Abstract
Non-invasive genetic sampling can be used in research, monitoring, and conservation of wild animals to, for example, provide insights into diets, identify individuals and estimate population sizes. Non-invasive genetic sampling may be especially useful to monitor elusive species because DNA can be derived from materials such as hairs and faeces without handling individuals. However, the reliability of the results derived from this technique is dependent on the quality of DNA obtained from samples, which can deteriorate from exposure to environmental conditions and sample age. While freshness of the sample is an important factor, the combined effect of different field conditions on the genotyping success is not fully understood. To address this gap, we systematically investigated the effects of sample age and environmental conditions on genotyping success of faeces, in an experimental setting of four treatments that combined rain and temperature conditions typical of central European climates. We compared the performance of a microsatellite marker set and a SNP panel for red fox (Vulpes vulpes), as well as a microsatellite marker set for pine marten (Martes martes) from faeces resampled over 21 days. We found that genotyping success decreased significantly with sample age, however environmental treatments did not impact the success. Furthermore, the SNPs we used amplified more successfully over time than the respective microsatellites. Therefore, the use of SNPs instead of conventional microsatellites, when using faecal samples for analysis relying on correct amplifications, could be advantageous. We recommend to prioritise the collection of fresh faeces regardless of environmental conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Successful management of wildlife populations depends on the ability to reliably monitor populations and derive accurate measures of, among others, species abundance or density (Stephenson et al. 2022). Using camera trapping data to monitor wildlife populations has become common practice (Burton et al. 2015), but camera trapping does not always provide accurate estimates, particularly for populations of elusive species, or for species where individuals are visually indistinguishable (Sollmann et al. 2013; Amburgey et al. 2021). For such populations, the use of genetics for capture-recapture methods may solve this problem.
Genetic information is a valuable resource for the conservation and management of wildlife, and can be used to verify the presence of rare and elusive species, as well as to explore a range of parameters, such as gene flow, hybridization, and demographic population structure (DeYoung and Honeycutt 2005; Mills 2013). Obtaining genetic information from live animals requires the often labour- and cost-intensive process of capturing and handling individuals. However, improvements in genetic methods have made it possible to extract individual and population-level data from non-invasive sampling of materials left behind by animals such as faeces, which are commonly sampled for a wide range of taxa (Waits and Paetkau 2005; Broquet et al. 2007). Since non-invasive genetic sampling does not require individuals to be captured or marked, data collection can be less costly in terms of materials, expertise, time, and effort, and is more ethical as animals are not stressed or harmed in the data collection process (Zemanova 2021).
Main challenges to non-invasive genetic sampling are the low quality and quantity of target DNA available in the sampling materials (Waits and Paetkau 2005; Broquet et al. 2007). If the DNA is not well preserved or sufficiently abundant, genotyping success may vary, and genetic analysis may be inconsistent or unachievable. Genotyping success can be influenced by the age of the sample (Lucchini et al. 2002; Piggott 2004; Murphy et al. 2007; Santini et al. 2007), and the environmental conditions to which it has been exposed. Low temperatures tend to be favourable while humidity and precipitation can reduce amplification success (Piggott 2004; Hájková et al. 2006; Santini et al. 2007; Brinkman et al. 2010; Kubasiewicz et al. 2016).
The efficiency of non-invasive genetic sampling can also be affected by the length of the marker types used in genetic analysis: typically, multi-allelic microsatellite makers (“microsatellites”) or single-nucleotide polymorphisms (SNPs). While both can be used for individual identification, parentage analysis and population genetics (Vignal et al. 2002; Ogden 2011; von Thaden et al. 2017; Allendorf et al. 2022), each can perform differently due to the length of the DNA fragments required for genetic amplification. SNP assays generally utilize DNA fragments that are shorter than traditional microsatellites (Morin et al. 2004; Fitak et al. 2016). If the DNA in the sample is degraded, genotyping errors can increase with the length of amplified fragments (Bonin et al. 2004; Broquet and Petit 2004), which may mean that SNPs are better at producing data from lower quality DNA than microsatellites that require longer DNA fragments (Seddon et al. 2005; Carroll et al. 2018). When applied on faeces, some studies found that indeed SNPs can outperform the longer microsatellites by generating higher amplification rates and fewer genotyping errors (Fabbri et al. 2012; Fitak et al. 2016). However, few studies have directly compared the two methods in realistic settings that allow for reliable evaluation.
Despite the strong and potentially interactive effects of sample age, temperature, rainfall, and methodological approaches on the success of genotyping non-invasive samples, few studies have directly compared the influence of these factors. By identifying the individual and interactive effects of these factors on genotyping success, sampling techniques can be improved by focusing collection efforts on the conditions that yield the best results. Alternatively, uncertainties in results can be better accounted for. In this study, we investigated the effects of sample age and four different environmental conditions on genotyping success of faeces in a standardized experimental setting, and compared performance of a microsatellite marker set versus a SNP panel. Using faeces from two elusive species, in which individuals are visually indistinguishable, red fox (Vulpes vulpes) and pine marten (Martes martes), we tested the following hypotheses on the effect of environmental conditions and amplification method on genotyping success:
-
(1)
Genotyping success decreases with sample age, i.e., the duration the faeces have been exposed to environmental conditions;
-
(2)
Genotyping success varies between environmental treatments, i.e., amplification success will decrease in samples exposed to an increasing amount of rain and to warm temperatures;
-
(3)
Genotyping success of the SNP panel is better than that of the microsatellite marker set;
-
(4)
Genotyping success is consistent between our study species, regardless of environmental conditions and genotyping method.
With the result from this study, we aim to provide guidance to researchers and practitioners to develop effective and efficient sampling protocols for the genetic identification of individuals. Improving the reliability of the data on which further analyses such as estimates of genetic variation, connectivity, parentage, abundance, and vital rates are based. This is vital information for the monitoring of species and ultimately for their management.
Materials and methods
Study species
In this study, we use red fox and pine martens as model species. The red fox is a generalist, highly adaptable mammal with circumglobal distribution (Hoffmann and Sillero-Zubiri 2016). Red fox are commonly hunted with the intention to control population sizes and reduce predation on threatened prey species, especially in Australia where it is an invasive species (Lowe et al. 2000; Mahon 2009; Doherty et al. 2016). The European pine marten is a forest-dwelling mustelid widely distributed across most of Europe, the Caucasus, and northern Asia (Herrero et al. 2016). Throughout its range, the pine marten is subject to national and international conservation legislation (O’Mahony et al. 2017), which has increased the demand for monitoring (Manzo et al. 2012; Sheehy et al. 2014; O’Mahony et al. 2015, 2017; Weber et al. 2018; Croose et al. 2019), particularly under the Habitats Directive (Council Directive 92/43/EEC of 21 May 1992 on the conservation of natural habitats and of wild fauna and flora). Due to their elusiveness and widespread distribution, a range of monitoring programs are in place: Non-invasive genetic sampling methods have been used to estimate absolute population densities and population structures of red fox (e.g., Wegge et al. 2019; Walton et al. 2021; Lindsø et al. 2022), and to monitored population success of pine marten (e.g., Ruiz-González et al. 2008, 2013; Mullins et al. 2010; Natali et al. 2010; Balestrieri et al. 2016; Croose et al. 2016; Kubasiewicz et al. 2016). While non-invasive genetic sampling could be used as a tool in the field, as far as we are aware, regularly occurring monitoring programs have rarely adopted the method in practice, also due to the inconsistency in genotyping success of faeces collected in the field (e.g., Croose et al. 2019; Wegge et al. 2019; Walton et al. 2021; Mergey et al. 2023).
Experimental design
To investigate the effects of temperature and precipitation over time on the performance of genetic markers, we exposed red fox and pine marten faeces to different environmental treatments. We used captive animals to ensure that faeces were from known individuals and fresh, i.e., excluding unintentional effects of time and weather. We collected faecal samples from zoo enclosures from February to April 2021. Fox faeces came from seven individuals housed in four different enclosures: eight faeces from an enclosure with two individuals at Schwarzwaldzoo Waldkirch, eight from an enclosure with two individuals at Wildpark Eekholt, three from an enclosure with one individual and one faeces from an enclosure with two individuals both at Tierart Wild Animal Sanctuary Massweiler. Pine marten faeces originated from four individuals, all of which came from enclosures housing single individuals (one each from Schwarzwaldzoo Waldkirch and Wildpark Eekholt, two from Otterzentrum Hankensbüttel). All enclosures were cleaned daily, ensuring a sample age of less than 24 h. Samples were collected individually in plastic sample tubes, using the inside of its lid to move the sample into the tub and immediately stored at −28 °C until the beginning of the experiment. Additionally, we stored the faeces at −80 °C for one week prior to the experiment, to reduce the risk of a potential Echinococcus multilocularis infection for the staff.
We exposed the faeces to four different temperature and precipitation treatments: (1) cold & wet, (2) cold & intermediate, (3) warm & intermediate, and (4) warm & dry (Fig. 1). These treatments were designed to match typical field conditions during a central European spring and autumn, when red fox and pine marten densities tend to be monitored because this is when there is no snow as well as less dense ground vegetation, further the species are reproducing and dispersing during this time (e.g., Güthlin et al. 2012, 2013; Baines et al. 2013; Kämmerle et al. 2018; Croose et al. 2019). For each species, we placed five faeces per treatment in a respective plastic pan with drainage holes containing forest soil (20 faeces total per species; Fig. 1). In every treatment, we aimed to maximise the number of individuals from which the faeces originated. Each sample was marked with plastic signs providing its unique ID. We used the same soil in all samples to mimic realistic field conditions as closely as possible, avoiding potential differences in sample degradation due to artificial substrate compared to natural soil (Santini et al. 2007), and to avoid potential effects of different local microbiota. We performed the two ‘cold’ treatments (8.9 °C [3.1–24.4 °C]) outdoors, sheltered from rain, in the Black Forest between May 10 and May 31, 2021 (Muggenbrunn; 47.858568, 7.918341). To maximise the contrast in temperatures, the ‘warm’ treatments (26.0 °C [14.0–42.0 °C]) were conducted a month later, June 08 to June 29, 2021, in a greenhouse (Freiburg; 48.013670, 7.833566). For all treatments, temperatures were continuously recorded using Onset HOBO Pendant MX2201 temperature loggers (accuracy ± 0.5 °C).
We simulated different degrees of precipitation by watering the treatments at different frequencies. We watered the wet treatment every second day (eleven times over the experimental period of 21 days), the intermediate treatment twice a week (six times), and the dry treatment once a week (three times). Each time the treatments were watered, we used 4.2 mm of pre-collected rainwater, which is the average daily precipitation in Germany between 1990 and 2020 (791 mm p.a.; Deutscher Wetterdienst 2021) multiplied by the duration of the experiment (21 days) and then equally divided across eleven rainfall events (the maximum number of times the wet treatment got watered). Thus, over the course of the experiment, the wet treatments received 46 mm of water, intermediate treatments received 25 mm of water, and dry treatments received 12 mm of water. We made sure that rainwater was evenly distributed across all samples. From each faeces, we collected an aliquot at five points in time: day 0 (start of experiment), 3, 7, 14 and 21, to account for potential changes in DNA quality over time. For each aliquot, we sampled a fifth of the faeces’ length with surgical scissors, which we sterilised after every use. We stored the aliquots immediately at −28 °C until genetic analyses.
Experimental design: (a) temperature profiles over the course of the experiment (21 days) for both cold (left) and warm treatments (right). (b) For both species, six faeces were placed on forest soil and exposed to the following treatment combinations respectively: cold & wet, cold & intermediate, warm & intermediate, and warm & dry. (c) Five aliquots were taken from five of the six faeces after 0, 3, 7, 14, and 21 days, and one faecal sample remained untouched in order to observe visual progression of ageing (d) Aliquots of fox faeces were analysed using eight microsatellite markers as well a panel of 90 SNPs for individual identification, those from marten faeces were analysed using 16 microsatellite markers
Genetic analysis
DNA was isolated from faecal samples using a commercial kit (NucleoSpin Stool Kit, Macherey-Nagel, Baesweiler, Germany). Fox faeces were genotyped using microsatellite markers as well as a SNP panel, while pine marten faeces were only genotyped using microsatellite markers since a SNP panel is not yet available.
Microsatellite genotyping – red fox and pine marten
To genotype red fox and pine marten faeces, we used eight and 17 dinucleotide microsatellite markers respectively for individual identification. For both species, all PCRs were carried out in three replicates. For the red fox, the eight markers were combined and co-amplified in two separate multiplex PCRs (Manivannan 2013). For the pine marten, the 17 microsatellites were amplified in three multiplex PCRs reactions (O’Mahony et al. 2017; Modorov et al. 2020). PCR protocols and fragment length determination for both species are described in detail in appendices 1–3.
SNP genotyping – red fox
We analysed the obtained isolates from aliquots of day 0, 7, 14, 21 from red fox faeces containing a sufficient amount of DNA (7 faeces: 3 cold & wet, 1 cold & intermediate, 1 warm & intermediate, 2 warm & dry). For cost reasons, we chose to not analyse aliquots of day 3, since we expected that declines in successful amplifications of SNPs would not be apparent until later in time. For the analysis, we used a SNP panel consisting of a total of 95 SNPs (90 somatic SNPs for individual identification, three Y-chromosomal SNPs for sex determination, and two mitochondrial SNPs for species recognition). The somatic SNPs were originally selected from Illumina’s® CanineHD™ 170 K Whole-Genome Genotyping BeadChip by their frequency in red foxes and first used for individual identification by Walton et al. (2021) utilising a microarray platform. The selection of the Y-chromosomal SNPs was based on Sacks et al. (2021) while the mitochondrial SNPs were determined by comparing the mitochondrial DNA of twelve habitat-sharing carnivores with the red fox. All SNPs were analysed using a matrix assisted laser desorption/ionisation time-of-flight mass spectrometer specially designed for nucleic acid analysis (detailed methods in appendix and Alef & Ebert et al., in prep.).
Quantifying sample performance
For microsatellites, we deduced consensus genotypes from all the triplicate results of day 0 for each individual. For SNPs, we deduced them from the replicates over the course of the experiment for each individual faeces. For all marker sets, we calculated the rate of amplifications leading to a positive PCR result, i.e. the amplification success rate (ASR), the rate of allelic dropouts (ADR) according to \(p={\stackrel{-}{p}}_{w}={\sum }_{j=1}^{L}{D}_{j}/{\sum }_{j=1}^{L}{A}_{{het}_{j}}\) and the rate of false alleles (FAR) according to \(f={\stackrel{-}{f}}_{w}={\sum }_{j=1}^{L}{F}_{j}/{\sum }_{j=1}^{L}{A}_{{het}_{j}}\) relative to the consensus (Broquet and Petit 2004). In addition, we defined two quality metrics reflecting sample performance, with the explicit goal of individual identification. First, we processed the ASR into the rate of amplifications that were both successful and correct (ASRc). For microsatellites, this entailed all amplifications leading to a PCR result matching the consensus genotype; for the SNPs, we calculated the proportion of all calls that were both successful and produced the correct result (i.e., matching the consensus). Secondly, for the microsatellite markers we also defined a more applied measure, expressed as a rate that we hence call the correct consensus rule (CCR). This rule indicates if the three amplifications at a given locus would have led to a reliably identified locus, even if the correct consensus were unknown (as would be the case in practice, i.e. samples are collected in the field and thus the individual is unknown). We defined a reliably identified locus in accordance with laboratory practice and the literature: homozygote loci were only accepted as correct when the allele was present in all three amplifications or in two, but only if the third amplification resulted in no amplification, while heterozygote loci were accepted when the alleles were present in at least two out of three amplifications (Taberlet et al. 1996; Piggott 2004; Arandjelovic et al. 2009; Lampa et al. 2013; Agetsuma-Yanagihara et al. 2017). Lastly, for each marker set, we summarised ASRc and CCR as ratios for each treatment at every given point in time.
Statistical analysis
We tested the effects of sample age and treatment (i.e., environmental conditions) on genotyping success for both target species separately. We fitted mixed-effect beta regression models with ASRc as the response and the treatment conditions as fixed effects. Due to the nature of ratios, the values of the response variable were squeezed by applying y’ = [y(N-1) + 0.5]/N (with N being the sample size) to avoid 0 and 1 values (Smithson and Verkuilen 2006). We included a random intercept term for sample ID to account for the longitudinal nature of the data (i.e., resampling on the same sample during the experiment). Given the limited sample size, we assessed the effect of treatment conditions and age by creating a full model for each species. Further, we assessed differences in ASRc between methods by Kruskal-Wallis rank sum test.
To investigate the differences in performance between the two red fox genotyping methods, we modelled the performance as a function of method (microsatellite vs. SNPs) instead of treatment, to explore the change in performance over time given the method. Here, we compared only those aliquots that were analysed by both methods (i.e., those from red fox).
All analyses were performed using R v. 4.2.2 (R Core Team 2022), using the “glmmTMB” function of the glmmTMB package (Brooks et al. 2017).
Results
We exposed 20 red fox faeces and 20 pine marten faeces to four environmental treatments. One fox faeces from the cold & intermediate treatment was excluded from all analyses due to cross-contamination. All other treatments consisted of 5 faeces from each species. For the pine marten marker set, one of the markers (Ma2) did not amplify in a single aliquot, thus we excluded it from further analysis. The extracts from seven of the red fox faeces contained a sufficient amount of DNA to run both marker sets, and ran a SNP panel consisting of 90 SNPs for individual identification, in addition to the microsatellite analysis.
ASRc and CCR (Table 1) were highly positively correlated in the red fox (τ = 0.905, p < 0.001) and the pine marten data sets (τ = 0.926, p < 0.001). Since ASRc is made up of two error sources, FAR and ADR, we did not analyse them separately, but summarised them across treatments in Table 2.
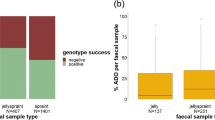
ASRc of microsatellites decreased significantly over time for both species (p < 0.001; Table 3). ASRc of microsatellites did not differ among treatments (Red Fox: Kruskal-Wallis chi-squared = 3.25, df = 3, p = 0.35; Pine Marten: Kruskal-Wallis chi-squared = 5.83, df = 3, p = 0.12). For the SNP panel, ASRc did not differ with treatment (Kruskal-Wallis chi-squared = 4.13, df = 3, p = 0.25) or time (p > 0.05, Table 3). Finally, ASRc from red fox SNPs was significantly higher than ASRs from red fox microsatellites (p < 0.001, Fig. 2).
Discussion
Successful DNA extraction using non-invasive genetic sampling methods (i.e., from faeces) can help to streamline efforts to monitor wildlife populations and estimate population densities, particularly for rare or elusive species. To investigate the role of time, environmental conditions and genotyping method on the success of DNA amplification, we exposed fresh faecal samples from red foxes and pine martens to four different environmental conditions in an experimental setting. Our study shows that the age of the sample was the most important driver in reducing DNA amplification success, regardless of the environmental conditions to which the faeces were exposed. We also found that, over the 21-day experimental period, the SNP assay used in this study was significantly more reliable than the respective microsatellites.
Time effects
In accordance with our hypotheses, genotyping success by microsatellites decreased over time (hypothesis 1) and there was consistency in the success of genotyping samples from both study species (hypothesis 4; Table 3). This temporal decline in correct genotyping rate is consistent with previous studies (e.g., red fox, Piggott 2004; pine marten, Kubasiewicz et al. 2016; grey wolf (Canis lupus), Lucchini et al. 2002 & Santini et al. 2007; and brown bear (Ursus arctos), Murphy et al. 2007). Interestingly, the amplification success rate of the SNP panel did not decrease significantly with increasing sample age (Table 3). While these results might have changed if we had continued the experiment beyond 21 days, the absence of a significant age effect is in contrast with hypothesis 1. Our results match one other study, that demonstrated that 14-day old faeces can still be used to reliably identify individuals when SNP markers were used (Schultz et al. 2018). When genotyping samples using microsatellites, samples collected in the field should be as fresh as possible to enable a high genotyping success. The SNP assay used in this study on the other hand is more robust than the microsatellite we used and reliably genotyped even older faeces.
Environmental effects
Contrary to our second hypothesis, the different tested environmental conditions had no significant effect on DNA quality and consequently the measured genotyping success for either species or methods. While in our experiment the effects of precipitation and temperature could offset each other, it is unlikely, since there was no difference within the two cold nor the two warm treatments either. This absence of environmental effects is in contrast with previous studies, which found, e.g., higher amplification success in winter compared to summer conditions (Lucchini et al. 2002; Piggott 2004), and a negative effect of warmer temperatures (Hájková et al. 2006). Moreover, other studies found that rain had a strong negative effect compared to sheltered samples (e.g., Brinkman et al. 2010; Wedrowicz et al. 2013; Kubasiewicz et al. 2016; Agetsuma-Yanagihara et al. 2017), while in the absence of rain, amplification success did not differ with environmental conditions (Santini et al. 2007). However, in these examples, the experimental conditions tended to investigate extremes in temperature (winter vs. summer) or precipitation (rain vs. dry), whereas in our study, we investigated intermediate conditions with different levels of rainfall, similar to central-European climate conditions. To our knowledge, Murphy et al. (2007) is the only other study comparing more realistic weather conditions, and the authors found that wet and cool microclimate reduce amplification success rate only slightly compared to warm dry microclimate, when the study area received approximately 15 mm of rain (comparable to our warm & dry treatment). Our experiment showed that the amount of rain as well as the temperature does not affect genotyping quality, and thus faecal DNA quality. Thus, it is likely that the initial rainfall reduces DNA quality but subsequent ones have little to no significant additional effect on DNA quality (at least up to 46 mm distributed over several rainfall events).
Marker type
We were able to directly compare the genotyping success of a microsatellite marker set and a SNP panel by using the same DNA extract for both methods. The SNP panel generally resulted in a higher rate of correct amplifications than the microsatellites (Fig. 2), which supports our third hypothesis. Additionally, our results match other experiments designed to compare the performances of the two different marker types on field-collected faeces (wolf, Fabbri et al. 2012; puma (Puma concolor), Fitak et al. 2016; European wildcat (Felis silvestris), wolf, & brown bear, von Thaden et al. 2017). While we only tested one specific SNP and one microsatellite method, our results provide further evidence that SNPs can perform better than commonly applied, i.e., longer, microsatellites when used to analyse degraded samples, such as faeces. The advantage of the SNPs is due to the size differences of the DNA section that are targeted by our marker sets (SNPs 72–120 bp, microsatellites 154–410 bp). Recently developed microsatellite techniques that result in marker lengths similar to SNPs (e.g., Liu et al. 2024) could potentially perform as well as SNPs.
While the SNPs used in this study performed better in the given experiments, there are potential significant drawbacks of using SNPs compared to microsatellites. SNP data alone does not allow the detection of cross-contaminated samples, i.e., samples containing DNA from more than one individual. In microsatellite genotypes, this can be detected due to the presence of surplus alleles. For faecal sampling, failing to detect cross-contamination can become a problem: several carnivore species tend to urinate or defecate on faeces deposited by conspecifics or heterospecific carnivores (DeMatteo et al. 2018), or because the targeted individuals consume the faeces of another species, e.g., wild red foxes may consume dog faeces (Waggershauser et al. 2022). For instance, we detected a cross-contamination in one of our red fox faeces (and thus excluded it from further analyses) through the microsatellite analysis which would be missed by the SNP panel. When this contamination remains undetected in sampled faeces, it could result in a novel but untrue SNP genotype and subsequently affect further analysis, e.g., leading to an overestimation of population size in a capture-recapture analysis. One solution to detecting samples containing DNA from more than one individual could be to integrate a subset of microsatellites, solely for said purpose, into the SNP panel.
An important consideration in choosing an amplification method can be costs and processing time, or simply availability of existing marker sets. Even though SNPs require larger marker sets than microsatellites (Fabbri et al. 2012; Walton et al. 2021), SNP analyses are not necessarily more expensive than microsatellites (von Thaden et al. 2017) and allow for comparisons across different laboratories (Fitak et al. 2016). However, recent developments using high-throughput microsatellite genotyping have high potential to resolve the well-known drawbacks of microsatellites against SNPs, by making analysis processes faster and more efficient as well as easier standardized (e.g., Suez et al. 2016; De Barba et al. 2017; Zhan et al. 2017; Liu et al. 2024). Thus, as this field continues to develop, it is likely that new solutions and recommendations emerge.
Applicability
In studies that genotype faeces of wild animals, the true genotypes are rarely known. Therefore, we wanted to ensure that the conclusions we draw, based on the effects on the ASRc, hold true in real life situations, e.g., using faecal DNA for capture-recapture. Thus, we correlated ASRc with CCR, a measure indicating if the genotype would have been determined correctly, without knowledge of the true correct genotype. We found very high correlations between both measures, which indicates that our results would hold true in field situations with unknown genotypes. While we explored faeces from mesocarnivores, previous research suggests microsatellite amplification success seems not to be influenced by the diet type (Broquet et al. 2007). Thus, these conclusions are likely applicable in a broader context and may transfer to other terrestrial mammals.
Limitations
While our experiment allows certain conclusions, there are some limitations. First, we detected strong variability for the different quality measures (ASRc, FAR and ADR) even within treatments (Tables 1 and 2), which might have masked some treatment effects and a potential negative effect of time on the SNP panel. Second, there were also some apparent variations in amplification success on day 0, when samples were fresh and untreated. Which could be partially, due to the sampling location within the faeces (Stenglein et al. 2010). Future studies with an extensive sample size could bring clarity to these discrepancies and determine if they are due to a limited sample size only or simply due to the variable nature of faecal samples.
Recommendation
Reliable genotyping is essential for non-invasive genetic sampling. While different laboratory procedures, such as extraction and storage can have an effect on the quality of the amplified DNA (Santini et al. 2007; Walton et al. 2021), the sampling itself plays a major role in increasing the probability of obtaining faeces with a high genotyping success rate and thus is a factor that should be notably considered both during planning and implementation. The principle “the fresher the better”, continues to be one of the main factors ensuring correct genotyping, especially when using microsatellites. However, in practice this can pose a problem. Fresh faeces are harder to find and, unless they are very fresh, a visual determination of their age can be challenging (Piggott 2004). Thus, there may be no way to control for the age of collected samples unless defined transects are sampled repeatedly, which might be unfeasible and reduces the yield per sampling round. Fortunately, our study demonstrates that older faeces can be genotyped successfully and thus are still usable for analysis (Table 1; Fig. 2). It is important to establish the right balance between increased effort from repeated sampling, lower sample size, and lower genotyping success for the given circumstances. When using microsatellites (that require longer DNA fragments), we advise short intervals between resampling (fewer than 21 days). When faecal samples are not fresh, SNP panels will likely perform better than conventional microsatellites. Given the trade-offs in collection efforts, genotyping success, and costs, again, SNPs can be more suitable for non-invasive genetic sampling than commonly used microsatellites are, when reliable genotyping is the aim. However, as discussed, as genetic techniques continue to develop, new solutions may become available and suitable.
In conditions similar to central European climate, weather during the faeces sampling period plays, in many cases, a subordinate role on genotyping success. Since collecting exclusively fresh samples is often unfeasible, the chance that faeces have been exposed to rain before being sampled is high. Since we demonstrated that additional rain after the initial rainfall has little impact on genotyping success, we suggest there is no need to adjust the collection period in accordance with predicted rain. However, if the collection of faeces coincides with a prolonged period without precipitation or is preceded by an extended period of drought, sampling ought to be completed before a forecasted rainfall.
Conclusions
Monitoring of wild populations is often a result of the trade-off between effort, costs, time, and access. Non-invasive genetic sampling may provide a means by which effort, costs and time are minimized, thus streamlining monitoring efforts. Given these developments, as well as improvements to genetic methods and falling costs, the use and value of non-invasive genetic sampling is likely going to increase. In order to use non-invasive genetic sampling to adequately monitor wild population, it is important that the techniques are used reliable in field and that samples are collected under the optimal conditions to allow for successful genotyping. From the results of this study, we suggest that monitoring programs prioritize short successive resampling efforts, that are no longer than 21 days in between collections. When conventional microsatellite genotyping techniques are to be used, we recommend the interval between consecutive sampling to be even shorter. Where short sampling intervals cannot be maintained, monitoring programs could utilize the advantages of SNP genotyping or potentially contemporary microsatellite protocols. Non-invasive genetic sampling shows great promise, and understanding its limitations and strengths helps to improve our collective knowledge of its use, improving the probability that non-invasive genetic sampling can be further integrated into monitoring and management programs for rare and elusive species.
Appendix
Microsatellite genotyping
For red fox, the amplification reactions of eight microsatellites were performed in triplicates (altogether three repeats of the two multiplex PCRs, i.e. six PCRs per sample) in a total volume of 12 µl using the Qiagen Multiplex PCR kit (Qiagen, Hilden, Germany). The PCR protocol is provided in Appendix 1 and primer concentrations in Appendix 2. We separated fluorescently labelled DNA fragments on an ABI3730 DNA analyser and determined allele size using the ABI GS500LIZ size ladder (Applied Biosystems, Darmstadt, Germany). We included two negative PCR controls in every PCR set to detect potential contamination. Raw data for all samples were evaluated manually using the Genemapper software (Version 5, Applied Biosystems).
For pine marten amplification of 17 microsatellites were performed in three multiplexes in a total volume of 10 µL with 5 µl SuperHot Master Mix (two times) (Genaxxon Bioscience), 3 µl DNA extract and primers in concentrations of 0.12–0.9 µM (Appendix 3). Details of the PCR protocol are described in Appendix 1. We ran three replicates for each sample (altogether three repeats of the three multiplex PCRs, i.e. nine PCRs per sample).
For fragment length analysis we mixed 1 µl PCR product with 14 µl Hi-Di formamide (Applied Biosystems) and GeneScan-500 LIZ size standard (Applied Biosystems) and ran it on an Applied Biosystems SeqStudio genetic analyser under standard run conditions. Allele lengths were scored manually using Genemapper software (Version 6, Applied Biosystems).
SNP genotyping details
The multiplex reactions were performed in a 96-well plate including an initial PCR, a Shrimp-Alkaline-Phosphatase (SAP) and an extension reaction using the iPLEX Pro® Reagent Set (#10160F, Agena Bioscience, Hamburg, Germany) according to the manufacturer’s protocol USG-CUS − 071 Rev03 (Agena Bioscience). 2 µL DNA input (20 ng absolute) in a total PCR mix of 5 µL was used on a Thermocycler (Labcycler Basic with Thermoblock 96, SensoQuest, Göttingen, Germany). To remove surplus nucleotides each PCR reaction was treated with SAP mix. 2 µL extension reaction mix composed of mass-modified terminator nucleotides was added to elongate the amplified DNA strands at the nucleotide position of interest. Each sample was treated with 41 µL nanopure water (> 18 mΩ) and transferred into a Chip Prep Module (Agena Bioscience) to process post extend PCR samples for desalting in 15 mg clean resin (#8040, Agena Bioscience), spotting on a matrix-precoated Spectro-CHIP (Agena Bioscience) and analysis in a matrix assisted laser desorption/ionisation time-of-flight mass spectrometer (MassARRAY Analyzer 4, Agena Bioscience). After manually checking the raw data, a final report (GenotypeArea Report v1, Agena Bioscience) was generated within the commercial software (MassARRAY Typer Analyzer Software v5.0.2, Agena Bioscience), which listed all samples with their corresponding genotypes. We included a negative control in every PCR to avoid cross contamination of samples.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Agetsuma-Yanagihara Y, Inoue E, Agetsuma N (2017) Effects of time and environmental conditions on the quality of DNA extracted from fecal samples for genotyping of wild deer in a warm temperate broad-leaved forest. Mammal Res 62:201–207. https://doi.org/10.1007/s13364-016-0305-x
Allendorf FW, Funk WC, Aitken SN et al (2022) Conservation and the Genomics of populations, 3rd edn. Oxford University Press, USA
Amburgey SM, Yackel Adams AA, Gardner B et al (2021) Evaluation of camera trap-based abundance estimators for unmarked populations. Ecol Appl 31. https://doi.org/10.1002/eap.2410
Arandjelovic M, Guschanski K, Schubert G et al (2009) Two-step multiplex polymerase chain reaction improves the speed and accuracy of genotyping using DNA from noninvasive and museum samples. Mol Ecol Resour 9:28–36. https://doi.org/10.1111/j.1755-0998.2008.02387.x
Baines D, Aebischer N, Macleod A, Woods J (2013) Pine marten Martes martes and red fox Vulpes vulpes sign indices in Scottish forests: Population change and reliability of field identification of scats. Wildl Biol 19:490–495. https://doi.org/10.2981/13-030
Balestrieri A, Ruiz-González A, Vergara M et al (2016) Pine marten density in lowland riparian woods: a test of the Random Encounter Model based on genetic data. Mamm Biol 81:439–446. https://doi.org/10.1016/j.mambio.2016.05.005
Bonin A, Bellemain E, Eidesen PB et al (2004) How to track and assess genotyping errors in population genetics studies. Mol Ecol 13:3261–3273. https://doi.org/10.1111/j.1365-294X.2004.02346.x
Brinkman TJ, Schwartz MK, Person DK et al (2010) Effects of time and rainfall on PCR success using DNA extracted from deer fecal pellets. Conserv Genet 11:1547–1552. https://doi.org/10.1007/s10592-009-9928-7
Brooks ME, Kristensen K, van Benthem KJ et al (2017) glmmTMB balances speed and flexibility among packages for zero-inflated generalized linear mixed modeling. R J 9:378–400. https://doi.org/10.32614/rj-2017-066
Broquet T, Petit E (2004) Quantifying genotyping errors in noninvasive population genetics. Mol Ecol 13:3601–3608. https://doi.org/10.1111/j.1365-294X.2004.02352.x
Broquet T, Ménard N, Petit E (2007) Noninvasive population genetics: a review of sample source, diet, fragment length and microsatellite motif effects on amplification success and genotyping error rates. Conserv Genet 8:249–260. https://doi.org/10.1007/s10592-006-9146-5
Carroll EL, Bruford MW, DeWoody JA et al (2018) Genetic and genomic monitoring with minimally invasive sampling methods. Evol Appl 11:1094–1119. https://doi.org/10.1111/eva.12600
Croose E, Birks JDS, O’Reilly C et al (2016) Sample diversity adds value to non-invasive genetic assessment of a pine marten (Martes martes) population in Galloway Forest, Southwest Scotland. Mammal Res 61:131–139. https://doi.org/10.1007/s13364-015-0257-6
Croose E, Birks JDS, Martin J et al (2019) Comparing the efficacy and cost-effectiveness of sampling methods for estimating population abundance and density of a recovering Carnivore: the European pine marten (Martes martes). Eur J Wildl Res 65. https://doi.org/10.1007/s10344-019-1282-6
De Barba M, Miquel C, Lobréaux S et al (2017) High-throughput microsatellite genotyping in ecology: improved accuracy, efficiency, standardization and success with low-quantity and degraded DNA. Mol Ecol Resour 17:492–507. https://doi.org/10.1111/1755-0998.12594
DeMatteo KE, Blake LW, Young JK, Davenport B (2018) How Behavior of Nontarget species affects Perceived Accuracy of Scat Detection dog surveys. Sci Rep 8:13830. https://doi.org/10.1038/s41598-018-32244-1
Deutscher Wetterdienst (2021) Time series and trends. https://www.dwd.de/EN/ourservices/zeitreihen/zeitreihen.html#buehneTop. Accessed 9 Dec 2021
DeYoung RW, Honeycutt RL (2005) The molecular toolbox: genetic techniques in wildlife ecology and management. J Wildl Manage 69:1362–1384. https://doi.org/10.2193/0022-541X(2005)69[1362:TMTGTI]2.0.CO;2
Doherty TS, Glen AS, Nimmo DG et al (2016) Invasive predators and global biodiversity loss. Proc Natl Acad Sci 113 :11261 LP – 11265. https://doi.org/10.1073/pnas.1602480113
Fabbri E, Caniglia R, Mucci N et al (2012) Comparison of single nucleotide polymorphisms and microsatellites in non-invasive genetic monitoring of a wolf population. Arch Biol Sci 64:320–336. https://doi.org/10.2298/ABS1201321F
Fitak RR, Naidu A, Thompson RW, Culver M (2016) A new panel of SNP markers for the individual identification of north American pumas. J Fish Wildl Manag 7:13–27. https://doi.org/10.3996/112014-JFWM-080
Güthlin D, Kröschel M, Küchenhoff H, Storch I (2012) Faecal sampling along trails: a questionable standard for estimating red fox Vulpes vulpes abundance. Wildl Biol 18:374–382. https://doi.org/10.2981/11-065
Güthlin D, Storch I, Küchenhoff H (2013) Landscape variables associated with relative abundance of generalist mesopredators. Landsc Ecol 28:1687–1696. https://doi.org/10.1007/s10980-013-9911-z
Hájková P, Zemanová B, Bryja J et al (2006) Factors affecting success of PCR amplification of microsatellite loci from otter faeces. Mol Ecol Notes 6:559–562. https://doi.org/10.1111/j.1471-8286.2006.01269.x
Herrero J, Kranz A, Skumatov D et al (2016) Martes martes. IUCN Red List Threat Species 2016 8235:e.T12848A45199169
Hoffmann M, Sillero-Zubiri C (2016) Vulpes vulpes. IUCN Red List Threat Species 2016(8235):eT23062A46190249
Kämmerle J-L, Corlatti L, Harms L, Storch I (2018) Methods for assessing small-scale variation in the abundance of a generalist mesopredator. PLoS ONE 13:1–16. https://doi.org/10.1371/journal.pone.0207545
Kubasiewicz LM, Minderman J, Woodall LC et al (2016) Fur and faeces: an experimental assessment of non-invasive DNA sampling for the European pine marten. Mammal Res 61:299–307. https://doi.org/10.1007/s13364-016-0276-y
Lampa S, Henle K, Klenke R et al (2013) How to overcome genotyping errors in non-invasive genetic mark-recapture population size estimation - A review of available methods illustrated by a case study. J Wildl Manage 77:1490–1511. https://doi.org/10.1002/jwmg.604
Lindsø LK, Dupont P, Rød-Eriksen L et al (2022) Estimating red fox density using non-invasive genetic sampling and spatial capture–recapture modelling. Oecologia 198:139–151. https://doi.org/10.1007/s00442-021-05087-3
Liu P, Wilson P, Redquest B et al (2024) Seq2Sat and SatAnalyzer toolkit: towards comprehensive microsatellite genotyping from sequencing data. Mol Ecol Resour 24:e13929. https://doi.org/10.1111/1755-0998.13929
Lowe S, Browne M, Boudjelas S, De Poorter M (2000) 100 of the World’s worst invasive alien species a selection from the global invasive species database. Aukland, New Zealand
Lucchini V, Fabbri E, Marucco F et al (2002) Noninvasive molecular tracking of colonizing wolf (Canis lupus) packs in the western Italian Alps. Mol Ecol 11:857–868. https://doi.org/10.1046/j.1365-294X.2002.01489.x
Mahon PS (2009) Targeted control of widespread exotic species for biodiversity conservation: the Red Fox (Vulpes vulpes) in New South Wales, Australia. Ecol Manag \& Restor 10:S59–S69. https://doi.org/10.1111/j.1442-8903.2009.00455.x
Manivannan A (2013) Population Genetic Analysis of Red Foxes (Vulpes vulpes) in Hedmark Country, Norway - A Pilot Study. Hedmark University College
Manzo E, Bartolommei P, Rowcliffe JM, Cozzolino R (2012) Estimation of population density of European pine marten in central Italy using camera trapping. Acta Theriol (Warsz) 57:165–172. https://doi.org/10.1007/s13364-011-0055-8
Mergey M, Kaerle C, Helder R (2023) Variations in density of pine marten Martes martes populations according to forest fragmentation. Mammal Res 68:507–519. https://doi.org/10.1007/s13364-023-00704-3
Mills LS (2013) Conservation of Wildlife populations: demography, Genetics, and management, 2nd edn. John Wiley & Sons, Ltd., Oxford, UK
Modorov M, Monakhov V, Mikryukov V et al (2020) Microsatellite multiplex assay for sable (Martes zibellina) and pine marten (Martes martes). Mammal Res 65:855–862. https://doi.org/10.1007/s13364-020-00529-4
Morin PA, Luikart G, Wayne RK (2004) SNPs in ecology, evolution and conservation. Trends Ecol Evol 19:208–216. https://doi.org/10.1016/j.tree.2004.01.009
Mullins J, Statham MJ, Roche T et al (2010) Remotely plucked hair genotyping: a reliable and non-invasive method for censusing pine marten (Martes martes, L. 1758) populations. Eur J Wildl Res 56:443–453. https://doi.org/10.1007/s10344-009-0332-x
Murphy MA, Kendall KC, Robinson A, Waits LP (2007) The impact of time and field conditions on brown bear (Ursus arctos) faecal DNA amplification. Conserv Genet 8:1219–1224. https://doi.org/10.1007/s10592-006-9264-0
Natali C, Banchi E, Ciofi C et al (2010) Characterization of 13 polymorphic microsatellite loci in the European pine marten Martes martes. Conserv Genet Resour 2:397–399. https://doi.org/10.1007/s12686-010-9282-6
O’Mahony DT, Turner P, O’Reilly C (2015) Pine marten (Martes martes) abundance in an insular mountainous region using non-invasive techniques. Eur J Wildl Res 61:103–110. https://doi.org/10.1007/s10344-014-0878-0
O’Mahony DT, Powell C, Power J et al (2017) Non-invasively determined multi-site variation in pine marten Martes martes density, a recovering Carnivore in Europe. Eur J Wildl Res 63:48. https://doi.org/10.1007/s10344-017-1108-3
Ogden R (2011) Unlocking the potential of genomic technologies for wildlife forensics. Mol Ecol Resour 11:109–116. https://doi.org/10.1111/j.1755-0998.2010.02954.x
Piggott MP (2004) Effect of sample age and season of collection on the reliability of microsatellite genotyping of faecal DNA. Wildl Res 31:485–493. https://doi.org/10.1071/WR03096
R Core Team (2022) R: a Language and. Environment for Statistical Computing
Ruiz-González A, Rubines J, Berdión O, Gómez-Moliner BJ (2008) A non-invasive genetic method to identify the sympatric mustelids pine marten (Martes martes) and stone marten (Martes foina): preliminary distribution survey on the northern Iberian Peninsula. Eur J Wildl Res 54:253–261. https://doi.org/10.1007/s10344-007-0138-7
Ruiz-González A, Madeira MJ, Randi E et al (2013) Non-invasive genetic sampling of sympatric marten species (Martes martes and Martes foina): assessing species and individual identification success rates on faecal DNA genotyping. Eur J Wildl Res 59:371–386. https://doi.org/10.1007/s10344-012-0683-6
Sacks BN, Lounsberry ZT, Rando HM et al (2021) Sequencing Red Fox Y chromosome fragments to develop phylogenetically informative SNP markers and Glimpse Male-Specific Trans-Pacific Phylogeography. Genes (Basel) 12. https://doi.org/10.3390/genes12010097
Santini A, Lucchini V, Fabbri E, Randi E (2007) Ageing and environmental factors affect PCR success in wolf (Canis lupus) excremental DNA samples. Mol Ecol Notes 7:955–961. https://doi.org/10.1111/j.1471-8286.2007.01829.x
Schultz AJ, Cristescu RH, Littleford-Colquhoun BL et al (2018) Fresh is best: accurate SNP genotyping from Koala scats. Ecol Evol 8:3139–3151. https://doi.org/10.1002/ece3.3765
Seddon JM, Parker HG, Ostrander EA, Ellegren H (2005) SNPs in ecological and conservation studies: a test in the scandinavian wolf population. Mol Ecol 14:503–511. https://doi.org/10.1111/j.1365-294X.2005.02435.x
Sheehy E, O’Meara DB, O’Reilly C et al (2014) A non-invasive approach to determining pine marten abundance and predation. Eur J Wildl Res 60:223–236. https://doi.org/10.1007/s10344-013-0771-2
Smithson M, Verkuilen J (2006) A better lemon squeezer? Maximum-likelihood regression with beta-distributed dependent variables. Psychol Methods 11:54–71. https://doi.org/10.1037/1082-989X.11.1.54
Sollmann R, Mohamed A, Samejima H, Wilting A (2013) Risky business or simple solution - relative abundance indices from camera-trapping. Biol Conserv 159:405–412. https://doi.org/10.1016/j.biocon.2012.12.025
Stenglein JL, De Barba M, Ausband DE, Waits LP (2010) Impacts of sampling location within a faeces on DNA quality in two Carnivore species. Mol Ecol Resour 10:109–114. https://doi.org/10.1111/j.1755-0998.2009.02670.x
Suez M, Behdenna A, Brouillet S et al (2016) MicNeSs: genotyping microsatellite loci from a collection of (NGS) reads. Mol Ecol Resour 16:524–533. https://doi.org/10.1111/1755-0998.12467
Taberlet P, Griffin S, Goossens B et al (1996) Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res 24:3189–3194. https://doi.org/10.1093/nar/24.16.3189
Vignal A, Milan D, SanCristobal M, Eggen A (2002) A review on SNP and other types of molecular markers and their use in animal genetics. Genet Sel Evol 34:275–305. https://doi.org/10.1051/gse:2002009
von Thaden A, Cocchiararo B, Jarausch A et al (2017) Assessing SNP genotyping of noninvasively collected wildlife samples using microfluidic arrays. Sci Rep 7:10768. https://doi.org/10.1038/s41598-017-10647-w
Waggershauser CN, Taberlet P, Coissac E et al (2022) Interspecific coprophagia by wild red foxes: DNA metabarcoding reveals a potentially widespread form of commensalism among animals. Ecol Evol 12:e9029. https://doi.org/10.1002/ece3.9029
Waits LP, Paetkau D (2005) J Wildl Manage 69:1419–1433. https://doi.org/10.2193/0022-541x(2005)69[1419:ngstfw]2.0.co;2 Noninvasive Genetic Sampling Tools for Wildlife Biologists: a Review of Applications and Recommendations for Accurate Data Collection
Walton Z, Hagenlund M, Østbye K et al (2021) Moving far, staying close: red fox dispersal patterns revealed by SNP genotyping. Conserv Genet 22:249–257. https://doi.org/10.1007/s10592-021-01332-7
Weber D, Roth T, Tesini C, Thiel D (2018) Widespread distribution of Pine martens (Martes martes) in a fragmented suburban landscape. Mammal Res 63:349–356. https://doi.org/10.1007/s13364-018-0363-3
Wedrowicz F, Karsa M, Mosse J, Hogan FE (2013) Reliable genotyping of the koala (Phascolarctos cinereus) using DNA isolated from a single faecal pellet. Mol Ecol Resour 13:634–641. https://doi.org/10.1111/1755-0998.12101
Wegge P, Bakke BB, Odden M, Rolstad J (2019) DNA from scats combined with capture–recapture modeling: a promising tool for estimating the density of red foxes—a pilot study in a boreal forest in southeast Norway. Mammal Res 64:147–154. https://doi.org/10.1007/s13364-018-0408-7
Zemanova MA (2021) Noninvasive Genetic Assessment Is an Effective Wildlife Research Tool When Compared with Other Approaches. Genes (Basel) 12:. https://doi.org/10.3390/genes12111672
Zhan L, Paterson IG, Fraser BA et al (2017) Megasat: automated inference of microsatellite genotypes from sequence data. Mol Ecol Resour 17:247–256. https://doi.org/10.1111/1755-0998.12561
Acknowledgements
We would like to thank the staff at Schwarzwaldzoo Waldkirch, Otterzentrum Hankensbüttel, Wildpark Eekholt, and Tierart Wild Animal Sanctuary Massweiler that kindly supported this research by providing faecal samples. We furthermore thank Ilir Avduli for his help with the development of the lab protocols.
Funding
LS and the genetic analyses of pine marten samples were funded by a grant from by the Ministry of Food, Rural Affairs and Consumer Protection Baden-Wuerttemberg, Germany. MB was supported by the German Science Foundation (DFG), Research Training Group ConFoBi (GRK 2123).
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
LS led investigation, formal analysis and writing, and supported conceptualization, methodology. HT co-led investigation, data collection, genetic analysis, and supported conceptualization, methodology, writing (editing). CE co-led genetic analysis and supported methodology, data collection, and writing (editing). TA supported methodology, data collection, genetic analysis, and writing (editing). JLK led funding acquisition, conceptualization, methodology, and supported investigation, supervision and writing (editing). IS co-led funding acquisition, and supported conceptualization, methodology, investigation and writing (editing). MB led supervision, co-led formal analysis, and supported writing (original and editing). All authors reviewed the manuscript and approved its submission.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose. However, we wish to clarify that two of the authors (CE & TA) work for a commercial laboratory that provides commercial services such as analysis with both the red fox microsatellite and SNP panel.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Scholz, L.G., Tripke, H., Ebert, C. et al. Environmental effects on faecal genotyping success in mesocarnivores. Conservation Genet Resour (2024). https://doi.org/10.1007/s12686-024-01363-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12686-024-01363-5