Abstract
Seven isolates of Trichoderma spp. (T1 to T7) from Egypt were evaluated in vitro by bioassay for their potential to antagonize Fusarium oxysporum f.sp. lycopersici (FOL, the causal pathogen of tomato wilt disease). The highest percentage of inhibition against the tested pathogenic isolate were obtained with Trichoderma isolate (T7) followed by Trichoderma isolate (T3). In greenhouse experiments, the application of the highly antagonistic isolates of Trichoderma spp. (T3 and T7) led to a significant decrease of disease severity compared to the untreated control treatment. The lowest severity was achieved with the T3 isolate (24.8%) followed by isolate T7 (34.6%) compared with the other tested isolates. To understand the ability of Trichoderma isolates to protect against wilt disease, its induced systemic resistance in tomato plants has been studied. The expression of a defense-related gene (β-1,3-glucanase gene) was assessed by real-time RT-PCR in tomato plants to test the accumulation kinetics of transcripts encoding PR proteins in the roots of tomato in control (only with the pathogen), T3&FOL, and T7&FOL treatments. The highest degree of gene expression was found in tomato plants which were treated with T3&FOL compared with control (pathogen only). Two species of antagonistic Trichoderma (T3& T7) were characterized based on molecular tools using internal transcribed spacers (ITS1 and ITS4). The results of genetic characterization identified two different species of Trichoderma (T. atroviride and T. longibrachiatum).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tomato (Lycopersicon esculentum Mill.) is one of the most important vegetable crops worldwide [1]. Several serious pathogens attack tomato plants and cause a significant reduction in tomato production and productivity. One of these main pathogens of both greenhouse and field grown tomatoes is the soil-borne and host-specific pathogen Fusarium oxysporum f.sp. lycopersici (FOL) [2]. Fusarium wilt of tomato is a very difficult disease to treat because the pathogen spreads within the vascular tissues causing the plant to die. It also causes a high loss in yield in many tomato growing regions worldwide [3, 4]. There are two main approaches to control the disease; chemical application and biocontrol agents. Chemical application is a widely applied method to control soil-borne diseases, however, it has a potential risk to human health and increases environmental pollution, such as affecting the beneficial functions of microorganisms living in the soil and root ecosystem [5, 6]. Biocontrol agents (BCAs) have been used as an eco-friendly alternative method to control the disease instead of synthetic fungicides. Trichoderma spp. such as T. longibrachiatum and T. atroviride are common saprophytic fungi inhabitants in the soil with no inverse effect on the environment and non-toxic to human health [3, 7,8,9]. Many earlier studies focused on the ability of Trichoderma to reduce the incidence of Fusarium wilt disease of tomato [4, 10,11,12]. It can enhance plant growth, crop productivity, and induce both systemic and localized resistance to several plant pathogens [8]. Root colonization with Trichoderma strains can increase levels of defense-related plant enzymes and pathogenesis-related proteins (PR) [13, 14]. β 1,3-glucanase (PR-2) is one of the most important PR proteins which causes a direct inhibition of growth of several plants pathogens [15, 16]. Due to the ecological importance of Trichoderma spp. and its application as a biocontrol agent in the field, it is important to identify strains of Trichoderma spp. that can be used for controlling many plant diseases. The molecular and identification tools based on sequence analysis of multiple genes can be useful to identify each Trichoderma isolate and characterize it as a putative new species [17,18,19]. The internal transcribed spacer (ITS) regions of the rDNA are the most widely sequenced DNA regions in fungi. It has typically been used for molecular systematic study among and within species [19,20,21,22]. The objectives of this study were to (1) evaluate the efficacy of various isolates of Trichoderma species as a biocontrol agent against Fusarium oxysporum f.sp. lycopersici (FOL) for control of tomato wilt disease in vitro and under greenhouse conditions, (2) quantify the effect of Trichoderma isolates on the expression of genes controlling Pr-proteins production as a defense mechanism in plants using a qRT-PCR technique and (3) identify and characterize the most effective Trichoderma isolates using ITS.
Materials and methods
Plant material and pathogenic isolates
Seeds from Super Marmande tomato cultivar were procured from the Department of Vegetables, Faculty of Agriculture, Assiut Uni., Egypt and used in this study. This tomato cultivar is a susceptible to Fusarium wilt disease [23].
The virulent isolate of Fusarium oxysporum f. sp. lycopersici (FOL) was obtained from the Department of Plant Pathology, Faculty of Agriculture, Assiut University, Egypt.
Isolation and purification of Trichoderma isolates
Seven different isolates of Trichoderma spp. were isolated from the soil rhizosphere of a tomato field in Egypt. For the isolation of Trichoderma spp., soil samples were diluted into different concentrations and well vortexed. The supernatant was poured into plates of PDA (potato dextrose agar, with a 10 mg/ml stock solution of chloramphenicol which is used to prevent bacterial growth in the medium) and incubated at 28 °C ± 1 °C for 4 days. After incubation period colonies appearing on the plates were transferred into a new plate containing PDA media for 7 days at 28 °C ± 1 °C. Then, the single spore colonies were obtained by subculturing at 28 °C ± 1 °C to get a homogenous fungal culture. Isolates were maintained on PDA medium at 4 °C.
Effect of Trichoderma isolates against FOL pathogen in vitro
The antagonistic capability of the seven fungal isolates of Trichoderma spp. was tested in vitro against FOL by a dual culture technique. Isolates of Trichoderma spp. and FOL were grown on PDA medium for 6 days and used as an inoculum. Disks from each isolate of Trichoderma spp. (5 mm in diameter) were inoculated on PDA medium on one side in the Petri plate and the opposite side was plated by FOL inoculum (5 mm in diameter). Three replicates were used for each treatment and inoculated plates with FOL only were used as a control treatment. After a five days incubation period at 28 °C ± 1, the linear growth of the tested pathogen was recorded. Percentage of growth inhibition was calculated using the following formula [24]:
Effect of Trichoderma spp. against FOL pathogen under greenhouse conditions
Preparation of FOL inocula and soil infestation
Inocula of the FOL isolate was prepared by growing it 15 days on a Barley grain medium which consisted of 100 g of barley grains and 80 ml water. Then, the media was autoclaved at 121 °C for 20 min two times (two consecutive days). Sterilized pots (20 cm.) filled with three kg sterilized sandy loam soil (soil: sand w/w) were used in this experiment. The soil was infested with the FOL (2% w/w) and kept for 7 days with regular irrigation. Then, tomato plants were transplanted to the infested soil. The highest antagonistic isolates of Trichoderma spp. (T7 and T3) were selected from the in vitro test to be used in further genetic analyses. The inoculums of the selected antagonistic isolates were multiplied by transferring the disc of each isolate (5 cm diameter culture) into Erlenmeyer flasks containing100 ml of moist wheat bran. Then, the inoculated substrates were incubated at 28 °C ± 1 for 2 weeks (until all substrates were covered by Trichoderma isolates) [25]. Twenty-eight days old tomato seedlings were transplanted into pots after 7 days from soil infestation with the pathogen. Each pot contained three tomato seedlings [23]. The antagonistic isolates were added to the infested soil (at the rate of 1% w/w) at the time of transplanting tomato plants to the pots. Pots without the Trichoderma spp. were used as control (only pathogen). Three replicates (three pots) were used in each treatment. Three tomato seedlings (1-month old) were transplanted in each pot
Measurement of disease severity
Wilt severity from tomato seedlings was scored after 60 days using a scale suggested by [26]. This was based on the wilt severity rated as follows; (% of shoot wilted, using a scale of 0–5 where, 0 = no symptoms, 1 = one leaf wilted (1–25%), 2 = 2 or 3 leaves wilted (26–49%), 3 = half plant wilted (50–74%), 4 = all leaves wilted (75–100%), 5 = dead plan).
where n refers to the number of plants with in each infection category, V is the numerical values of infection categories, N is total number of plants examined, and 5 is a constant and the highest numerical value.
Expression analysis of the PR gene by RT-PCR in tomato roots
Roots samples (2 g) were collected 15 days after inoculation from each treatment (the infested plants with FOL only, FOL& T3 and FOL& T7) at greenhouse experiment, washed, and briefly dried. Harvested plant tissue were immediately chilled in liquid nitrogen, crushed with a glass rod, then stored in liquid nitrogen until buffer was added. RNA was extracted following a protocol adapted from the Extract-A-Plant RNA Isolation kit (Agilent Plant RNA Mini Protocol, Agilent Technologies, USA). Total cellular RNA was extracted for (qRT-PCR) by removal of genomic DNA from RNA preparations and generation of cDNA were performed using a modified protocol from Thermo Scientific Revert Aid, First Strand cDNA Synthesis Kit. The gene-specific PCR primers used in this study were designed for the β-1,3-glucanase gene of tomato. Tomato actin gene (LOC100147713) was used as a constitutive control in all gene expression experiments. The reaction included an initial 5 min denaturation at 95◦C, followed by 39 cycles of PCR (95 °C, 15 s 58 °C, 30 s, and 72 °C, 45 s) and a final extension period at 65 °C, 0.05 s and 95 °C, 5 s. PCR products were separated on 1% agarose gels and stained with ethidium bromide to determine the size of the PCR products. Primer sequences were checked with GenEx software from multi D analyses.
Molecular characterization of different Trichoderma isolates
DNA extraction and amplification
For DNA extraction, each tested isolate of Trichoderma spp. was grown on potato dextrose agar. Fresh mycelia samples were collected from three days old cultures. Mycelia were kept in the refrigerator before genomic DNA isolation. The extraction of total genomic DNA of each isolate of Trichoderma spp. was made according to [27].
Amplification of isolated DNA using ITS
The internal transcribed spacer (ITS) regions of the rDNA were amplified using the two primers, ITS-1 (TCC GTA GGT GAA CCT GCG G) and ITS4 (TCC TCC GCT TAT TGA TAT GC) (Table 1) which were synthesized on the basis of conserved regions of the eukaryotic rRNA gene [28]. The PCR-amplification reactions were performed in a total volume of 50 µl with extraction kit using a protocol of PCR amplification by PEQGOLD Cycle-Pure Kit (PEQLAB Biotechnologie GmbH, Erlangen) and sequenced with the forward primers (Source Bioscience, Berlin). The cycle start step of 30 s at 98 °C, followed by 35 cycles of denaturation for 10 s at 98 °C, annealing for 20 s at 54 °C, and elongation for 35 s at 72 °C, and then a finishing step of 10 min at 72 °C. Amplified products were separated on 1.2% agarose gel in TAE buffer, pre-stained with ethidium bromide (1 μg/ml) and electrophoresis was carried out at 60 V for 3 h in TAE buffer. GeneRuler 1 kb DNA ladder was used as a marker. The gel was observed in a transilluminator over ultraviolet light. The desired bands were cut from the gel with a minimum quantity of gel portion using the QIAGEN gel extraction kit. A pair of universal ITS primers ITS-1 (forward) and ITS-4 (reverse) was used for sequencing of the amplified product and this step was carried out at the Source Bioscience Sequencing (Berlin, Germany). The band size was determined in the gel using GenAlEx 6.5 software [29].
Statistical analysis
Data was analyzed by statistical variance (ANOVA) using MSTAT-C program version 2.10 (1991). The mean values of tested treatments were compared according to the procedures described by [30]. The least significant differences (LSD) at P ≤ 0.05 were used to test the significance of the differences among the mean values of tested treatments.
Results
Effect of Trichoderma antagonists against FOL pathogen in vitro
The percentage of inhibition for the different Trichoderma isolates against FOL pathogen is presented in Table 2. The results indicated that the tested isolates of Trichoderma spp. had different percentages of inhibition against the tested pathogenic isolate with a range extended from 51.83 (T6) to 66.80% (T7). The tested isolates of Trichoderma spp. No.7 followed by No. 3 had the greatest percentage of inhibition. Therefore, these two isolates were used for further experiments.
Effect of Trichoderma spp. against FOL pathogen under greenhouse experiment
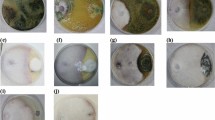
The effect of selected highly antagonistic isolates of Trichoderma spp. (No. T3 and T7) soil on the pathogenic fungi (Fig. 1) was found effective in suppressing wilt disease incidence of tomato compared with control (80.7%). Notably, the application of the isolate T3 to infested soil caused the highest reduction in disease severity percentage (24.8%) followed by isolate T7 (34.6%).
Expression analysis of the PR gene by RT-PCR in tomato roots
The expression profile of the defense-related β-1,3-glucanase gene was tested in the control (infected with FOL) and treated plants (FOL&T3 and FOL&T7) using RT-PCR (Fig. 2). The gene was expressed in the three different treatments. As expected, the gene expression was higher in the treated plants with FOL&T3 and FOL&T7 than in control plants. Noticeably, the gene expression in FOL&T3 was much higher than in FOL&T7. The analysis of variance for the amount of gene expression among the three treatments was tested (Table 3). High significant differences in the amount of gene expression (P >0.01) among the different treatments were observed.
Real-time RT-PCR analysis of PR gene expression in tomato roots upon a treatment with either a pathogenic only (control (F)) or FOL (F) with Trichoderma spp. (T). Means in each column designated by the same letter(s) are not significantly different at 0.01% probability according to Duncan’s multiple range test
Molecular characterization among different Trichoderma isolates
The internal transcribed spacer (ITS) regions for ITS1 and ITS4 have been successfully used in this study to test and identify Trichoderma spp. (T3 and T7). Specific primers can differentiate closely related fungal species. The PCR results of T3 and T7 are presented in Fig. 3. A band size of 636 was amplified in T7, while, a band size of 657 was amplified in T3. The result of molecular characterization indicated that the candidate isolates for T3 and T7 were Trichoderma longibrachiatum and Trichoderma atroviride, respectively (Table 4).
Discussion
Seven unidentified isolates of Trichoderma spp., collected from Egypt were tested for their ability to inhibit the growth of FOL. This effect was tested by a dual culture experiment. The dual culture experiment as described by many earlier studies has been widely used in antagonistic activity experiments [31,32,33]. The results of this study revealed that all tested fungal isolates varied in inhibiting FOL growth due to the different mycoparasitic capacity of the various isolates. The diversity observed in vitro might be due to the variability of fungal genotypes with differences in growth, sporulation, and gene-environment interaction possibly explaining differences in their capacity for FOL inhibition. The growth rate of the seven Trichoderma isolates was faster than the FOL in culture tests, and a quick overpowering of the pathogen was observed. The same observation was also reported by [34]. Of the seven isolates, T3 and T7 displayed the greatest percentage of inhibition. The inhibitory effect of these bioagents against the tested pathogen may be due to the direct effect of antagonistic fungi against the pathogens through the cloning of their hyphae around the hyphae of the pathogens to prevent their continued growth [35] and/or produce an antagonistic substance which can have an important role in lysis of cell wall components of the pathogenic fungi to help the antagonists to penetrate the host hyphae and grow on it as a hyperparasite [36].
Under greenhouse conditions, we applied the two most antagonistic fungi (T3 and T7), as selected from in vitro tests, to soil infested with FOL and observed a reduction of disease severity percentage. Trichoderma isolate T3 had the lowest disease severity percentage. The greenhouse experiment was very useful for evaluating the efficiency of different isolates of Trichoderma species in suppressing fusarium wilt disease and improving plant health. Moreover, the ability of Trichoderma to induce systemic resistance in plants has been extensively studied [4, 37]. Occasionally, the antifungal activity of Trichoderma seen in vitro assay may be different from in vivo in controlling the diseases caused by pathogenic fungi due to many other factors. The factors that affect the antifungal activity of Trichoderma in vivo are plant species, soil type, soil temperature, moisture and nutrient availability [37, 38]. Trichoderma spp. is a nonpathogenic microorganism that provides protection to many crops against fungal diseases caused by the Fusarium genera [39]. Using biocontrol agents may prove to be a convenient alternative method to control plant disease, avoiding the adverse effects accompanying chemical control [8]. They have been commercialized as biopesticides, biofertilizers, and soil enhancers [39].
The expression of the PR (β-1,3-glucanase) gene was tested by RT-PCR in tomato roots (plants infested with FOL only, FOL&T3 and FOL&T7). PR proteins were found to be highly expressed under biotic stress conditions because they have direct antifungal activity [40]. Moreover, these proteins can induce host resistance by letting out oligomers from microbial cell walls, which work as elicitors for a defense response [41]. Therefore, they play a vital role in the biological control of soil-borne diseases [42]. Compared to control (plants infected with FOL), treatment of infected plants with Trichoderma increased the expression of PR proteins. Significant differences were found in the expression of PR proteins in the roots of plants treated with the two Trichoderma isolates. The expression of the PR gene was higher in FOL&T3 than those in FOL&T7. The results of gene expression are in full agreement with the percentage of disease severity. FOL&T3 had the lowest percentage of disease severity because this treatment had increased the expression of β-1,3-glucanase gene, leading to a high amount of PR protein accumulation. On the other hand, plants treated with FOL&T7 had a higher percentage of disease severity with a corresponding lower expression of the β-1,3-glucanase gene.
The efficacy of the induced resistance varied according to the biocontrol strains [5]. The degree of resistance depended not only on the qualitative differences in the activated defense genes but also on differences in the timing and magnitude of their expression [43]. Induction of PR genes plays an essential and indispensable role in the protection against root pathogens [44]. β-1,3-glucanases are considered key plant defense enzymes acting on fungal cell wall degradation and constitute several of the PR gene families (PR2, PR3, PR4, PR8, and PR11). The difference in PR gene expression between FOL&T3 and FOL&T7 could be due to different isolates of Trichoderma. To investigate this assumption, we conducted further experiments to genetically distinguish and characterize the two isolates. DNA markers have been used for species identification because culture and disease score data did not discriminate Trichoderma species isolates [45].
In this study, we used a PCR assay to identify the species of the antagonistic Trichoderma isolates using primers amplifying the ITS region of ribosomal genes. The internal transcribed spacer (ITS) is one of the most reliable loci to generate specific primers that can differentiate closely related fungal species [21]. According to amplification of the rDNA region in both isolates (T3 and T7), two polymorphic bands appeared at 636 and 657 bp in T7 and T3. The results of the molecular characterization of these two bands revealed that T7 and T3 are different isolates and were identified as Trichoderma atroviride (636 bp) and Trichoderma longibrachiatum (657 bp), respectively. Trichoderma longibrachiatum was identified using ITS1 and ITS4 by [46]. They found an amplified PCR product with band size at 700 bp which is close to the band size found in this study for the same Trichoderma longibrachiatum. Species identification using molecular characterization tools is very useful for answering the question of whether a particular taxon is present on particular hosts or plants [47]. This will result in reducing the severity of wilt disease on tomato plants by using the appropriate bioagent.
In conclusion, plants interact with bioagents which induce resistance to serious fungal diseases. Trichoderma has been considered as an internationally important biocontrol fungus due to its significant effect on wilt disease in plants. Among the seven Trichoderma isolated in this study, we identified and characterized Trichoderma longibrachiatum as a very useful bioagent that can reduce the severity of wilt disease to 24.8%. Moreover, the expression of PR genes is very important and useful to support morphological and culture data.
References
Barari H (2016) Biocontrol of tomato Fusarium wilt by Trichoderma species under in vitro and in vivo conditions. Cercet Agron Mold 49(1):91–98. https://doi.org/10.1515/cerce-2016-0008
Jarvis W (1988) Fusarium crown and root rot of tomatoes. Phytoprotection 69:49–64
Ghazalibiglar H, Hampton JG, van ZijlldeJong E, Holyoake A (2016) Evaluation of Paenibacillus spp. isolates for the biological control of black rot in Brassica oleracea var. capitata (cabbage). Biocontrol Sci Technol 26:504–515. https://doi.org/10.1080/09583157.2015.1129052
Verma V, Ravindran P, Kumar PP (2016) Plant hormone-mediated regulation of stress responses. BMC Plant Biol 16:86. https://doi.org/10.1186/s12870-016-0771-y
Harman GE (2006) Overview of mechanisms and uses of Trichoderma spp. Phytopathology 96:190–194. https://doi.org/10.1094/PHYTO-96-0190
Akrami M, Yousefi Z (2015) Biological control of Fusarium wilt of tomato (Solanum lycopersicum) by Trichoderma spp as Antagonist fungi. Biol Forum 7:887–892
Boureghda H, Bouznad Z (2009) Biological control of Fusarium wilt of chickpea using isolates of Trichoderma atroviride, T. harzianum and T. longibrachiatum. Acta Phytopathol Entomol Hungarica 44:25–38. https://doi.org/10.1556/APhyt.44.2009.1.4
Kareem T, Ugoji O, Aboaba O (2016) Biocontrol of Fusarium wilt of cucumber with Trichoderma longibrachiatum NGJ167 (Rifai). Br Microbiol Res J 16:1–11. https://doi.org/10.9734/BMRJ/2016/28208
Galarza L, Akagi Y, Takao K et al (2015) Characterization of Trichoderma species isolated in Ecuador and their antagonistic activities against phytopathogenic fungi from Ecuador and Japan. J Gen Plant Pathol 81:201–210. https://doi.org/10.1007/s10327-015-0587-x
Bell DK, Wells HD, Markham CR (1982) In vitro antagonism of Trichoderma species against six fungal pathogens. Phytopathology 72(4):379–382
Elad Y, Kapat A (1999) The role of Trichoderma harzianum protease in the biocontrol of Botrytis cinerea. Eur J Plant Pathol 105:177–189. https://doi.org/10.1023/A:1008753629207
Ramezani H (2011) Efficacy of some fungal and bacterial bioagents against Fu sarium oxysporum f. sp. ciceri on chickpea
Harman GE, Howell CR, Viterbo A et al (2004) Trichoderma species—opportunistic, avirulent plant symbionts. Nat Rev Microbiol 2:43–56. https://doi.org/10.1038/nrmicro797
Yedidia I, Benhamou N, Chet I (1999) Induction of defense responses in cucumber plants (Cucumis sativus L.) By the biocontrol agent Trichoderma harzianum. Appl Environ Microbiol 65:1061–1070
Leah R, Tommerup H, Svendsen I, Mundy J (1991) Biochemical and molecular characterization of three barley seed proteins with antifungal properties. J Biol Chem 266:1564–1573
Kauffmann S, Legrand M, Geoffroy P, Fritig B (1987) Biological function of;pathogenesis-related’ proteins: four PR proteins of tobacco have 1,3-beta-glucanase activity. EMBO J 6:3209–3212
Druzhinina IS, Kopchinskiy AG, Komoń M et al (2005) An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genet Biol 42:813–828. https://doi.org/10.1016/j.fgb.2005.06.007
Samuels GJ (2006) Trichoderma: systematics, the sexual state, and ecology. Phytopathology 96:195–206. https://doi.org/10.1094/PHYTO-96-0195
Kubicek CP, Komon-Zelazowska M, Druzhinina IS (2008) Fungal genus Hypocrea/Trichoderma: from barcodes to biodiversity. J Zhejiang Univ Sci B 9:753–763. https://doi.org/10.1631/jzus.B0860015
Ospina-Giraldo MD, Royse DJ, Thon MR et al (1998) Phylogenetic relationships of Trichoderma harzianum causing mushroom green mold in Europe and North America to other species of Trichoderma from world-wide sources. Mycologia 90:76–81. https://doi.org/10.1080/00275514.1998.12026881
Kullnig-Gradinger CM, Szakacs G, Kubicek CP (2002) Phylogeny and evolution of the genus Trichoderma: a multigene approach. Mycol Res 106:757–767. https://doi.org/10.1017/S0953756202006172
Lee C-F, Hseu T-H (2002) Genetic relatedness of Trichoderma sect. Pachybasium species based on molecular approaches. Can J Microbiol 48:831–840
Selim ME, Khalifa E, Amer G et al (2015) Evaluation and characterization of some Egyptian Fusarium oxysporum isolates for their virulence on tomato and PCR detection of (SIX) effector genes. J Bioprocess Biotech 05:1–6. https://doi.org/10.4172/2155-9821.1000204
Elrazik AAA, Hassan M, Koch E (2009) Powder formulations of Bacillus subtilis, Trichoderma spp and Coniothyrium minitans for biocontrol of Onion White Rot. Arch Phytopathol Plant Protect 42(2):142–147. https://doi.org/10.1080/03235400600982675
Frommel MI, Pazos GS, Nowak J (1991) Plant-growth stimulation and biocontrol of Fusarium wilt (Fusarium oxysporum f.sp. lycopersici) by co-inoculation of tomato seeds with Serratia plymuthica and Pseudomonas sp. Fitopatologia 26:66–73
Waudo SW, Owino PO, Kuria M, (Kenyatta UN (Kenya). D of B (1995) Control of Fusarium wilt of tomatoes using soil amendments. East African Agric For J 60(4):235–245
Liu YJ, Whelen S, Hall BD (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol Biol Evol 16:1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092
White TJ, Bruns T, Leem S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Gelfandm MADH, Gelfand J, Sninsky J, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, New York, pp 315–322
Peakall R, Smouse PE (2006) genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Gomez KA, Gomez AA, Gomez KA (1984) Statistical procedures for agricultural research. Wiley, Hoboken
Nakkeeran S, Renukadevi P, Marimuthu T (2005) Antagonistic potentiality of Trichoderma viride and assessment of its efficacy for the management of cotton root rot. Arch Phytopathol Plant Prot 38:209–225. https://doi.org/10.1080/03235400500094472
Singh BN, Singh A, Singh BR, Singh HB (2014) Trichoderma harzianum elicits induced resistance in sunflower challenged by Rhizoctonia solani. J Appl Microbiol 116:654–666. https://doi.org/10.1111/jam.12387
Srivastava RK, Singh RK, Kumar N, Singh S (2010) Management of macrophomina disease complex in jute (Corchorus olitorius) by Trichoderma viride. J Biol Control 24:77–79. https://doi.org/10.18311/JBC/2010/3578
El-Komy MH, Saleh AA, Eranthodi A, Molan YY (2015) Characterization of Novel Trichoderma asperellum isolates to select effective biocontrol agents against tomato Fusarium Wilt. Plant Pathol J 31:50–60. https://doi.org/10.5423/PPJ.OA.09.2014.0087
Adekunle A, Ikotun T, Florini D, Cardwell K (2002) Field evaluation of selected formulations of Trichoderma species as seed treatment to control damping-off of cowpea caused by Macrophomina phaseolina. Afr J Biotechnol 5:419–424
Papavizas GC (1984) Liquid fermentation technology for experimental production of biocontrol fungi. Phytopathology 74:1171. https://doi.org/10.1094/Phyto-74-1171
Mayo S, Gutiérrez S, Malmierca MG et al (2015) Influence of Rhizoctonia solani and Trichoderma spp. in growth of bean (Phaseolus vulgaris L.) and in the induction of plant defense-related genes. Front Plant Sci 6:685. https://doi.org/10.3389/fpls.2015.00685
Juroszek P, von Tiedemann A (2011) Potential strategies and future requirements for plant disease management under a changing climate. Plant Pathol 60:100–112. https://doi.org/10.1111/j.1365-3059.2010.02410.x
Tsegaye Redda E, Ma J, Mei J et al (2018) Antagonistic potential of different isolates of Trichoderma against Fusarium oxysporum, Rhizoctonia solani, and Botrytis cinerea. Eur J Exp Biol 08:212. https://doi.org/10.21767/2248-9215.100053
Zehra A, Meena M, Kumar Dubey M et al (2017) Activation of defense response in tomato against Fusarium wilt disease triggered by Trichoderma harzianum supplemented with exogenous chemical inducers (SA and MeJA). Braz J Bot 40:651–664. https://doi.org/10.1007/s40415-017-0382-3
Ashutosh Rai PU, Rai A, Kumar R et al (2014) Differential expression of pathogenesis related protein genes in tomato during inoculation with A. Solani. J Plant Pathol Microbiol 05:1–7. https://doi.org/10.4172/2157-7471.1000217
Javed S, Ahmad M, Ahmad M et al (2013) Chitinases: an update. J Pharm Bioallied Sci 5:21. https://doi.org/10.4103/0975-7406.106559
Khraiwesh B, Zhu J-K, Zhu J (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta 1819:137–148. https://doi.org/10.1016/j.bbagrm.2011.05.001
Kavroulakis N, Ehaliotis C, Ntougias S et al (2005) Local and systemic resistance against fungal pathogens of tomato plants elicited by a compost derived from agricultural residues. Physiol Mol Plant Pathol 66:163–174. https://doi.org/10.1016/J.PMPP.2005.06.003
Chaverri P, Castlebury LA, Overton BE, Samuels GJ (2003) Hypocrea/Trichoderma: species with conidiophore elongations and green conidia. Mycologia 95:1100–1140
Shahid M, Srivastava M, Sharma A et al (2013) Molecular characterization of Trichoderma longibrachiatum 21PP isolated from rhizospheric soil based on universal ITS primers. Afr J Microbiol Res 7:4902–4906. https://doi.org/10.5897/AJMR2013.5761
Abd-Elsalam KA, Almohimeed I, Moslem MA, Bahkali AH (2010) M13-microsatellite PCR and rDNA sequence markers for identification of Trichoderma (Hypocreaceae) species in Saudi Arabian soil. Genet Mol Res 9:2016–2024. https://doi.org/10.4238/vol9-4gmr908
Acknowledgements
The authors wish to thank Prof. Dr. Ralf T. Vögele and his research team, Department of Phytopathology, Agricultural Science Faculty, University of Hohenheim, Germany for supporting this work. We also appreciate Dr. Javed Siddique, Wheat Breeding and Training Director GRAIN Project of the Michigan State University in Afghanistan and Nathan Abshir, University of Nebraska-Lincoln, USA for discussing the results of this study. This work was supported and funded by Cultural Affairs & Mission Sector in Egypt.
Funding
This work was supported and funded by Cultural Affairs & Mission Sector in Egypt.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sallam, N.M.A., Eraky, A.M.I. & Sallam, A. Effect of Trichoderma spp. on Fusarium wilt disease of tomato. Mol Biol Rep 46, 4463–4470 (2019). https://doi.org/10.1007/s11033-019-04901-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-04901-9