Abstract
Web blight/wet root rot caused by Rhizoctonia solani is one of the major constraints for mung bean (Vigna radiata) production. Growing of resistant varieties and use of biocontrol agents are the feasible options available to manage the disease. The present study was conducted to determine the variation in the expression of various defense-related genes in susceptible and resistant mung bean varieties in response to biocontrol agent Trichoderma virens and R. solani interactions. The primers were designed using sequences of defense-related genes, namely PR 10, epoxide hydrolase (EH), catalase and calmodulin available in NCBI database and evaluated against cDNA obtained from both susceptible and resistant mung bean plants at 1–4 days post-inoculation (dpi) with the test pathogen R. solani and biocontrol agent T. virens using conventional PCR and qPCR analyses. R. solani inoculation upregulated the mean expression of PR 10 and calmodulin in susceptible and resistant varieties, respectively, whereas downregulated in the rest of the treatments. Quantitative PCR analysis showed that except catalase in the susceptible variety, which is downregulated, the expression of PR 10, EH, catalase and calmodulin was upregulated in both resistant and susceptible varieties in response to T. virens alone and in the presence of R. solani. In general, the expression of PR 10 and calmodulin was highest at 1 dpi whereas EH and catalase expression were maximum at 4 dpi. The application of T. virens suppressed the development of disease in the presence of R. solani in both susceptible and resistant varieties with more pronounced effect in resistant variety. Thus, the application of biocontrol agent T. virens upregulated the expression of defense-related genes and reduced disease development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mung bean [Vigna radiata (L.) Wilczek] is one of the most important pulse crops in India and considered as a major source of dietary protein for the vegetarian population (Khattak et al. 2002). As a legume crop, mung bean also sustains soil fertility by improving soil physical properties and fixing atmospheric nitrogen. The crop is susceptible to various diseases, of which, wet root rot or web blight caused by Rhizoctonia solani Kühn [Thanatephorus cucumeris (Frank) Donk] is one of the most devastating soil- and seed-borne diseases (Dubey 2003; Dubey et al. 2011). The management of diseases caused by R. solani is difficult using fungicides, which provide inadequate protection to the expanding root zone of the plants hence ineffective in controlling the root-rot phase of the pathogen (Abawi 1989).
In response to pathogen invasion or association with biocontrol agents, plants are induced to express genes associated with plant defense responses, which include pathogenesis-related (PR) and antimicrobial genes involved in biological activities related to disease resistance. The PR10 proteins are typically intracellular (Liu and Ekramoddoullah 2006) and their homologues are found in both dicotyledonous and monocotyledonous plant species (Walter et al. 1990). They have various functions including antimicrobial and RNase activity which play a potential role in defense against pathogenic infections (Liu and Ekramoddoullah 2006). The pathogens (viruses, bacteria and fungus) trigger a PR-10 response and PR 10 is also reported to act against fungal invasion (Jacobs et al. 1999; Dowd et al. 2004; Liu and Ekramoddoullah 2006). Enzymes such as superoxide dismutase, peroxidase, catalase and ascorbate peroxidase are included in the antioxidant system of plants to counteract the toxicity of reactive oxygen species (Foyer et al. 1994). The susceptibility of plants to pathogens was positively correlated with the activities of catalase (Conrath et al. 1995). Epoxide hydrolase is involved in the biosynthesis of cutin (Pinot et al. 1992) and repair of damaged cuticle caused by pathogens. Epoxy fatty acids, the products of epoxide hydrolase, are also known to inhibit spore germination and germ tube growth of Pyricularia oryzae causing rice blast (Kato et al. 1983). Calmodulin (CaM) is a major calcium-binding protein in plants (Means and Dedman 1980).
A seed dressing formulation Pusa5SD and soil application formulations Pusa Biogranule 6 (PBG6) and Pusa Biopellet 16G (PBP16G) developed from Trichoderma virens (IARI-P3) alone and in combination were found effective in increasing seed germination, shoot and root lengths, grain yield and reducing wet root rot (R. solani) incidence in mung bean (Dubey et al. 2011), but the mechanism of interaction related to induction of defense was not studied. Nogueira-Lopez et al. (2018) reported that T. virens has the capacity to behave as an opportunistic plant endophyte and colonized the maize roots under hydroponic conditions. The proteins secreted by T. virens were mainly involved in cell wall hydrolysis, scavenging of reactive oxygen species and secondary metabolism, as well as putative effector-like proteins. Thus, T. virens may be used to manipulate host immune responses. Therefore, the aim of the present study was to investigate the differences in expression of defense-related genes in the resistant and susceptible mung bean varieties in the presence of T. virens alone and in the presence of R. solani infection.
Materials and methods
Plant material
Mung bean seeds of variety Ratna, susceptible and HUM-1, resistant to R. solani, used in the present study were obtained from Pulse Laboratory, Division of Plant Pathology, ICAR-IARI, New Delhi. Seeds were surface sterilized in 2.0% sodium hypochlorite solution for 1 min and thoroughly washed twice with sterile distilled water. Seeds were sown in 15-cm pot (10 seeds/pot) containing sterile (autoclaved) ProMix soil (peat moss: sand: vermiculite; 1:1:1) in a controlled environmental condition (Phytotron, ICAR-IARI, New Delhi) with 32 °C temperature and 80% humidity.
Trichoderma virens and Rhizoctonia solani cultures
The culture of T. virens (IARI-P3) was obtained from Pulse Laboratory, Division of Plant Pathology, ICAR-IARI, New Delhi. The isolate of T. virens (IARI-P3) used in the present study has been proved effective against wet root rot of mung bean caused by R. solani (Dubey et al. 2011). Highly virulent isolate of R. solani obtained from Pulse Laboratory, Division of Plant Pathology, ICAR-IARI, New Delhi, was used for seedling inoculation. The fungal culture was grown in potato dextrose broth (PDB) at 25 ± 1 °C for 5 days. The broth containing culture was macerated and 25 ml of inoculum having 105 cfu/ml was added in the pot for root drenching inoculation. The culture of T. virens was multiplied on PDB at 25 ± 1 °C for 7 days and 108 cfu/ml was used for inoculation.
Pre-treatment/inoculation
Mung bean seedlings of both the varieties at two- to three-leaf stage were drenched with T. virens having 108 cfu/ml (25 ml/pot) 4 days prior to R. solani inoculation. The mycelial suspension of R. solani multiplied on PDB was prepared by gently macerating 5-day-old mycelial mat along with the medium with sterilized pestle and mortar and used at 25 ml/pot (105 cfu/ml) for inoculation. Four treatments as T. virens alone, with R. solani, R. solani alone and control (un-inoculated) were maintained for each variety. Plants (eight plants, four from each replication) were uprooted at 1–4 days post-inoculation (dpi) and stored at − 80 °C or further use.
Reverse transcription PCR
Total RNA was isolated using RNA sure plant kit (Nucleopore, UK). After quantification, 1 µg of total RNA was used for cDNA synthesis using Verso cDNA kit (Thermo Scientific, UK) according to the manufacturer’s instructions. These cDNAs served as templates in PCRs using 10 pmol gene-specific primers, 0.2 mM dNTPs and 1 U Taq polymerase (Bangalore Genei, India). The most common defense-related genes were selected for primer designing. The primers for various defense-related genes were designed using the sequences of mung bean defense genes available in the NCBI database. The gene-specific primers (Table 1) were used for conventional as well as quantitative PCR analysis.
Conventional PCR
Conventional PCR was performed in 25 µl reaction mixture consisting of 30 ng cDNA, 2.5 mM MgCl2, 150 µM dNTPs, 5 pmol of each gene-specific primer, and 1.5 U Taq polymerase (Bangalore Genei, India) in 1 × Taq buffer. The PCR conditions were as follows: initial denaturation at 95 °C for 5 min followed by 35 cycles of denaturation at 94 °C for 45 s, annealing at 60 °C for 45 s for PR 10, epoxide hydrolase and catalase, whereas 62 °C for 45 s for calmodulin, extension at 72 °C for 30 s and final extension at 72 °C for 5 min. The experiment was repeated twice.
Quantitative real-time PCR
PCRs were carried out in eight-well PCR strips (20 µl per well) in a reaction buffer containing 1 × Eva green supermix (BioRad Laboratories, USA), 2.5 pmol of each primer and 20 ng cDNA. Quantitative expression analysis was performed using real-time PCR system (Mini Opticon, BioRad Laboratories, USA). The real-time PCR conditions for PR10, epoxide hydrolase and catalase genes were as follows: initial denaturation at 95 °C for 3 min followed by 45 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 20 s, extension at 62 °C for 20 s and plate reading at 62 °C. For calmodulin gene, the reaction conditions were initial denaturation at 95 °C for 4 min followed by 45 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 30 s and plate read at 62 °C. Melting curves were performed from 65 to 95 °C with readings at every 0.5 °C and 5-s hold between readings for verification of single PCR product. Specificity of primers to mung bean genes was examined using DNA from T. virens, R. solani and their reverse-transcribed RNA as templates. The absence of primer dimer formation was checked in non-template control. Each treatment sample was used in two replications. The relative expression ratios of these defense genes were normalized using actin, a non-regulated reference gene (same copy number per cell and stable expression in every cell). The crossing point [C(T) values] differences of each sample versus a control sample were used to determine relative expression. The mock-inoculated samples (without any inoculation) from each mung bean variety were selected as the control samples, thus, comparing each sample’s C(T) to the control sample’s C(T). Differences in the mRNA abundance between various genes in the control and the corresponding treated samples were measured with CFX Manager (BioRad Laboratories, USA). The experiment was repeated twice.
Disease development
The disease incidence was recorded using the formula disease incidence (%) = number of plants showing symptoms × 100/total plants standing at 1, 2, 3, 4 and 20 dpi (Dubey and Singh 2013). Since development of the disease was less up to 4 dpi, the observation was continued up to 20 dpi.
Statistical analysis
Data were analyzed as per the procedure for a completely randomized design (CRD) factorial subjected to ANOVA. Significant differences were observed in Fisher’s Protected Least Significant Difference test (Gomez and Gomez 1984) performed using the SAS Software (SAS Institute, version 9.1, Cary, NC).
Results
In susceptible variety Ratna, PR10 mRNA expression level was induced in T. virens-treated plants both alone and in the presence of R. solani. However, only T. virens-inoculated plants showed significantly highest (around sevenfold) increase in PR10 expression at 1 dpi followed by 2 dpi which declined steeply at 3 dpi and again increased at 4 dpi. However, in the plants inoculated both with T. virens and R. solani, PR10 expression constantly decreased from day 1 to day 4 with the highest expression up to fourfold relative to control at 1 dpi. R. solani inoculation increased the expression of PR 10 at 1, 2 and 4 dpi. Considering the mean expression, T. virens-treated plants showed the highest upregulation of PR10 in variety Ratna followed by combined inoculation with T. virens and R. solani (Fig. 1). In resistant variety HUM 1, the PR10 expression was comparatively low as compared to the susceptible variety. Initially, around four- and fivefold up-regulation was observed in T. virens alone and T. virens in the presence of R. solani, respectively. In both the treatments, PR10 expression was induced at 1 dpi and the second increase was observed at 3 dpi. The highest level of expression was observed in T. virens + R. solani-inoculated plants at 3 dpi (more than eightfold increase) followed by 1 dpi. T. virens-treated plants showed the maximum expression of PR10 at 1 dpi (fourfold increase) followed by 3 dpi. The mean expression was the highest in the case of T. virens + R. solani-inoculated plants followed by only-T. virens-inoculated plants (Fig. 2).
In susceptible variety Ratna, T. virens-treated plants showed the highest expression of epoxide hydrolase (EH) at 3 dpi (more than threefold increase), followed by 4 and 2 dpi. While T. virens-treated and challenged (with R. solani) plants showed upregulation at all times of observations, with the maximum at 4 dpi followed by 2, 1 and 3 dpi. R. solani-inoculated plants showed the highest expression at 1 dpi and further, it declined till 3 dpi and again increased at 4 dpi. Considering mean expression, the highest expression was observed in T. virens + R. solani-inoculated plants followed by T. virens and R. solani (Fig. 3). In resistant variety HUM 1, T. virens alone followed by T. virens + R. solani-inoculated plants showed the highest expression (about eightfold) of EH at 1 dpi. The expression decreased at 2 and 3 dpi to a great extent and then further increased at 4 dpi. The expression of EH was downregulated at 3 dpi in all the treatments and at 1 and 2 dpi only in R. solani-inoculated plants, whereas the expression was upregulated in all the other observations. Mean expression was the highest in T. virens-treated plants followed by T. virens + R. solani (Fig. 4).
Trichoderma virens-treated plants of susceptible variety Ratna showed maximum catalase expression at 1 dpi followed by 4 and 2 dpi. T. virens along with R. solani-inoculated plants exhibited down-regulation in all 4 days of observations. Only R. solani-inoculated plants showed downregulation up to 3 dpi, which elevated significantly at 4 dpi. Mean expression indicated downregulation in all the treatments in comparison to the control (Fig. 5). In resistant variety HUM 1, the expression of catalase was upregulated in all the treatments at 1 dpi and 4 dpi. It was the highest (fourfold increase) in T. virens-treated plants at 1 dpi followed by T. virens + R. solani-inoculated plants at 4 dpi. At 2 and 3 dpi, its expression was downregulated in all the treatments. The mean expression was upregulated in T. virens alone and T. virens + R. solani-inoculated plants while it was downregulated in the R. solani-treated plants (Fig. 6).
Significantly, higher expression of calmodulin was observed in the susceptible variety Ratna at 1 dpi in response to T. virens treatment. Further, it was downregulated at 2 dpi and again gradually upregulated at 3 and 4 dpi. In contrast, T. virens-treated plants in the presence of R. solani showed upregulation only at 1 dpi while it was downregulated on the rest of the days. Plants inoculated with R. solani showed upregulation at 1 and 3 dpi while downregulation at 2 and 4 dpi. The mean expression was the highest in T. virens-treated plants followed by T. virens + R. solani-inoculated plants (Fig. 7). In resistant variety HUM 1, the plants treated with T. virens alone and in the presence of R. solani showed upregulation of calmodulin at all days of observations. It was significantly highest in T. virens + R. solani-inoculated plants (about 42-fold) at 3 dpi followed by 4 dpi. T. virens inoculation also upregulated the expression of calmodulin with the maximum at 4 dpi (about 11-fold) followed by 3 dpi and 1 dpi. Only R. solani-treated plants exhibited upregulation at 2 dpi and 3 dpi while downregulation at 1 dpi and 4 dpi. The highest mean expression was observed in T. virens + R. solani followed by T. virens and R. solani-treated plants (Fig. 8).
Disease development
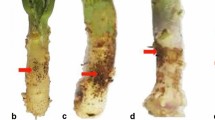
Both susceptible (Ratna) and resistant (HUM 1) varieties did not show any disease symptoms under T. virens and un-inoculated (control) conditions up to 20 dpi. R. solani-inoculated plants of susceptible variety Ratna showed 2.8, 11.1, 19.4, 33.3 and 94.5% disease incidence at 1, 2, 3, 4 and 20 dpi, respectively, whereas resistant variety HUM 1 did not show any infection at 1 dpi. The disease incidences recorded in HUM 1 variety at 2, 3, 4 and 20 dpi were 5.6, 8.3, 13.9 and 16.7%. In the presence of T. virens, R. solani-inoculated plants did not show any infection at 1 dpi in both susceptible and resistant varieties. Resistant variety HUM 1 did not show any infection at 2 dpi. At 3, 4 and 20 dpi, the disease incidences were 2.8, 2.8 and 53% in HUM 1. In case of susceptible variety, disease incidences were 2.8, 5.6, 8.3 and 27.8% at 2, 3, 4 and 20 dpi, respectively.
Discussion
The expression of defense genes, namely PR 10, EH, catalase and calmodulin, induced in response to T. virens and R. solani was measured in the resistant and susceptible varieties of mung bean. In the present study, the expression of PR 10 was upregulated in both susceptible and the resistant varieties in response to T. virens excluding 2 and 4 dpi in the resistant variety. Although T. virens alone induced upregulation in the susceptible variety at all dpi, in case of resistant variety, it was upregulated only at 1 and 2 dpi. PR10 expression was upregulated at 1, 2 and 4 dpi in the susceptible variety while downregulated in the resistant variety in response to R. solani. T. virens in the presence of R. solani upregulated PR 10 in both susceptible and resistant varieties at all days of observations. Similarly, Pulla et al. (2010) observed an increase in PR10 transcripts in ginseng leaves compared to the untreated control after challenging with Colletotrichum gloeosporioides, Phytophthora capsici, and Alternaria solani. Starting from as early as 3 h after challenge, there was a continuous increase in the expression of PR10 transcript up to 48 h of challenge. Inoculation of maize ears with Aspergillus flavus significantly upregulated the expression of PR10 gene as early as 1 dpi up to 10 dpi as compared to non-inoculated control and reached the highest level of 3.1-fold induction at 2 dpi. The present results are in contrast with the expression rate of defense gene PR10 in 2-week-old seedlings of resistant and susceptible rice varieties inoculated with a virulent isolate of R. solani AG-I-1A. The expression rate of PR10 gene in the resistant cultivar was higher than that of the susceptible cultivar (Sayari et al. 2014). In the present findings, the mean expression in the resistant variety (HUM 1) of mung bean was downregulated in response to R. solani, whereas it was upregulated in the presence of T. virens. Contrary to our results, in partially field-resistant cotton variety, PR10 was overexpressed after inoculation with Fusarium oxysporum f. sp. vasinfectum but not in the susceptible cultivar (Zambounis et al. 2012). Similarly, PR10 was induced in infected hypocotyls from 2 dpi to 4 dpi in cotton after infection with F. oxysporum f. sp. vasinfectum (Dowd et al. 2004). The elevated level of PR-10 in rice plants in response to M. grisea was also observed (Kim et al. 2004). The present study is also in accordance with the results of Perazzolli et al. (2011) who observed that Plasmopara viticola infection induced the expression of PR10 in grape wine. In addition, Trichoderma harzianum T39 increased the upregulation of PR-10 threefold.
Catalase is one of the most important scavengers of reactive oxygen species, which catalyzes the decomposition of hydrogen peroxide (H2O2). The oxidative burst may escort to the cross-linking of cell wall proteins making the cell wall more resistant to attack by fungal enzymes (Bradley et al. 1992). The active oxygen species may act directly as toxins against the pathogens (Mehdy 1994) and they may act as second messengers for the activation of a variety of defense genes (Lamb and Dixon 1997). In the present findings in response to R. solani and T. virens separately, the activity of catalase increased at 1 dpi and 4 dpi in the resistant variety, whereas it was increased only at 4 dpi in the susceptible variety, but the combined response of these two upregulated the expression of catalase only in the resistant variety at 1 dpi and 4 dpi. The present findings are supported by the earlier observations that the catalase activity significantly increased in rice leaf sheaths at 2 days after inoculation with R. solani and reached maximum at 5 days after inoculation (Paranidharan et al. 2003). Similarly, increased catalase activity was observed in inoculated apple fruits in combination of T. virens and Penicillium expansum in comparison with healthy control at all days of observations and the highest activity was noted at 6 days after inoculation (Bordbar et al. 2010). The present results are in accordance with the findings of Nogueira-Lopez et al. (2018). They reported that biocontrol fungus T. virens as an opportunistic plant endophyte colonized the maize roots and provided immune response due to the presence of secreted proteins which are mainly involved in cell wall hydrolysis, scavenging of reactive oxygen species and secondary metabolism, as well as act as putative effector-like proteins.
Epoxide hydrolase (EH) is another important defense-related gene expressed in both resistant and susceptible varieties of mung bean at 4 dpi in response to R. solani and T. virens alone and in combination. Except 3 dpi in resistant variety, its expression was upregulated at all days of observations in both the varieties in combined application of R. solani and T. virens. T. virens alone induced more expression in the resistant variety as compared to plants treated with T. virens + R. solani. Similar to the present findings, induced expression of EH was observed in rough lemon in response to non-pathogenic strains of Alternaria alternata (Gomi et al. 2003).
Calcium ion (Ca2+) influx is involved in the activation of defense response through the production of phytoalexin, induction of defense-related genes and hypersensitive cell death (Levine et al. 1996). In the present study, T. virens alone and in combination with R. solani induced the expression of calmodulin gene at all days after inoculation in the resistant variety of mung bean with the highest expression at 4 dpi. In case of the susceptible variety inoculated with R. solani, it was induced at 1 dpi and 3 dpi whereas in the resistant variety it was induced at 2 and 3 dpi. The present findings are in accordance with the earlier observations that the treatment of soybean cell suspension cultures with a non-specific fungal elicitor prepared from F. solani increased mRNA more than tenfold. The mRNA level peaked at 1 h and then slowly declined to basal level by 12 h (Heo et al. 1999).
In general, the expression of the defense-related genes in response to T. virens was more as compared to R. solani alone which may be due to the production of enzymes and toxins by R. solani during the process of infection to bypass the defense mechanisms. It is also evident that the disease development was less in the plants treated with T. virens in the presence of R. solani as compared to R. solani alone. R. solani did not cause any infection up to 1 dpi in the susceptible variety and up to 2 dpi in the resistant variety in the presence of T. virens. This might be due to overexpression of defense-related genes particularity PR 10 and calmodulin at 1 dpi. The present findings are in accordance with the earlier observations that T. virens spores or cell-free culture filtrate significantly reduced disease incidence in tomato caused by F. oxysporum f. sp. lycopersici and induced jasmonic acid and salicylic acid signalling cascades for the elicitation of wilt resistance in tomato (Jogaiah et al. 2018). T. virens as an opportunistic plant endophyte also induced immune response in maize (Nogueira-Lopez et al. 2018). Seed treatment with T. hamatum induced systemic and durable immunity in pearl millet against downy mildew under greenhouse and field conditions. RT-PCR analysis revealed differentially expressed transcripts of the defense enzymes and PR proteins in treated and untreated checks. PR-1, PR-5, and cell wall defense-related genes were significantly overexpressed in T. hamatum-treated seedlings as compared to controls (Siddaiah et al. 2017). In the present study, the level of expression of defense-related genes was higher in case of bio-control agent T. virens. The expression of defense genes was higher in the resistant variety, which may be due to the inbuilt mechanism of the pathway related to these genes. The present results clearly indicated that the expression of defense-related genes in response to T. virens alone and in the presence R. solani is accelerated both in resistant and susceptible varieties of mung bean which is an important factor to protect the plants at early stages of infection by R. solani. The findings may be further utilized for the management of the disease by applying the bio-formulations of T. virens and understanding of defense pathway mediated by these genes as well as manipulation of defense genes during breeding programmes.
References
Abawi GS (1989) Root rots. In: Schwartz HF, Pastor-Corrales MA (eds) Bean production problems in the tropics. CIAT, Cali, pp 105–157
Bordbar FT, Etebarian HR, Navazollah S, Hamid R (2010) Control of postharvest decay of apple fruit with Trichoderma virens isolates and induction of defense responses. J Plant Prot Res 50:146–152
Bradley DJ, Kjellbom P, Lamb CJ (1992) Elicitor-and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid defense response. Cell 70:21–30
Conrath U, Chert Z, Ricigliano JW, Kiessig DF (1995) Two inducers of plant defense responses, 2-6-dichloroisonicotinic acid and salicylic acid, inhibit catalase activity in tobacco. P Natl Aca Sci USA 92:7143–7147
Dowd C, Wilson IW, McFadden H (2004) Gene expression profile changes in cotton root and hypocotyl tissues in response to infection with Fusarium oxysporum f. sp. vasinfectum. Mol Plant Microbe Interact 17:654–667
Dubey SC (2003) Integrated management of web blight of urd/mung bean by bio seed treatment. Indian Phytopathol 56:34–38
Dubey SC, Singh B (2013) Integrated management of major diseases of mungbean by seed treatment and foliar application of insecticide, fungicides and bioagent. Crop Prot 47:55–60
Dubey SC, Bhavani R, Singh B (2011) Integration of soil application and seed treatment formulations of Trichoderma species for management of wet root rot of mungbean caused by Rhizoctonia solani. Pest Manag Sci 67:1163–1168
Foyer CH, Descourvieress P, Kunert KJ (1994) Protection against oxygen radicals: an important defense mechanism studied in transgenic plants. Plant Cell Environ 17:507–523
Gomez KA, Gomez AA (1984) Statistical procedures for agricultural research. Wiley, Singapore, pp 39–153
Gomi K, Yamamato H, Akimitsu K (2003) Epoxide hydrolase: a mRNA induced by the fungal pathogen Alternaria alternate on rough lemon (Citrus jambhiri Lush). Plant Mol Biol 53:189–199
Heo WD, Lee SH, Kim MC, Kim JC, Chung WS, Chun HJ, Lee KJ, Park CY, Park HC, Choi JY, Cho MJ (1999) Involvement of specific calmodulin isoforms in salicylic acid-independent activation of plant disease resistance responses. Proc Natl Acad Sci USA 96:766–771
Jacobs AK, Dry IB, Robinson SP (1999) Powdery mildew infection and ethephon treatment induce different pathogenesis related cDNAs in grapevine. Plant Pathol 48:325–336
Jogaiah S, Abdelrahman M, Tran LS, Ito S-I (2018) Different mechanisms of Trichoderma virens-mediated resistance in tomato against Fusarium wilt involve the jasmonic and salicylic acid pathways. Mol Plant Pathol 19:870–882
Kato T, Yamaguchi Y, Uehara T, Yokoyama T, Namai T, Yamanaka S (1983) Defense mechanism of rice plants against rice blast disease. Naturwissenschaften 70:200–201
Khattak GSS, Haq MA, Shraf M, Tahir GR (2002) Triple test cross analysis for some morphological traits in mungbean (Vigna radiata (L.) Wilczek). Euphytica 123:413–420
Kim ST, Kim SG, Hwang DH, Kang SY, Kim HJ, Lee BH, Lee JJ, Kang KY (2004) Proteomic analysis of pathogen-responsive proteins from rice leaves induced by rice blast fungus, Magnaporthe grisea. Proteomics 4:3569–3578
Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48:251–275
Levine A, Pennell RI, Alvarez ME, Palmer R, Lamb C (1996) Calcium-mediated apoptosis in a plant hypersensitive disease resistance response. Curr Biol 6:427–437
Liu JJ, Ekramoddoullah AKM (2006) The family 10 of plant pathogenesis-related proteins: their structure, regulation, and function in response to biotic and abiotic stresses. Physiol Mol Plant Pathol 68:3–13
Means AR, Dedman JR (1980) Calmodulin-an intracellular calcium receptor. Nature 285:73–77
Mehdy MC (1994) Active oxygen species in plant defense against pathogens. Plant Physiol 105:467–472
Nogueira-Lopez G, Greenwood DR, Middleditch M, Winefield C, Eaton C, Steyaert JM, Mendoza-Mendoza A (2018) The Apoplastic Secretome of Trichoderma virens during interaction with maize roots shows an inhibition of plant defence and scavenging oxidative stress secreted proteins. Front Plant Sci 9:409. https://doi.org/10.3389/fpls.2018.00409
Paranidharan V, Palaniswami A, Vidhyasekaranand Velazhahan PR (2003) Induction of enzymatic scavengers of active oxygen species in rice in response to infection by Rhizoctonia solani. Acta Physiol Plant 25:91–96
Perazzolli M, Roatti B, Bozza E, Pertot I (2011)) Trichodermaharzianum T39 induces resistance against downy mildew by priming for defense without costs for grapevine. Biol Control 58:74–82
Pinot F, Salaun JP, Bosch H, Lesot A, Mioskowski C, Durst F (1992) W-hydroxylation of Z9-octadecenoic, Z9, 10-epoxystearic and 9, 10-dihydroxystearic acids by microsomal cytochrome P450 systems from Vicia sativa. Biochem Biophys Res Commun 184:183–193
Pulla RK, Lee OR, Jun-Gyo I, Yu-Jin K, Senthil K, Yang DC (2010) Expression and functional characterization of pathogenesis-related protein family 10 gene, PgPR10-2, from Panax ginseng C.A. Meyer. Physiol Mol Plant Pathol 74:323–329
Sayari M, Babaeizad V, Ghanbari MAT, Rahimian H (2014)) Expression of the pathogenesis related proteins, NH-1, PAL, and lipoxygenase in the Iranian Tarom and Khazar rice cultivars, in reaction to Rhizoctonia solani—the causal agent of rice sheath blight. J Plant Prot Res 54:36–43
Siddaiah CN, Satyanarayana NR, Mudili V, Gupta VK, Gurunathan S, Rangappa S, Huntrike SS, Srivastava RK (2017) Elicitation of resistance and associated defense responses in Trichoderma hamatum induced protection against pearl millet downy mildew pathogen. Sci Rep 7:43991. https://doi.org/10.1038/srep43991
Walter MH, Liu JW, Grand C, Lamb CJ, Hess D (1990) Bean pathogenesis-related (PR) proteins deduced from elicitor-induced transcripts are members of a ubiquitous new class of conserved PR proteins including pollen allergens. Mol Gen Genet 222:353–360
Zambounis AG, Kalamaki MS, Tani EE, Paplomatas EJ, Tsaftaris AS (2012) Expression analysis of defense-related genes in cotton (Gossypium hirsutum) after Fusarium oxysporum f. sp. vasinfectum infection and following chemical elicitation using a salicylic acid analog and methyl jasmonate. Plant Mol Biol Rep 30:225–234
Acknowledgements
The financial support provided through ICAR-NFBSFARA project is duly acknowledged by the authors.
Funding
The financial support provided through ICAR-NFBSFARA project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No potential conflict of interest was reported by the authors.
Rights and permissions
About this article
Cite this article
Dubey, S.C., Tripathi, A. & Tak, R. Expression of defense-related genes in mung bean varieties in response to Trichoderma virens alone and in the presence of Rhizoctonia solani infection. 3 Biotech 8, 432 (2018). https://doi.org/10.1007/s13205-018-1453-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-018-1453-2