Abstract
Excess nitrogen input into water bodies can cause eutrophication and affect the community structure and abundance of the nitrogen-transforming microorganisms; thus, it is essential to remove nitrogen from eutrophic water bodies. Aquatic plants can facilitate the growth of rhizosphere microorganisms. This study investigated the impact of ammonium pollution on the anammox and denitrifying bacteria in the rhizosphere of a cultivated submerged macrophyte, Potamogeton crispus (P. crispus) by adding three different concentrations of slow-release urea (0, 400, 600 mg per kg sediment) to the sediment to simulate different levels of nitrogen pollution in the lake. Results showed that the ammonium concentrations in the interstitial water under three pollution treatments were significantly different, but the nitrate concentration remained stable. The abundance of anammox 16S rRNA and nitrite reductase (nirS) gene in rhizosphere sediments exhibited no significant differences under the three pollution conditions. The increase in the nitrogen pollution levels did not significantly affect the growth of anammox bacteria and nirS denitrifying bacteria (denitrifiers). The change trend of the abundance ratio of (anammox 16S rRNA)/nirS in different nitrogen treatment groups on the same sampling date was very close, indicating that this ratio was not affected by ammonium pollution levels when P. crispus existed. The redundancy analysis showed that there was a positive correlation between the abundance of anammox 16S rRNA and nirS gene and that the abundance of these bacteria was significantly affected by the mole ratio of NH4+/NO3−. This study reveals that submerged plants weaken the environmental changes caused by ammonia pollution in the rhizosphere, thereby avoiding strong fluctuation of anammox bacteria and nirS denitrifiers.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The removal of nitrogen in aquatic ecosystems is mainly accomplished by plant absorption and nitrogen-transforming microorganisms capable of nitrogen conversion processes such as ammonia oxidation, nitrite oxidation, and nitrate reduction. However, excessive accumulation of nitrogen will destroy the stability of the community structure and abundance of these nitrogen-transforming microorganisms, thereby breaking the balance of nitrogen input and output in water bodies (Le et al. 2010), finally causing environmental problems such as water quality decline and eutrophication.
Submerged plants are an important part of the aquatic ecosystem. They can provide habitat and food for aquatic animal, absorb pollutant (Brisson and Chazarenc 2009), inhibit algae growth (Zhao et al. 2021), and reduce sediment resuspension (Tang et al. 2018). Anammox bacteria and denitrifying bacteria are two important nitrogen-transforming microorganisms. They can convert the ammonium (NH4+) and nitrite (NO2−), nitrate (NO3−) into nitrogen gas, respectively, thereby removing the excessive nitrogen from lakes and maintaining the nitrogen balance in water bodies (Pajares et al. 2017). These two microorganisms are both cooperative and competitive. The denitrification process consumes organic matter, thus slowing down the inhibitory effect of organic matter on the growth of anammox bacteria (Yang et al. 2019). Moreover, in the denitrification process of converting NO3− to nitrite (NO2−), NO2− produced by denitrifying bacteria can be used by anammox bacteria as a reaction substrate, thus facilitating the growth of anammox bacteria (Wang et al. 2017; Koop-Jakobsen and Giblin 2009). However, in the denitrification process of converting NO2− to NO (nitric oxide), nirS denitrifiers with Cu-containing enzymes and nirK denitrifiers with cytochrome cd1 enzymes may compete with anammox bacteria for NO2− (Wang et al. 2020).
In shallow lakes, aquatic plants can reduce the nitrogen level in the lake by directly absorbing NH4+ and NO3− either from water bodies or sediments (Zhou et al. 2016; Zhao et al. 2014). Aquatic plants can also facilitate the growth of rhizosphere microorganisms by providing them nutrients through the root system, leading to a much higher abundance of rhizosphere microorganisms, compared to the non-rhizosphere environment, thus showing the rhizosphere effect (Christensen et al. 1994). Plant roots can release oxygen to the rhizosphere and form diverse micro-environments such as aerobic, anoxic, and anaerobic conditions in the rhizosphere (Niu et al. 2015). This provides different living environment for the coexistence of anaerobic microorganisms such as anammox and denitrifying bacteria in the rhizosphere (Wang et al. 2020) and more possibilities for combining denitrification with anammox (Kumar and Lin 2010). It has been reported that the activity and the abundance of anammox bacteria in the rhizosphere were higher than those in the non-rhizosphere (Nie et al. 2015). The coexistence of anaerobic and aerobic environments in the rhizosphere was also considered to be beneficial to the growth of anammox bacteria (Wang et al. 2015; Vazquez-Padin et al. 2010).
The growth of anammox and denitrifying bacteria in sediments can be affected by the changes in water pollutants. Some studies have shown that the abundance of nirS gene in sediments was decreased with the increase in lake nutrients (Wan et al. 2019; Guo et al. 2014), while eutrophication accelerated the anammox bacteria growth (Zhao et al. 2019b). However, there is still a lack of understanding of how these two bacteria respond to the increased nitrogen content in the presence of submerged plants in lakes. Plants have been reported to be able to maintain their growth environment, especially the stability of the physical and chemical properties of rhizosphere environment (Hussain et al. 2011). Based on it, we hypothesized that submerged plants could provide a stable environment for anammox and denitrifying bacteria in the rhizosphere, thus alleviating the rapid impact of increased pollution on these two bacteria in the rhizosphere. To test this hypothesis, the abundance of anammox and denitrifying bacteria in the rhizosphere of a submerged macrophyte, Potamogeton crispus (P. crispus), under three ammonium pollution levels in the lake sediment were investigated. The most popular nitrogen fertilizer today is urea, which can dissolve in water within a few seconds, and the nitrite produced during the oxidation process of the ammonia produced by its hydrolysis can become the substrate of the anammox reaction. Therefore, we choose water-dissolved slow-release urea as ammonium pollution levels.
Materials and methods
Experiment design
The sediment used to cultivate P. crispus in the experiment was sampled from Lake Liangzi (114° 38′ 23″ N, 30° 14′ 28″ E), a mesoeutrophic lake located in Yangtze River catchment, Hubei Province, China. The total nitrogen content in the sediment was 0.50 g kg−1, and the concentrations of NH4+-N and NO3−-N in the fresh sediment interstitial water were 4.39 mg L−1 and 0.22 mg L−1, respectively (Wang et al. 2018). The submerged plant P. crispus was selected from Liangzi Lake as a cultivated plant which is one of the dominant plants in the middle reaches of the Yangtze River. It is characterized by rapid growth and well-developed root systems. A three-compartment with multiple interlayers rhizobox design was used to conduct rhizosphere experiments in the present study (Wang et al. 2018). The size of the rhizobox was 175×175×115 (length × width × height) mm (Fig. 1). The 20-mm width in the center of the rhizobox chamber was the central root chamber. Submerged plants were planted only in this part, and the 1–5-mm area on both sides of the central root chamber was the rhizosphere. Six sheets of nylon mesh (pore size <25 μm, 1 mm each) were inserted on rhizosphere to restrain the plant root to grow within the central root compartment only. The rhizobox was, therefore, divided into three main compartments and a total of seven layers, i.e. the central root compartment (20 mm in width, 1 layer), rhizosphere (1–5 mm from the root compartment, 5 layers), and non-rhizosphere (>5 mm from the root compartment, 1 layer) compartments. The rhizobox were filled with air-dried and sieved (1 mm) Lake Liangzi sediments in all layers.
The schematic diagram of the rhizobox (modified from Wang et al. 2018)
In November 2014, six P. crispus turions with similar growth characteristics were transplanted into each central root compartment to start the cultivation. After transplanting, the whole rhizobox and P. crispus were immersed under the distilled water to simulate the growth conditions of submerged plants. After 5 months, the central root compartment was filled with the root system in April 2015. Three concentrations of water-dissolved slow-release urea (Luxi Chemical Co., Ltd., Liaocheng, Shandong, China) were injected into sediment to simulate three nitrogen-contaminated sediment conditions (oligotrophic, mesoeutrophic, and hypereutrophic) in freshwater lakes. Before injection, the urea was dissolved in ultrapure water, and then the urea solution was injected into the sediment with a sterile syringe. Each rhizobox was arranged with 10 injection points, and the injection depth interval was 3 cm at each point to ensure that the urea solution could be evenly distributed in the sediment. After injection, the urea contents in the sediment were 0 mg kg−1 (N0), 400 mg kg−1 (N400), and 600 mg kg−1 (N600), respectively. On the 14, 28, and 42 days after urea injection, sediment samples from three replicate rhizobox were collected by destructive sampling. At each rhizobox, a total of seven layers of sediments were collected from three compartments: root compartment (R), rhizosphere compartment (N1-N5), and non-rhizosphere compartment (Non). Plant cultivation and sediment collection followed the procedures described by Wang et al. (2018).
Sediment chemical properties analysis
Before the samples were collected, the dissolved oxygen (DO) and pH of the interstitial water in each layer of the rhizobox were measured by using a microelectrode system (Unisense, Aarhus, Denmark). Each sediment was centrifuged at 4000 rpm for 15 min to obtain interstitial water, and then the interstitial water was filtered with a 0.45-μm pore filter membrane. The contents of NH4+-N and NO3−-N in the interstitial water were measured by using a flow injection analyzer (SEAL Analytical AA3; SEAL Analytical, Norderstedt, Germany). All samples were individually tested for physical and chemical indicators, as well as anammox 16S rRNA and nirS gene abundance.
DNA extraction and PCR amplification
Genomic DNA of each sediment sample was extracted using Fast DNA Spin Kit for Soil (MP Biomedicals, Solon, OH, USA) according to the manufacturer’s protocol. The purity and concentration of the extracted DNA samples were checked by super differential spectrophotometer (NanoPhotometer-N60; Implen, Munich, Germany), and then the qualified DNA samples were used for PCR amplification. The amplification primers for nirS gene were nirS1F/nirS6R (Braker et al. 1998). Anammox 16S rRNA amplification consisted of two steps with the amplification of primers pla46f/630r as the first step (Juretschko et al. 1998; Neef et al. 1998) and that of primers Amx368f/Amx820r as the second step (Schmid et al. 2005). PCR amplification of anammox 16S rRNA and nirS gene was performed in a total volume of 25 mL system containing 1 mL of each primer (10 mM), 12.5 mL of I-5™ 2X High-Fidelity Master Mix, and 9.5 mL of double-distilled water (ddH2O), with 1 mL of DNA as templates (20–50 ng). The anammox 16S rRNA and nirS gene amplification process is shown in Table 1. PCR products were tested by using agarose gel (1.0%) electrophoresis.
Cloning, sequencing, and phylogenetic analysis
The DNA samples of the same layer were mixed in equal amounts and used for sequencing. Then, the PCR products of anammox 16S rRNA and nirS gene were purified with magnetic bead purification kits (GeneOn BioTech, China). After purification, these products were cloned with the pClone 007 Vector Linker Kit (TSING KE, Beijing, China) and transformed into competent Escherichia coli cells according to the manufacturer’s instructions. Approximately 59 anammox 16S rRNA and 61 nirS positive clones were randomly picked and checked by agarose gel. The re-amplified PCR conditions of the anammox 16S rRNA and nirS gene in every clone are shown in Table 1. A 20-μL aliquot of the PCR product was digested with restriction endonuclease Msp I, separated by 110 mV electrophoresis on a 15% polyacrylamide gel for about 1 h and then stained with ethidium bromide (0.5 μg mL−1). Images of the gel were taken using a Kodak Gel Logic 100 system (Eastman Kodak, Rochester, NY, USA) to store their fingerprint of each clone. For each clone library, anammox 16S rRNA and nirS gene were sequenced by ABI 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA). Sequences with 97% identity were classified into same OTUs with anammox 16S rRNA and nirS gene using MOTHUR software (Wise et al. 2020; Schloss et al. 2009). Alignment of DNA sequences was carried out using the ClustalW 1.6, and a neighbor joining phylogenetic tree was constructed using MEGA 6.0 software. Bootstrap values for each branch were determined using 1000 iterations.
The sequences obtained in this study were deposited in the GenBank database with accession numbers of MK641594-MK641650 for anammox 16S rRNA and MK783879-MK783936 for nirS, respectively.
Real-time quantitative PCR (qPCR)
Quantitative analysis of anammox 16S rRNA and nirS genes was performed by real-time quantitative PCR by using the SYBR Green method with the QuantStudio™ 6 Flex quantitative PCR instrument (Thermo Fisher Scientific, Singapore). The anammox 16S rRNA and nirS genes qPCR primers (Tsushima et al. 2007; Throback et al. 2004) and amplification procedures are shown in Table 1. Each qPCR reaction system contained 10 μL mixture as follows: 5 μL of SybrGreen qPCR Master Mix, 0.5 μL of each primer (10 mM), 3 mL of ddH2O, and 1 μL of DNA template. The ddH2O was set as negative control, and the known copy number of plasmid DNA subjected to ten-fold serial dilution was set as positive control. Six standard plasmid DNA samples were obtained for quantitative amplification with three replicates each to generate a standard curve. The amplification efficiency of anammox 16S rRNA and nirS genes was 92.4% and 94.7%, respectively. The correlation coefficient (r2) was greater than 0.99, and the melting curve showed a single peak.
Statistical analysis
The physicochemical properties (pH, and NH4+-N and NO3−-N concentrations) of the interstitial water and the abundance of anammox 16S rRNA and nirS gene were investigated under three nitrogen pollution concentrations through one-way ANOVA and Tukey’s test (P<0.05) with SPSS 20.0 software. Detrended correspondence analysis (DCA) (Canoco 4.5 software) was used to examine the correlation between microbial abundance and its environmental factors. Since the length of the gradient value obtained by DCA in this study was less than three, the redundancy analysis (RDA) method was selected from the Canoco software. The significance test of Monte Carlo permutations (999) was used to explore the environmental factors related to the abundance of anammox 16S rRNA and nirS gene.
Results
Physicochemical properties of interstitial water in the rhizosphere
The concentration of NH4+-N in the sediment interstitial water differed significantly with the average concentration of 1.87, 16.42, and 20.16 mg L−1 for the three nitrogen treatments (N0, N400, and N600), respectively (P<0.05). The NH4+-N concentration in the urea addition treatment (N400 and N600) was significantly higher than that in the non-addition treatment (N0) during the entire experiment of up to 42 days (Fig. 2A). On day 28, the average of NH4+-N concentrations in urea addition treatments (N400: 20.94 mg L−1, N600: 28.01 mg L−1) was significantly higher than that on day 14 (N400: 15.54 mg L−1, N600: 16.20 mg L−1) and day 42 (N400: 12.79 mg L−1, N600: 16.28 mg L−1) after the urea addition.
NH4+-N concentration (A) and NO3−-N concentration (B) in sediment interstitial water under different nitrogen pollution conditions. Different letters for the same gene indicated the significant difference between different distances from root at P < 0.05 (Tukey’s test). Error bars indicated standard deviation
The NO3−-N concentration in the root compartment and non-rhizosphere showed no significant difference. During the entire experiment, the NO3−-N concentration in the rhizosphere interstitial water reached the peak at 14 d after the urea addition and then was gradually decreased with the extended time of urea addition (Fig. 2B). In contrast, the NO3−-N concentrations in the root compartment and non-rhizosphere interstitial water remained stable with the extended time of urea addition. The average interstitial water NO3−-N concentrations were 1.18, 1.21, and 1.34 mg L−1, respectively, for three urea addition treatments of N0, N400, and N600, and there was no significant difference between different treatments (P>0.05). The average pH values in the rhizosphere (1–5 mm) for three treatments of N0, N400, and N600 ranged from 7.46 to 7.63, 7.60 to 7.73, and 7.60 to 7.64, respectively. There was no significant difference in oxygen concentration at the sediment-water interface under each treatment, and the average value was 11.86 μmol L−1.
Nitrogen-transforming bacterial abundance in the rhizosphere of P. crispus under ammonium pollution
The gene abundance of anammox 16S rRNA in the rhizosphere was generally higher than that in the root compartment and non-rhizosphere, but there was no significant difference between them (P>0.05) (Fig. 3). The anammox 16S rRNA gene abundance in the rhizosphere for each treatment group (0, 400, 600 mg kg−1) was 1.96×107, 1.82×107, and 1.83×107 copies g−1 (P>0.05) (Fig. 3), respectively. The gene abundance of anammox 16S rRNA (2.01×107 copies g-−1) on day 14 was significantly higher than that on day 28 (1.69×107 copies g−1), but it was closer to that on day 42 (1.89×107 copies g−). During the entire experiment, the anammox 16S rRNA abundance for each treatment group (0, 400, 600 mg kg−1) was 1.96×107, 1.82×107, and 1.83×107 copies g−1, respectively. And there was no significant difference between different groups (P>0.05, Fig. 4).
Average concentrations of NH4+-N and NO3−-N in interstitial water and the average abundance of anammox 16S rRNA and nirS gene in sediment at different slow-release urea concentrations during the whole experiment. Different letters above the bars indicated significant differences (P<0.05) (Tukey’s test) between different distances from root. Error bars indicated standard deviation
Under N0 treatment, the abundance of nirS gene in rhizosphere showed significant difference on days 14 and 28 (P<0.05) (Fig. 3). The nirS gene abundance in the rhizosphere for each treatment group (0, 400, 600 mg kg−1) was 2.25×105, 2.12×105, and 1.99×105 copies g−1 (P>0.05) (Fig. 3), respectively. Under N400 and N600 treatments, the abundance of nirS gene in the rhizosphere, root compartment, and non-rhizosphere showed no significant difference (P>0.05). Throughout the entire experiment, the average abundance of nirS gene under three nitrogen treatments (0, 400, 600 mg kg−1) was 2.25×105, 2.12×105, and 1.99×105 copies g−1, respectively, and no significant difference was observed between them (P>0.05, Fig. 4). The fluctuation trend of (anammox 16S rRNA)/nirS ratio was very close on the same sampling date under three nitrogen treatments.
Impact of key physicochemical factors on rhizosphere bacterial abundance
The first two RDA axes together explained 62.2% of the variance (Fig. 5). RDA analysis showed that the abundance of the anammox 16S rRNA and nirS gene was significantly correlated with the concentration of NO3−-N (P=0.001, F=14.725, 999 Monte Carlo permutations) and the mole ratio of NH4+-N/NO3−-N in the interstitial water (P=0.001, F=7.655, 999 Monte Carlo permutations). The abundance of the anammox 16S rRNA gene was positively correlated with the abundance of the nirS gene. The abundance of the anammox 16S rRNA gene was negatively correlated with the concentration of NO3−-N and pH in the interstitial water. The abundance of nirS gene was negatively correlated with pH and the concentrations of NH4+-N and NO3−-N in the interstitial water. The abundance of anammox 16S rRNA and nirS gene was positively correlated with the ratio of NH4+-N/NO3−-N in the interstitial water.
Diversity of anammox and nirS denitrifiers in the rhizosphere
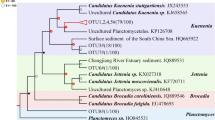
A total of 57 anammox 16S rRNA gene sequences were obtained from the constructed clone library. A total of five OTUs were obtained with a similarity of 97% (Fig. 6). The sequences of OTU1 and OTU2 accounted for 61.4% and 17.5% of the cloned library, respectively. Anammox 16S rRNA gene sequences in the sediments of phylogenetic trees were mainly clustered to the genus Brocadia and Kuenenia. OTU1, OTU2, OTU3, and OTU5 were all assigned to the genus Brocadia. The sequences of this genus accounted for 93% of all the sequences of anammox 16S rRNA. Therefore, genus Brocadia was the dominant species of anammox in this study, and the similar sequences were from freshwater lake sediments, dehydrated aluminum sludge, suspended sediments of the Yellow River, and rhizosphere sediment of Potamogeton. OTU4 belonged to the genus Kuenenia and had a high similarity with the sequences from freshwater sediment, but its sequences accounted for only 7% of the total number of sequences.
Phylogenetic tree of representative anammox 16S rRNA sequences and reference sequences from GenBank. The number in parentheses indicates that the sequence in the OTU out of the total number of sequences. The numbers at the nodes represent percentages that indicate the levels of bootstrap support based on neighbor-joining analysis of 1000 resampled datasets (bootstrap values greater than 50% are shown). Branch lengths represent sequence differences indicated by the scale bar
Among the 58 nirS sequences, 84.5% belonged to Betaproteobacteria (β-Proteobacteria) class, 5.2% belonged to Gammaproteobacteria (γ-Proteobacteria) class, and 10.3% belonged to Bacilli class (Fig. 7). The β-Proteobacteria class was the most dominant group in the nirS gene clone library with 24 OTUs belonging to this class. The β-Proteobacteria class was divided into three clusters, namely uncultured cluster, Thauera cluster, and Cupriavidus cluster, respectively. Uncultured cluster (24 sequences) contained more sequences than Thauera cluster (13 sequences) and Cupriavidus cluster (12 sequences) in the clone library. OTU1 exhibited the highest abundance, which belonged to Cupriavidus cluster including 9 sequences, and these 9 sequences exhibited high similarity with those in the sediments of submerged plants. Three OTUs (OTU23, OTU24, OTU28) belonging to Thiothrix cluster, γ-Proteobacteria class, exhibited the similar sequences with those from Yangtze lake sediment and Jiulong river estuary sediment. OTU4 and OTU6 belonged to Bacillus cluster, Bacilli class.
Phylogenetic tree of representative nirS sequences and reference sequences from GenBank. The number in parentheses indicates that the sequences in the OTU out of the total number of sequences. The numbers at the nodes represent percentages that indicate the levels of bootstrap support based on neighbor-joining analysis of 1000 resampled datasets (bootstrap values greater than 50% are shown). Branch lengths represent sequence differences indicated by the scale bar
Discussion
Many studies have shown that changes in nitrogen content in the environment will significantly change the abundance of anammox and denitrifying bacteria in sediments (Zhao et al. 2020; Fu et al. 2019; Kim et al. 2016). Guo et al. (2014) found that compared with those in mesoeutrophic lakes, denitrifying bacteria in hypereutrophic lakes exhibited the lowest abundance and evenness. However, the eutrotrophic Dianchi Lake had greater anammox bacterial abundance than mesotrophic Erhai Lake (Yang et al. 2017). The similar phenomenon was also found in the sediment of Yangcheng Lake, that is, the maximum abundance of anammox bacteria was in months with high ammonia concentration (Zhang et al. 2016). Therefore, a high nitrogen input was likely to be able to stimulate the growth of anammox bacteria (Nie et al. 2019). The content of ammonium also affected the composition of anammox bacteria and the types of denitrifying bacteria. Some studies found that in eutrophic Taihu Lake and Kitaura Lake, community structure analysis showed that anammox bacteria Ca. Brocadia sp. were the dominant genus (Qin et al. 2018; Zhu et al. 2015), and the species richness of denitrifying bacteria decreased with the decrease of ammonium concentrations from surface flow wetland to ditch wetland (Chen et al. 2020a). In the present study, the NH4+-N concentrations in the sediment interstitial water for each nitrogen pollution treatment were significantly different during the whole experiment (Fig. 4) in the presence of P. crispus. Especially, the maximum NH4+-N concentration of 600 mg kg−1 urea treatment was 20.2 mg L−1, while the minimum NH4+-N concentration of 0 mg kg−1 urea treatment was 0.9 mg L−1. However, the abundance of anammox 16S rRNA and nirS gene showed no significant difference under the three nitrogen pollution conditions. Therefore, the presence of submerged plants provided a stable environment for anammox bacteria and nirS denitrifiers to avoid rapid fluctuations in the abundance of these two bacteria in the rhizosphere.
The ratio of the abundance of two microorganisms can reflect which microorganism has better adaptability to the environment (Kent et al. 2019; Liu et al. 2015). Our previous study found that the ratio of the abundance of anammox 16S rRNA to nirS gene in the rhizosphere could quickly respond to the damage of P. crispus leaves at day 10 after the damage treatment (Hu et al. 2020). However, in the present study, during the 42 days of pollution treatment, no significant changes in the gene abundance ratio were found between different nitrogen pollution treatments, although NH4+-N concentrations differed significantly (Fig. 3). Our results indicated that the ecological relationship between these two bacteria was not impacted by different pollution levels in the presence of submerged plant. This further confirms that submerged plants provided a stable environment for anammox bacteria and nirS denitrifiers to avoid the rapid fluctuations in the abundance of these two bacteria in the rhizosphere.
One reason why plants can provide a stable environment might be the radial oxygen loss (ROL) from plants. When there were no plants in the lake, the sediment-water interface gradually becomes anoxic, and nitrification can only occur in the oxygen-containing area of the surface sediments (Satoh et al. 2007). However, oxygen secretion from plant roots can provide oxygen to ammonium-oxidizing microorganisms in deep rhizosphere sediments (Okabe et al. 2012). Therefore, the amount of oxygen secreted from the root system is an important rate-limiting factor for ammonia oxidation in rhizosphere sediments (Wang et al. 2012; Lam et al. 2007). Different macrophytes may show different ROL (Tian et al. 2015); however, there might be a certain limit on the amount of oxygen loss. For example, the ROL values of Juncus effusus L. and Juncus inflexus L. were (9.5 ± 1)×10−7 and (4.5 ± 0.5)× 10−7 mol O2 h−1 root−1 (Sorrell 1999), respectively. The limited oxygen loss of the root system prevents the ammonium around the rhizosphere from being rapidly oxidized to nitrate, thus avoiding the impact of the rapid rise in nitrate concentrations on anammox and denitrifying bacteria activities. Our results also showed that although the ammonium concentration under each pollution treatment was significantly different, the nitrate concentration was relatively stable (Fig. 4). However, the concentration of nitrate only increased significantly in the rhizosphere on 14 days and remained stable in the subsequent 28 and 42 days (Fig. 2). This indicated that the presence of plants can stabilize the concentration of nitrate in the rhizosphere and provide a more stable growth environment for anammox bacteria and denitrifying bacteria.
Another reason why plants can provide a stable environment wasthat plants may also increase their NH4+-N absorption capacity when NH4+-N content increases in the sediment, thereby alleviating the ammonium pollution. Most macrophytes have been confirmed to prefer ammonium rather than nitrate due to their energy-saving strategies (Fang et al. 2007; Tylova-Munzarova et al. 2005). Therefore, plants can absorb ammonium in the interstitial water faster than nitrate (Zhao et al. 2019a). The ammonium concentration in sediment has been detected to be significantly reduced after planting Phragmites australis (Toyama et al. 2016). In the present experiment, the concentration of urea added in N600 treatment was 1.5 times as much as that in N400 treatment, but the average ammonium concentration in N600 treatment was detected to be only 1.2 times as much as that in N400 treatment (Fig. 4), indicating that when the ammonium content in the interstitial water was increased, P. crispus also increased its capacity of ammonium absorption to avoid the damage to the micro-habitat caused by the rapid rise of ammonium in the rhizosphere to a certain extent.
The third reason why plants can provide a stable environment may be that plant roots increase the absorption of nitrate in interstitial water. On 14 days, nitrate in rhizosphere interstitial water showed an upward trend (Fig. 2). It may be that ammonia-oxidizing archaea and ammonia-oxidizing bacteria (AOB) in the rhizosphere oxidized the ammonia released by urea, which caused the concentration of nitrate to rise. Studies also found that the application of urea and fertilizer stimulated the enrichment of AOB (Chen et al. 2020b), and it also significantly increased the abundance and diversity of AOB genes (Tao et al. 2021). The enrichment of AOB accelerates ammonia oxidation speed. However, on 28 and 42 days, the concentration of nitrate in rhizosphere interstitial water remained relatively low when the ammonia concentration did not decrease significantly. This indicated that when the abundance of two nitrate consumers, anammox bacteria and denitrifying bacteria, did not increase significantly, plant roots were likely to increase the absorption of nitrate after short-term adaptation.
Nie et al. (2019) found that the ratio of NH4+/NO3− in the soil was a key factor regulating the anammox process. In the present study, the gene abundance of anammox 16S rRNA was negatively correlated with NO3−-N content in the interstitial water, but it was positively correlated with the ratio of NH4+/NO3-, which might be due to the fact that at high ammonium concentrations, anammox bacteria might be more inclined to use NO2− produced by nitrification as reaction substrate compared to NO3− or NH4+ (Nie et al. 2019). In the present study, the abundance of nirS gene was also negatively correlated with NO3−, which was consistent well with the results of our previous study (Hu et al. 2020). The research on neutralized used acid biofilter and reeds rhizosphere sediment also found that the abundance of nirS gene was not positively correlated with NO3− (Xu et al. 2020; Wang et al. 2017). This might be attributed to the interference of the anaerobic ammonia oxidation process, since the intermediate products from nitrification and denitrification were used as reaction substrates, these intermediate products might interfere with the second step of the denitrification process dominated by nirS denitrifiers (Wang et al. 2017).
Five genera of anammox bacteria have been identified (Zhou et al. 2018). The present study found that Candidatus Brocadia and Candidatus Kuenenia of anammox bacteria could coexist in sediments, and these two genera were reported to be more adapted to the growth in freshwater systems (Sun et al. 2014; Zhu et al. 2013). In our study, Ca. Brocadia was dominant in anammox bacteria, and this genus was also found to be dominant in the sediments of rivers and eutrophic lakes in previous studies (Hu et al. 2012; Yoshinaga et al. 2011; Zhang et al. 2007). Candidatus Brocadia prefers a rich ammonium environment (Oshiki et al. 2016). The high ammonium content in the present study might be the reason for Ca. Brocadia predominance. Candidatus Brocadia was also reported to coexist with other genera of anammox bacteria (Sonthiphand et al. 2014). Our study also found that a small amount of Ca. Kuenenia coexisted with Ca. Brocadia. Candidatus Kuenenia showed lower affinity constants for NH4+ and NO2− which might make it more suitable for a low ammonium environment (Oshiki et al. 2016). The rich ammonium environment in the present study might have an inhibitory effect on the growth of these bacteria.
This study indicated that β-Proteobacteria and γ-Proteobacteria accounted for 84.5% and 5.2% of the total number of nirS sequences, respectively. Both the β-Proteobacteria class and the γ-Proteobacteria class belonged to the Proteobacteria phylum. Previous study has found that the Proteobacteria phylum has an advantage in the river environment (Zwart et al. 2002), which is in line with our findings for this phylum. The present study found that Thauera cluster contained 13 sequences, which has already been confirmed to be more suitable for survival in an eutrophic environment in previous study (Yang et al. 2013). Some studies have also reported that the nitrate addition will increase the denitrification activity, but this nitrate addition cannot change the community composition of the denitrifying bacteria (Wallenstein et al. 2006).
Conclusions
In this study, three different concentrations of slow-release urea were added to the rhizosphere sediments of P. crispus to investigate their impacts on anammox and denitrifying bacteria. Our results showed that the ammonium concentrations in the interstitial water were significantly increased after the addition of the slow-release urea, but the concentrations of nitrate remained stable. The abundance of the anammox 16S rRNA and nirS gene showed no significant difference among three nitrogen pollution treatments. And the change trend of the ratio of (anammox 16S rRNA)/nirS in different nitrogen treatment groups on the same sampling date was very close. These results support our hypothesis that under increased nitrogen pollution conditions, submerged plants weaken the environmental changes caused by ammonia pollution in the rhizosphere, thereby avoiding strong fluctuations anammox bacteria and nirS denitrifiers.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Braker G, Fesefeldt A, Witzel KP (1998) Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl Environ Microbiol 64:3769–3775
Brisson J, Chazarenc F (2009) Maximizing pollutant removal in constructed wetlands: should we pay more attention to macrophyte species selection? Sci Total Environ 407:3923–3930
Chen XY, Lu JJ, Zhu J, Liu CQ (2020a) Characteristics of denitrifying bacteria in different habitats of the Yongding River wetland, China. J Environ Manag 275:111273
Chen ZM, Wang Q, Zhao J, Chen YD, Wang HY, Ma JW, Zou P, Bao L (2020b) Restricted nitrous oxide emissions by ammonia oxidizers in two agricultural soils following excessive urea fertilization. J Soils Sediments 20:1502–1512
Christensen PB, Revsbech NP, Sand-Jensen K (1994) Microsensor analysis of oxygen in the rhizosphere of the aquatic macrophyte Littorella uniflora (L.) Ascherson. Plant Physiol 105:847–852
Fang YY, Babourina O, Rengel Z, Yang XE, Pu PM (2007) Ammonium and nitrate uptake by the floating plant Landoltia punctata. Ann Bot 99:365–370
Fu LL, Chen YY, Li SQ, He H, Mi TZ, Zhen Y, Yu ZG (2019) Shifts in the anammox bacterial community structure and abundance in sediments from the Changjiang Estuary and its adjacent area. Syst Appl Microbiol 42:383–396
Guo LY, Hu ZX, Fang F, Liu T, Chuai XM, Yang LY (2014) Trophic status determines the nirS-denitrifier community in shallow freshwater lakes. Ann Microbiol 64:999–1006
Hu B, Shen L, Du P, Zheng P, Xu X, Zeng J (2012) The influence of intense chemical pollution on the community composition, diversity and abundance of anammox bacteria in the Jiaojiang Estuary (China). PLoS One 7:e33826
Hu JL, Zhou YH, Lei ZY, Liu GL, Hua YM, Zhou WB, Wan XQ, Zhu DW, Zhao JW (2020) Effects of Potamogeton crispus decline in the rhizosphere on the abundance of anammox bacteria and nirS denitrifying bacteria. Environ Pollut 260:114018
Hussain Q, Liu YZ, Jin ZJ, Zhang AF, Pan GX, Li LQ, Crowley D, Zhang XH, Song XY, Cui LQ (2011) Temporal dynamics of ammonia oxidizer (amoA) and denitrifier (nirK) communities in the rhizosphere of a rice ecosystem from Tai Lake region, China. Appl Soil Ecol 48:210–218
Juretschko S, Timmermann G, Schmid M, Schleifer KH, Pommerening-Roser A, Koops HP, Wagner M (1998) Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol 64:3042–3051
Kent TR, Sun Y, An Z, Bott CB, Wang ZW (2019) Mechanistic understanding of the NOB suppression by free ammonia inhibition in continuous flow aerobic granulation bioreactors. Environ Int 131:105005
Kim H, Bae HS, Reddy KR, Ogram A (2016) Distributions, abundances and activities of microbes associated with the nitrogen cycle in riparian and stream sediments of a river tributary. Water Res 106:51–61
Koop-Jakobsen K, Giblin AE (2009) Anammox in tidal marsh sediments: the role of salinity, nitrogen loading, and marsh vegetation. Estuar Coasts 32:238–245
Kumar M, Lin JG (2010) Co-existence of anammox and denitrification for simultaneous nitrogen and carbon removal-strategies and issues. J Hazard Mater 178:1–9
Lam P, Jensen MM, Lavik G, McGinnis DF, Muller B, Schubert CJ, Amann R, Thamdrup B, Kuypers MMM (2007) Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc Natl Acad Sci U S A 104:7104–7109
Le C, Zha Y, Li Y, Sun D, Lu H, Yin B (2010) Eutrophication of lake waters in China: cost, causes, and control. Environ Manag 45:662–668
Liu S, Hu BL, He ZF, Zhang B, Tian GM, Zheng P, Fang F (2015) Ammonia-oxidizing archaea have better adaptability in oxygenated/hypoxic alternant conditions compared to ammonia-oxidizing bacteria. Appl Environ Microbiol 99:8587–8596
Neef A, Amann R, Schlesner H, Schleifer KH (1998) Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology 144:3257–3266
Nie SA, Li H, Yang XR, Zhang ZJ, Weng BS, Huang FY, Zhu GB, Zhu YG (2015) Nitrogen loss by anaerobic oxidation of ammonium in rice rhizosphere. ISME J 9:2059–2067
Nie SA, Zhu GB, Singh B, Zhu YG (2019) Anaerobic ammonium oxidation in agricultural soils-synthesis and prospective. Environ Pollut 244:127–134
Niu CZ, He ZX, Ge Y, Chang J, Lu ZM (2015) Effect of plant species richness on methane fluxes and associated microbial processes in wetland microcosms. Ecol Eng 84:250–259
Okabe S, Nakamura Y, Satoh H (2012) Community structure and in situ activity of nitrifying bacteria in Phragmites root-associated biofilms. Microbes Environ 27:242–249
Oshiki M, Satoh H, Okabe S (2016) Ecology and physiology of anaerobic ammonium oxidizing bacteria. Environ Microbiol 18:2784–2796
Pajares S, Merino-Ibarra M, Macek M, Alcocer J (2017) Vertical and seasonal distribution of picoplankton and functional nitrogen genes in a high-altitude warm-monomictic tropical lake. Freshw Biol 62:1180–1193
Qin HY, Han C, Jin ZW, Wu L, Deng H, Zhu GW, Zhong WH (2018) Vertical distribution and community composition of anammox bacteria in sediments of a eutrophic shallow lake. J Appl Microbiol 125:121–132
Satoh H, Nakamura Y, Okabe S (2007) Influences of infaunal burrows on the community structure and activity of ammonia-oxidizing bacteria in intertidal sediments. Appl Environ Microbiol 73:1341–1348
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Schmid MC, Maas B, Dapena A, van de Pas-Schoonen K, van de Vossenberg J, Kartal B, van Niftrik L, Schmidt I, Cirpus I, Kuenen JG, Wagner M, Damste JSS, Kuypers M, Revsbech NP, Mendez R, Jetten MSM, Strous M (2005) Biomarkers for in situ detection of anaerobic ammonium-oxidizing (anammox) bacteria. Appl Environ Microbiol 71:1677–1684
Sonthiphand P, Hall MW, Neufeld JD (2014) Biogeography of anaerobic ammonia-oxidizing (anammox) bacteria. Front Microbiol 5:399
Sorrell BK (1999) Effect of external oxygen demand on radial oxygen loss by Juncus roots in titanium citrate solutions. Plant Cell Environ 22:1587–1593
Sun W, Xia CY, Xu MY, Guo J, Wang AJ, Sun GP (2014) Diversity and distribution of planktonic anaerobic ammonium-oxidizing bacteria in the Dongjiang River, China. Microbiol Res 169:897–906
Tang X, Zhang XL, Cao T, Ni LY, Xie P (2018) Reconstructing clear water state and submersed vegetation on behalf of repeated flocculation with modified soil in an in situ mesocosm experiment in Lake Taihu. Sci Total Environ 625:1433–1445
Tao R, Li J, Hu BW, Chu GX (2021) Mitigating N2O emission by synthetic inhibitors mixed with urea and cattle manure application via inhibiting ammonia-oxidizing bacteria, but not archaea, in a calcareous soil. Environ Pollut 273:116478
Throback IN, Enwall K, Jarvis A, Hallin S (2004) Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol Ecol 49:401–417
Tian CC, Wang CB, Tian YY, Wu XQ, Xiao BD (2015) Root radial oxygen loss and the effects on rhizosphere microarea of two submerged plants. Pol J Environ Stud 24:1795–1802
Toyama T, Nishimura Y, Ogata Y, Sei K, Mori K, Ike M (2016) Effects of planting Phragmites australis on nitrogen removal, microbial nitrogen cycling, and abundance of ammonia-oxidizing and denitrifying microorganisms in sediments. Environ Technol 37:478–485
Tsushima I, Ogasawara Y, Kindaichi T, Satoh H, Okabe S (2007) Development of high-rate anaerobic ammonium-oxidizing (anammox) biofilm reactors. Water Res 41:1623–1634
Tylova-Munzarova E, Lorenzen B, Brix H, Votrubova O (2005) The effects of NH4+ and NO3- on growth, resource allocation and nitrogen uptake kinetics of Phragmites australis and Glyceria maxima. Aquat Bot 81:326–342
Vazquez-Padin J, Mosquera-Corral A, Campos JL, Mendez R, Revsbech NP (2010) Microbial community distribution and activity dynamics of granular biomass in a CANON reactor. Water Res 44:4359–4370
Wallenstein MD, Myrold DD, Firestone M, Voytek M (2006) Environmental controls on denitrifying communities and denitrification rates: insights from molecular methods. Ecol Appl 16:2143–2152
Wan Y, Ruan XH, Wang J, Shi XJ (2019) Spatial and seasonal variations in the abundance of nitrogen-transforming genes and the microbial community structure in freshwater lakes with different trophic statuses. Int J Environ Res Public Health 16:2298
Wang SY, Zhu GB, Peng YZ, Jetten MSM, Yin CQ (2012) Anammox bacterial abundance, activity, and contribution in riparian sediments of the Pearl River estuary. Environ Sci Technol 46:8834–8842
Wang SY, Peng YZ, Ma B, Wang SY, Zhu GB (2015) Anaerobic ammonium oxidation in traditional municipal wastewater treatment plants with low-strength ammonium loading: widespread but overlooked. Water Res 84:66–75
Wang F, Zhao Y, Xie SL, Li JY (2017) Implication of nitrifying and denitrifying bacteria for nitrogen removal in a shallow lake. Clean-Soil Air Water 45:7
Wang BH, Huang SS, Zhang LM, Zhao JW, Liu GL, Hua YM, Zhou WB, Zhu DW (2018) Diversity of NC10 bacteria associated with sediments of submerged Potamogeton crispus (Alismatales: Potmogetonaceae). PeerJ 6:e6041
Wang SY, Pi YX, Jiang YY, Pan HW, Wang XX, Wang XM, Zhou JM, Zhu GB (2020) Nitrate reduction in the reed rhizosphere of a riparian zone: from functional genes to activity and contribution. Environ Res 180:108867
Wise B, Roane TM, Mosier AC (2020) Community composition of nitrite reductase gene sequences in an acid mine drainage environment. Microb Ecol 79:562–575
Xu WW, Wang LM, Peng FQ, Zhang AG, Xie XG, Wang ZB, Wang X, Lian JJ, Ni LX, Cui YB, Zhang YM, Yang F, Zhu YM, Mao XH (2020) Spatiotemporal distribution and interaction of denitrifying functional genes in a novel DAS-NUA biofilter used for groundwater nitrate treatment. Sci Total Environ 712:136595
Yang JK, Cheng ZB, Li J, Miao LH (2013) Community composition of nirS-type denitrifier in a shallow eutrophic lake. Microb Ecol 66:796–805
Yang YY, Yu D, Li NN, Li BX, Xie SG, Liu Y (2017) Temporal and spatial dynamics of sediment anaerobic ammonium oxidation (anammox) bacteria in freshwater lakes. Microb Ecol 73:285–295
Yang JY, Li J, Zheng ZM, Hou LG, Liang DB, Sun YQ, Ma XR (2019) Effect of organic matters on anammox coupled denitrification system: when nitrite was sufficient. R Soc Open Sci 6:190771
Yoshinaga I, Amano T, Yamagishi T, Okada K, Ueda S, Sako Y, Suwa Y (2011) Distribution and diversity of anaerobic ammonium oxidation (anammox) bacteria in the sediment of a eutrophic freshwater lake, Lake Kitaura, Japan. Microbes Environ 26:189–197
Zhang Y, Ruan XH, Op den Camp HJ, Smits TJ, Jetten MS, Schmid MC (2007) Diversity and abundance of aerobic and anaerobic ammonium-oxidizing bacteria in freshwater sediments of the Xinyi River (China). Environ Microbiol 9:2375–2382
Zhang YP, Ruan XH, Wan Y, Li XW (2016) Effects of environmental factors on anammox bacterial community structure in sediments of a freshwater aquaculture farm, Yangcheng Lake. Geomicrobiol J 33:479–487
Zhao DY, Luo J, Zeng J, Wang M, Yan WM, Huang R, Wu QLL (2014) Effects of submerged macrophytes on the abundance and community composition of ammonia-oxidizing prokaryotes in a eutrophic lake. Environ Sci Pollut Res 21:389–398
Zhao CY, Liu SL, Jiang ZJ, Wu YC, Cui LJ, Huang XP, Macreadie PI (2019a) Nitrogen purification potential limited by nitrite reduction process in coastal eutrophic wetlands. Sci Total Environ 694:133702
Zhao ZS, Cao YL, Fan Y, Yang HL, Feng XW, Li L, Zhang HL, Xing L, Zhao MX (2019b) Ladderane records over the last century in the East China sea: proxies for anammox and eutrophication changes. Water Res 156:297–304
Zhao R, Summers ZM, Christman GD, Yoshimura KM, Biddle JF (2020) Metagenomic views of microbial dynamics influenced by hydrocarbon seepage in sediments of the Gulf of Mexico. Sci Rep 10:5527
Zhao DH, Chen C, Yang JQ, Zhou SY, Du J, Zhang M, An SQ (2021) Mutual promotion of submerged macrophytes and biofilms on artificial macrophytes for nitrogen and cod removal improvement in eutrophic water. Environ Pollut 277:116718
Zhou XH, Zhang JP, Li YM, Liu B, Chu JY, Wang MY, He ZL (2016) Distribution characteristics of ammonia oxidizing microorganisms in rhizosphere sediments of cattail. Ecol Eng 88:99–111
Zhou ZC, Wei QY, Yang YC, Li M, Gu JD (2018) Practical applications of PCR primers in detection of anammox bacteria effectively from different types of samples. Appl Microbiol Biotechnol 102:5859–5871
Zhu GB, Wang SY, Wang WD, Wang Y, Zhou LL, Jiang B, Op den Camp HJM, Risgaard-Petersen N, Schwark L, Peng YZ, Hefting MM, Jetten MSM, Yin CQ (2013) Hotspots of anaerobic ammonium oxidation at land-freshwater interfaces. Nat Geosci 6:103–107
Zhu GB, Xia C, Wang SY, Zhou LL, Lu L, Zhao SY (2015) Occurrence, activity and contribution of anammox in some freshwater extreme environments. Environ Microbiol Rep 7:961–969
Zwart G, Crump BC, Agterveld MPKV, Hagen F, Han SK (2002) Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat Microb Ecol 28:141–155
Funding
This work was supported by the National Natural Science Foundation of China (41371452), Major Science and Technology Program for Water Pollution Control and Treatment of China (2014ZX07203010).
Author information
Authors and Affiliations
Contributions
Yangfan Xu: Methodology, investigation, formal analysis, data curation, and writing-original draft. Jing Lu: Writing-review and editing. Shanshan Huang: Investigation, resources, and data curation. Jianwei Zhao: Conceptualization, visualization, supervision, and writing-review and editing, project administration, and funding acquisition.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
The authors confirm that the final version of the manuscript has been reviewed, approved, and consented for publication by all authors.
Competing interests
The authors declare no competing interests.
Additional information
Responsible editor: Diane Purchase
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Xu, Y., Lu, J., Huang, S. et al. Submerged plants alleviated the impacts of increased ammonium pollution on anammox bacteria and nirS denitrifiers in the rhizosphere. Environ Sci Pollut Res 28, 58755–58767 (2021). https://doi.org/10.1007/s11356-021-14715-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-021-14715-7