the condition of B. cinerea challenge inoculation. In addition, pretreatment with T. afroharzianum TM24 induced expression of many genes that associated with plant hormone signal transduction, MAPK signaling, plant-pathogen interaction, as well as flavonoid and phenylpropanoid biosynthesis in the tomato plants. Hence, we concluded that T. afroharzianum TM24 is a potential biocontrol candidate for grey mould management on tomato plants in facility cultivation.
Data Availability
The RNA-seq data were saved as FASTQ files, and deposited into National Center for Biotechnology Information under accession numbers of BioProject PRJNA666988 and BioSample SAMN16339768-16339785.
References
Liu S, Hai F, Jiang J (2017) Sensitivity to fludioxonil of Botrytis cinerea isolates from tomato in Henan province of China and characterizations of fludioxonil-resistant mutants. J Phytopathol 165:98–104
Liu SM, Che ZP, Chen GQ (2016) Multiple-fungicide resistance to carbendazim, diethofencarb, procymidone, and pyrimethanil in field isolates of Botrytis cinerea from tomato in Henan Province, China. Crop Prot 84:56–61
Xu SJ, Park DH, Kim JY, Kim BS (2016) Biological control of gray mold and growth promotion of tomato using Bacillus spp. isolated from soil. Trop Plant Pathol 41:169–176
Gao P, Qin J, Li D, Zhou S (2018) Inhibitory effect and possible mechanism of a Pseudomonas strain QBA5 against gray mold on tomato leaves and fruits caused by Botrytis cinerea. PLoS ONE 13:0190932
You JQ, Zhang J, Wu MD, Yang L, Chen WD, Li GQ (2016) Multiple criteria-based screening of Trichoderma isolates for biological control of Botrytis cinerea on tomato. Biol Control 101:31–38
Boukaew S, Prasertsan P, Troulet C, Bardin M (2017) Biological control of tomato gray mold caused by Botrytis cinerea by using Streptomyces spp. Bio Control 62:793–803
Teak SS, Nan HY, Jaeho L, Gyung JC, Jin CK, Chul SS (2017) Development of a biofungicide using a mycoparasitic fungus Simplicillium lamellicola BCP and its control efficacy against gray mold diseases of tomato and Ginseng. Plant Pathol J 33:337–344
Kumari S, Tayal P, Sharma E, Kapoor R (2014) Analyses of genetic and pathogenic variability among Botrytis cinerea isolates. Microbiol Res 169:862–872
Lopes FAC, Steindorff AS, Geraldine AM (2012) Biochemical and metabolic profiles of Trichoderma strains isolated from common bean crops in the Brazilian Cerrado, and potential antagonism against Sclerotinia sclerotiorum. Fungal Biol 116:815–824
El Komy MH, Saleh AA, Eranthodi A, Molan YY (2015) Characterization of novel Trichoderma asperellum isolates to select effective biocontrol agents against tomato Fusarium wilt. Plant Pathol J 31:50–60
Mwangi MW, Muiru WM, Narla RD, Kimenju JW, Kariuki GM (2019) Management of Fusarium oxysporum f. sp. lycopersici and root-knot nematode disease complex in tomato by use of antagonistic fungi, plant resistance and neem. Biocontrol Sci Technol 29:207–216
Zhang FL, Liu C, Wang Y, Dou K, Chen F, Pang L, Kong X, Shang C, Li Y (2020) Biological characteristic and biocontrol mechanism of Trichoderma harzianum T-A66 against bitter gourd wilt caused by Fusarium oxysporum. J Plant Pathol 102:1107–1120
Abo-Elyousr KAM, Abdel-Hafez SII, Abdel-Rahim IR (2014) Isolation of Trichoderma and evaluation of their antagonistic potential against Alternaria porri. J Phytopathol 162:567–574
Irene CC, José LTC, Concepción OG, Enrique M, Rosa H, Rafael MJD (2016) Trichoderma asperellum is effective for biocontrol of Verticillium wilt in olive caused by the defoliating pathotype of Verticillium dahlia. Crop Prot 88:45–52
Swain H, Adak T, Mukherjee AK, Mukherjee PK, Bhattacharyya P, Behera S, Bagchi TB, Patro RS, Khandual A, Bag MK, Dangar TK, Lenka S, Jena M (2018) Novel Trichoderma strains isolated from tree barks as potential biocontrol agents and biofertilizers for direct seeded rice. Microbiol Res 214:83–90
Abbey JA, Percival D, Abbey L, Asiedu SK, Prithiviraj B, Schilder A (2019) Biofungicides as alternative to synthetic fungicide control of grey mould (Botrytis cinerea)—prospects and challenges. Biocontrol Sci Technol 29:241–262
John RP, Tyagi RD, Prévost D, Brar SK, Pouleur S, Surampalli RY (2010) Mycoparasitic Trichoderma viride as a biocontrol agent against Fusarium oxysporum f. sp. adzuki and Pythium arrhenomanes and as a growth promoter of soybean. Crop Prot 29:1452–1459
Vinale F, Manganiello G, Nigro M, Mazzei P, Piccolo A, Pascale A, Ruocco M, Marra R, Lombardi N, Lanzuise S, Varlese R, Cavallo P, Lorito M, Woo SL (2014) A novel fungal metabolite with beneficial properties for agricultural applications. Molecules 19:9760–9772
Yuan J, Zhang W, Sun K, Tang MJ, Chen PX, Li X, Dai CC (2019) Comparative transcriptomics and proteomics of Atractylodes lancea in response to endophytic fungus Gilmaniella sp. AL12 reveals regulation in plant metabolism. Front Microbiol 10:1208
Zhang Y, Yang N, Zhao LH, Zhu HQ, Tang CM (2020) Transcriptome analysis reveals the defense mechanism of cotton against Verticillium dahliae in the presence of the biocontrol fungus Chaetomium globosum CEF-082. BMC Plant Biol 20:89
Coppola M, Diretto G, Digilio MC, Woo SL, Giuliano G, Molisso D, Pennacchio F, Lorito M, Rao R (2019) Transcriptome and metabolome reprogramming in tomato plants by Trichoderma harzianum strain T22 primes and enhances defense responses against aphids. Front Physiol 10:e745
Zhao J, Liu T, Liu WC, Zhang DP, Dong D, Wu HL, Zhang TT, Liu DW (2021) Transcriptomic insights into growth promotion effect of Trichoderma afroharzianum TM2-4 microbial agent on tomato plants. J Integr Agric 20:1266–1276
Qualhato T, Lopes F, Steindorff A, Brando R, Jesuino R, Ulhoa C (2013) Mycoparasitism studies of Trichoderma species against three phytopathogenic fungi: evaluation of antagonism and hydrolytic enzyme production. Biotechnol Lett 35:1461–1468
Sock J, Rohringer R, Kang Z (1990) Extracellular β-1, 3-glucanases in stem rust-affected and abiotically stressed wheat leaves. Plant Physiol 94:1376–1389
Yuan M, Huang YY, Ge WN, Jia ZH, Song SS, Zhang L, Huang YL (2019) Involvement of jasmonic acid, ethylene and salicylic acid signaling pathways behind the systemic resistance induced by Trichoderma longibrachiatum H9 in cucumber. BMC Genomics 20:144
Jiang CH, Wu F, Yu ZY, Xie P, Ke HJ, Li HW, Yu YY, Guo JH (2015) Study on screening and antagonistic mechanisms of Bacillus amyloliquefaciens against bacterial fruit blotch (BFB) caused by Acidovorax avenae subsp. citrulli. Microbiol Res 170:95–104
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-delta delta C (T)) method. Methods 25:402–408
Levy NO, Harel YM, Haile ZM, Elad Y, Rav-David E, Jurkevitch E, Katan J (2015) Induced resistance to foliar diseases by soil solarization and Trichoderma harzianum. Plant Pathol 64:365–374
Cruz AF, Barka GD, Sylla J, Reineke A (2018) Biocontrol of strawberry fruit infected by Botrytis cinerea: effects on the microbial communities on fruit assessed by next-generation sequencing. J Phytopathol 166:403–411
Saravanakumar K, Lu ZX, Xia H, Wang M, Sun JN, Wang SQ, Wang QQ, Li YQ, Chen J (2018) Triggering the biocontrol of Botrytis cinerea by Trichoderma harzianum through inhibition of pathogenicity and virulence related proteins. Front Agric Sci Eng 5:271–279
Pertot I, Giovannini O, Benanchi M, Caffi T, Rossi V, Mugnai L (2017) Combining biocontrol agents with different mechanisms of action in a strategy to control Botrytis cinerea on grapevine. Crop Prot 97:85–93
Balode A (2012) Biocontrol of grey mould in Lilies spp. by Trichoderma spp. and Bacillus spp. Acta Hortic 937:583–587
Jogaiah S, Abdelrahman M, Tran LSP, Ito SI (2018) Different mechanisms of Trichoderma virens-mediated resistance in tomato against Fusarium wilt involve the jasmonic and salicylic acid pathways. Mol Plant Pathol 19:870–882
Yu C, Luo X (2020) Trichoderma koningiopsis controls Fusarium oxysporum causing damping-off in Pinus massoniana seedlings by regulating active oxygen metabolism, osmotic potential, and the rhizosphere microbiome. Biol Control 150:e104352
Mukherjee PK, Horwitz BA, Herrera-Estrella A, Schmoll M, Kenerley CM (2013) Trichoderma research in the genome era. Annu Rev Phytopathol 51:105–129
Vos CMF, De Cremer K, Cammue BPA, De Coninck B (2015) The toolbox of Trichoderma spp. in the biocontrol of Botrytis cinerea disease. Mol Plant Pathol 16:400–412
Jiang CH, Huang ZY, Xie P, Gu C, Li K, Wang DC, Yu YY, Fan ZH, Wang CJ, Wang YP, Guo YH, Guo JH (2016) Transcription factors WRKY70 and WRKY11 served as regulators in rhizobacterium Bacillus cereus AR156 - induced systemic resistance to Pseudomonas syringae pv. tomato DC3000 in Arabidopsis. J Exp Bot 67:157–174
Murugesan C, Se CC (2016) Expression of PR protein genes and induction of defense-related enzymes by Bacillus subtilis CBR05 in tomato (Solanum lycopersicum) plants challenged with Erwinia carotovora subsp. carotovora. Biosci Biotechnol Biochem 80:2277–2283
Yu WJ, Mijiti G, Huang Y, Fan HJ, Wang YC, Liu ZH (2018) Functional analysis of eliciting plant response protein Epl1-Tas from Trichoderma asperellum ACCC30536. Sci Rep 8:7974
Voß U, Bishopp A, Farcot E, Bennett MJ (2014) Modelling hormonal response and development. Trends Plant Sci 19:311–319
Shang J, Liu BL, Xu Z (2020) Efficacy of Trichoderma asperellum TC01 against anthracnose and growth promotion of Camellia sinensis seedlings. Biol Control 143:e104205
Singh V, Upadhyay RS, Sarma BK, Singh HB (2016) Trichoderma asperellum spore dose depended modulation of plant growth in vegetable crops. Microbiol Res 193:74–86
Ben AM, Lopez D, Triki MA, Khouaja A, Chaar H, Fumanal B, Gousset-Dupont A, Bonhomme L, Label P, Goupil P, Ribeiro S, Pujade-Renaud V, Julien JL, Auguin D, Venisse JS (2017) Beneficial effect of Trichoderma harzianum strain Ths97 in biocontrolling Fusarium solani causal agent of root rot disease in olive trees. Biol Control 110:70–78
Jiang CH, Yao XF, Mi DD, Li ZJ, Yang BY, Zheng Y, Qi YJ, Guo JH (2019) Comparative transcriptome analysis reveals the biocontrol mechanism of Bacillus velezensis F21 against Fusarium wilt on watermelon. Front Microbiol 10:652
Chen F, Hu Y, Vannozzi A, Wu KC, Cai HY, Qin Y, Mullis A, Lin ZG, Zhang LS (2017) The WRKY transcription factor family in model plants and crops. Crit Rev Plant Sci 36:311–335
Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) AP2/ERF family transcription factors in plant abiotic stress responses. BBA-Gene Regul Mech 1819:86–96
Lee MH, Jeon HS, Kim HG, Park OK (2017) An Arabidopsis NAC transcription factor NAC4 promotes pathogen-induced cell death under negative regulation by microRNA164. New Phytol 214:343–360
Acknowledgements
This study was mainly funded by the Beijing Academy of Agriculture and Forestry Sciences (BAAFS), China.
Funding
This research was funded by the Youth Research Fund of Beijing Academy of Agriculture and Forestry Sciences (QNJJ201814), the Beijing Natural Science Foundation (6192008), the Innovative Capacity Construction of Beijing Academy of Agriculture and Forestry Sciences (KJCX20200110, KJCX20200426), and Beijing Key Laboratory of Green Control of Fruit Tree Diseases and Pests in the North China (BZ0432).
Author information
Authors and Affiliations
Contributions
JZ and TL conceived and designed the research. JZ and DPZ constructed the experiments and analyzed the data. JZ, HLW, TTZ, and DD conducted the experiments.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, J., Liu, T., Zhang, D. et al. Biocontrol Potential of Trichoderma afroharzianum TM24 Against Grey Mould on Tomato Plants. Curr Microbiol 78, 4115–4126 (2021). https://doi.org/10.1007/s00284-021-02671-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-021-02671-x