Abstract
Denitrification is a major biological process to reduce nitrate to molecular nitrogen (N2). In shallow eutrophic lakes, this process can remove the largest portion of fixed nitrogen and plays an important role in self-purification of this ecosystem. To understand the structure of denitrifying communities in a shallow eutrophic lake, denitrifier communities in four sub-lakes of East Lake in Wuhan, China, were explored by restriction fragment length polymorphisms (RFLP) analysis and sequencing of nirS gene clone libraries. nirS is a functional marker gene for denitrification encoding cytochrome cd 1-containing nitrite reductase, which catalyzes the reduction of nitrite to nitric oxide. Both RFLP fingerprints clustering analysis and phylogeny analysis based on the amino acid sequences of NirS revealed that NirS-type communities in East Lake sediment could be roughly divided into three clusters. Cluster I accounted for 74–82 % of clones from the moderately eutrophic sub-lakes Tuan, Tang Ling, and Guo Zheng. Cluster II accounted for 76 % of the communities in hypertrophic sub-lake Miao Lake and cluster III as a minor group (7 % of the total), mainly presented in Miao Lake. Phylogenetic analysis revealed that cluster I was related to the reference clones from a broad range of ecological environments, and clusters II and III were more phylogenetically related to the reference clones from entrophic environments. Canonical correspondence analysis indicated that total nitrogen, total phosphate, total organic carbon, and NH4–N and NO2–N were important environmental factors affecting the dispersion of NirS-type denitrifier in the sediments. Cluster I showed a weak relationship with the nutrient content, while cluster II and III were positively related with the nutrient content. Principal coordinates analysis indicated that NirS-type communities from Tuan Lake, Tang Ling Lake, and Guo Zheng Lake sediments were divergent from those found in river, estuary sediment, and forest soil but similar to communities in constructed wetland sediment despite large geographic distances. The communities from the hypertrophic sub-lake Miao Lake deviated from other sub-lakes and the reference communities and clustered independently. Our results support the argument that environmental factors regulate the composition and distribution of the functional bacterial groups.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Denitrification is a microbially facilitated process reducing nitrate or nitrite to molecular nitrogen (N2) under anaerobic conditions. It is the major biological pathway for the loss of fixed nitrogen in the ecosystem. It can remove more than 50 % of the nitrogen inputs into water systems and plays an important role in water quality control and self-purification of pollution water [11, 12]. The process of denitrification consists of four reaction steps to reduce nitrate into dinitrogen gas (N2), which go through a series of intermediate nitrogen oxide products including nitrite (NO2 −), nitric oxide (NO−), and nitrous oxide (N2O) catalyzed by nitrate reductase, nitrite reductase, nitric oxide reductase, and nitrous oxide reductase, respectively [2, 21, 33]. Nitrite reductases that reduce nitrite to nitric oxide are the main biocatalysts of the denitrification process [16]. This nitrite reductase consists of two different types, a cytochrome cd 1-containing nitrite reductase encoded by nirS (cdNIR) or a Cu-dependent nitrite reductase (CuNIR) encoded by nirK [13]. Recently, the functional nitrite reductase gene nirS and nirK have been proven as useful molecular markers to resolve the community structure of denitrifying microbes. The information gained from the sequences of nirS has provided a comprehensive measure of the community diversity and has now been extensively and effectively adopted to elucidate the community composition of denitrifiers in various environments [1, 6, 9, 18].
Due to the rapid industrialization and intensification of human activities, many lakes that have been drinking water sources, ecological regulators, and life sources in local areas are suffering serious eutrophication and deterioration. Denitrification is the major biological process for the quality control and self-purification of the lakes [15]. Although the chemical information on denitrification of the lakes has already been intensively studied by water chemists and limnologists [4, 5, 14, 28], the composition of the denitrifer community has not been properly assessed. Only recently have the communities in similar environments such as freshwater river sediment [17] and constructed wetland sediment [27] been elucidated. However, we still lack researches on denitrifying microbial community of eutrophic lake environments.
East Lake, which sits close to Wuhan city, China, is a medium-sized shallow freshwater lake. Since the late 1980s, due to the injection of sewage, aquaculture, and other reasons, this lake quickly deteriorated into a eutrophic lake [8, 26]. In this study, by using the nitrite reductase gene nirS as a molecular marker, we resolved the community composition and phylogenetic diversity of NirS-type denitrifying bacteria in the sediments of four sub-lakes of East Lake in Wuhan. We further comparatively analyzed the relationship between East Lake community and other typical ecological environments by principal coordinates analysis (PCoA), and explored the relationship between the denitrifying bacteria and the nutrient content in the sediment by canonical correspondence analysis (CCA) analysis. This study could enrich our understanding of the denitrifying bacterial community structure and provide references to the research, protection, and pollution control in freshwater lake environments.
Materials and Methods
Sample Collection and Environment Analysis
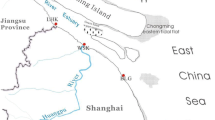
East Lake (114°23′E, 30°22′N) sits in the southeast of Wuhan city, China. Its watershed is about 187 km2 and consists of several sub-lakes (Fig. 1). The deepest water depth is 4.75 m and the average water depth is 2.21 m. Sediment samples were collected from four sub-lakes of East Lake in September 16–17, 2009. For every sub-lake, three or four sediment samples including one sample from the center of the sub-lake were collected and stored in sterile plastic bags at 4 °C for DNA extraction and environmental and nutrient parameter analysis, and then stored at −80 °C (Fig. 1).
Temperatures of bottom water were measured (three replicated readings) with reversing thermometers in situ. The pH value of sediment porewater was measured by a Mettler FE20 pH meter (Mettler-Toledo, Shanghai, China). The chlorophyll a (Chl-a) content and the chemical oxygen demand (COD) of the bottom water were measured by spectrophotometric method [32] and potassium dichromate oxidation method, respectively. The total organic carbon (TOC) of the sediment was determined by potassium dichromate oxidation spectrophotometry [29], and the total nitrogen (TN) of the sediment was determined by Kjeldahl method. Porewater dissolved nitrogen including nitrate (NO3–N), nitrite (NO2–N), and ammonia (NH4–N) were measured with nitrate–nitrogen ultraviolet spectrophotometer [7], Griess reagent colorimetric method [24], and Nessler’s reagent spectrophotometry, respectively. The total phosphorus (TP) in the sediment was determined by ammonium metamolybdate spectrophotometric method (Lachat method).
Sediment DNA Extraction, nirS Gene Amplification, and Clone Library Construction
DNA was extracted from 0.5 g sediments using a PowerSoil DNA isolation kit (Mo Bio Laboratories, CA, USA). The quality and quantity of DNA were checked with a Nanodrop device (Nanodrop, DE, USA). The primer set nirS1F (5′-CCTAYTGGCCGCCRCART-3′) and nirS6R (5′-CGTTGAACTTRCCGGT-3′) [3] were used to amplify the nirS gene fragments (∼890 bp) in an Eppendorf thermal cycler. The amplification condition was an initial denaturation at 94 °C for 5 min; 10 cycles of 94 °C for 50 s, 53 °C for 50 s with 0.5 °C decrease per cycle, and 72 °C for 50 s; another 25 cycles at 50 °C annealing temperature; and a final extension of 6 min at 72 °C. PCR products from three reactions were pooled together to minimize the bias caused by PCR and then purified by a gel extraction kit (Omega Bio-Tek, Doraville, GA, USA). The purified PCR products were then cloned into pMD18-T simple vector (Takara, Dalian, China). White colonies were randomly picked and cultured in the 96-well deep-well plate containing LB broth (100 μg/ml ampicillin). Two clone libraries were constructed for each sub-lake. The plasmid DNA (about 192 clones) were extracted by N96 Plasmid Kit (Tiangen Biotech, Beijing, China), checked by agarose gel, and stored at −80 °C for use.
RFLP Analysis and Sequencing of nirS Gene Clone Library
The nirS gene in every clone was re-amplified by using the above PCR conditions. Restriction endonuclease Msp I was used to digest an aliquot of 10 μl PCR product. The DNA fragments were separated electrophoretically on a 15 % polyacrylamide gel at 110 mV for approximately 1 h. After staining with ethidium bromide, the gel was photographed under UV illumination with a Kodak Gel Logic 100 gel image system (Eastman Kodak, Rochester, NY, USA) to store their fingerprint of every clone. For each clone library, the representative clone of every type of RFLP profile was selected for nirS gene sequencing by an ABI 3730 automatic sequencer (Applied BioSystems, Foster City, CA, USA) with M13F and M13R primers. DNA sequences were translated into conceptual protein sequences by DNAMAN software (Lynnon, QC, Canada). The nirS gene sequences obtained from this study was deposited in GenBank with the accession numbers JX852438–JX852569.
Clustering, Diversity, and Phylogeny Analyses
The nirS gene RFLP fingerprints of every clone were converted into a two-dimensional binary matrix through a binary scoring system (1 for the presence of a band and 0 for the absence), and then the similarity between every nirS clone was evaluated by a simple matching (SM) coefficient. A dendrogram was constructed from the distance matrix by the means of UPGMA algorithm using the NTSYS software package (Applied Biostatistic, Setauket, NY, USA).
The amino acid sequences of NirS translated from the nucleotides sequence were aligned and the phylogenetic analysis was performed using the PHYLIP package (http://www.phylip.com/). To compare the NirS-type denitrifier communities among different environments, the community similarity among nirS clones was determined by using weighted UniFrac environmental clustering and principal coordinates analyses (PCoA) [22, 23]. The reference sequences were extracted from GenBank with the accession number Pearl River sediment (JN016541–JN016591), Pearl River Estuary (HQ007353–HQ007643), temperate forest soil (AB456867–AB456954), and constructed wetland sediment (EF558372–EF558536). The correlation between the bacterial communities and environmental parameters was evaluated by canonical correspondence analysis (CCA) using the software Canoco (version 4.5; Microcomputer Power, USA) [30].
Results
Environmental Parameters
Environmental parameters in association with sediment samples from the sites of East Lake, Wuhan city were analyzed (Table 1). The eutrophication of the lake was evident at all sites by high COD value and high concentration of Chl-a, nitrogen, phosphate, and organic carbon in bottom water and sediments. Among these four sub-lakes, Tuan Lake was the least eutrophic, while sub-lake Miao Lake was hypertrophic.
RFLP Clustering of the nirS Clones
Nearly the entire 890-bp length of nirS fragments were amplified from the sediment samples to construct clone libraries of the four sub-lakes. The dendrogram of every sub-lake was depicted based on the RFLP fingerprints generated by digesting the amplified nirS fragment of every clone with endonuclease Msp I (Fig. 2). A complete dendrogram containing all the representative RFLP patterns of the four sub-lakes was also generated from the combined RFLP patterns using UPGMA algorithm (Fig. 3). As shown by these dendrograms, all nirS clones were roughly divided into three clusters. Cluster I accounted for 65.7 % of the whole community, while clusters II and III accounted for 29.8 % and 4.5 % of the clones, respectively. Cluster I consisted of clones from four sub-lakes, Tuan Lake (31 %), Tang Ling Lake (28 %), Guo Zheng Lake (30 %), and Miao Lake (11 %). In cluster II, the majority of this cluster consisted of clones from Miao Lake (52 %). The clones from Guo Zhen Lake were 21 % and the clones from the other two sub-lakes were less than 15 %. A similar phenomenon was found in cluster III; most of the clones in this cluster come from Miao Lake, and no clone was found coming from Guo Zheng Lake (Figs. 2 and 3).
Dendrograms generated from the RFLP fingerprints of nirS gene from four sub-lakes of East Lake by UPGMA method. Parenthesized are the numbers of clones having the same RFLP patterns. Pie charts show the ratio of cluster I, cluster II, and cluster III in every clone library. a Tuan Lake (TL); b Tang Ling Lake (TLL); c Guo Zheng Lake (GZL); d Miao Lake (ML)
A more detailed view of the dendrogram of each sub-lake found the nirS clones from cluster I were the majorities of communities from sub-lake Tang Ling Lake (74 %), Guo Zheng Lake (76 %), and Tuan Lake (82 %), and could be further divided into several sub-clusters in every sub-lake. While in Miao Lake, cluster II became the majority of the community (57 %), and cluster III, which was absent from Guo Zheng Lake, also accounted for 17 % of the clones in this site.
Phylogeny and Community Classification
A neighbor-joining tree was constructed based on the evolutionary distance between the amino acid sequences of nirS genes (Fig. 2). Coincident with the clustering analysis of RFLP fingerprints, all the NirS clone sequences were classified into three clusters. Cluster I (263 clones, 66 % of the total) was dominant in communities from three sub-lakes Tuan Lake, Tang Ling Lake, and Guo Zheng Lake with the ratio ranging from 74 % to 82 % (Fig. 3). This cluster could be further divided into three sub-clusters: IA, IB, and IC. Sequences in this cluster were related to the reference sequences originated from a broad range of environments such as terrestrial and greenhouse soil, constructed wetlands, and river and coastal sediments. Cluster II accounted for 29 % of the whole NirS communities, mainly consisting of clones from Miao Lake (52 %). This cluster could be further divided into two sub-clusters (II-A and II-B). The reference sequences of this cluster mainly come from river sediment, rice paddy soil, and constructed wetland sediment. Considering that Azospirillum brasilense, the pure culture reference of II-A, phylogenetically belongs to alpha-proteobacteria, and Azoarcus evansii, Alcaligenes faecalis, Thauera terpenica, and Thauera aromatica of II-B belong to beta-proteobacteria, we can speculated that sub-cluster II-A and II-B might be phylogenetically ascribed into alpha-proteobacteria and beta-proteobacteria, respectively. Cluster III was the minority among NirS-type community. Most clones of this cluster come from Miao Lake. Phylogeny analysis reflected that this cluster was related to references from the Pearl River and Changjiang Estuary sediment environment. The identified reference Methylomirabilis oxyfera is a type of bacteria participating in anaerobic methane oxidation during nitrite reduction [10]. This reflected that cluster III may be phylogenetically divergent from the main nitrite reduction communities and represent a cluster with special physiological and biochemical function and ecological adaptability. By comparing our sequence data with the references from other environments, we also found that this environment harbors unique denitrifying groups. As shown by the phylogeny tree (Fig. 4), several sub-clusters were found to specifically exist in this ecological environment without counterpart in other environments.
We comparatively analyzed the NirS-type denitrifier community in East Lake with those in similar environments by weighted PCoA analysis. Four regions with distinct environments, the constructed wetland, river sediment, forest soil, and Pearl River estuary, were included (Fig. 5). As shown in Fig. 5, our clones were closely related to those derived from the constructed wetland, and distantly divergent from the river sediment, forest soil, and estuary sediment communities, although the NirS-type communities in lake sediment may have a terrestrial origin (Fig. 5). Interestingly, the community from the eutrophic sub-lake Miao Lake was divergent from all the reference communities and clusters alone, which implied that nirS-type communities in this hypertrophic lake might contain unique denitrifying communities.
PCoA plots showing the similarity of NirS-type denitrifier communities from different geographic locations based on weighted UniFrac analysis. The plot was drawn basing on the first two principal coordinate axes, and the distribution of the bacterial communities in response to these axes is shown. EL East Lake, WL wetland sediment, PR Pearl River sediment, PRE Pearl River Estuary, TMF temperate mixed forest
Environmental Effect on the Community Classification of the NirS-Type Bacteria
CCA analysis was conducted to examine the NirS-type bacterial communities in response to the environmental variables at every site (Fig. 6). As shown in Table 1, Tuan Lake, Tang Lin Lake, and Guo Zheng Lake with relatively lower nutrient content were distributed in the same negative region, while the seriously eutrophic Miao Lake characterized by extremely high Chl-a, NH4–N, TN, TOC, and TP concentration and high COD value was distributed in the positive region by CCA1 (Fig. 6). The spatial distribution of cluster I, the majority in mild and moderately eutrophic sites with a relatively high ratio (74 % to 82 %) close to the center of the CCA plot, showed weak negative correlation (r value < 0.3) with the environment parameters Chl-a, NH4–N, TN, TP, TOC, and COD, but positively correlated with TOC/TN. Cluster II and cluster III had a positive correlation with the nutrient factors (r value ∼0.6), reflecting their adaptability in increasingly eutrophic environments.
Discussion
Due to rapid industrialization and urbanization, the East Lake in Wuhan, China, which has been a drinking water source, experienced intensified anthropogenic eutrophication. As reflected by the measured environmental parameters, this eutrophic site is similar with the constructed wetland, set up as a tertiary treatment of a wastewater treatment plant in Spain [27]. This site is ecologically divergent from the oligotrophic open ocean [6] and the polluted Pearl River sediment in China [17]. According to the description by Huang et al. [17], Pearl River has a high amount of nitrogen nutrient due to the injection of industry wastewater [17], while the East Lake sediment has a high value of TOC. We also noticed that the measured environmental parameters indicate that the sub-lake Miao Lake is suffering serious eutrophication. The nutrient levels in Miao Lake not only are far higher (∼20-fold) than other moderately eutrophic sub-lakes (Table 1) but also higher than the forest soil, and river and constructed wetland sediments [17, 20, 27]. In fact, this site is the catchment of sewage infusion. Because of the narrow area and shallow water, the water exchange of this sub-lake with neighboring Guo Zheng Lake is slow, the organic pollutants injected by sewage are mainly retained in the lake, and the organic particles are prone to precipitate into the bottom of lake to enrich the nutrient of sediment (Fig. 1). Although the content of NO2–N (0.044 mg/l) in Miao Lake is higher than other sub-lakes, this difference is not so distinguished as other nutrient contents (Table 1). We think that this is mainly because NO2–N is an unstable intermediate product in nitrification and denitrification process and subject to rapid degradation in surface sediment. Moreover, during the handing process of sediment samples, NO2–N can also be oxidized into NO3–N [25].
Two classes of enzymes participate in the reduction of nitrite. A nirS gene encoded cytochrome cd 1-containing nitrite reductase (cdNIR) and a nirK gene encoded copper containing nitrite reductases (CuNIR). Both nirS and nirK are useful molecular markers to resolve the diversity and community composition of denitrifying microbes [2, 13, 16]. As previously reported, nirS and nirK reflected different denitrifier communities [19, 31]. In our work, both nirS and nirK genes were checked initially in all samples. We could amplify nirS gene from all samples with high quality, but unfortunately, nirK could not be efficiently amplified from all the samples, although we have tried several pairs of primers such as nirK1F/nirK5R [3] and F1aCu/R3Cu [31]. This makes the comparative analysis the community composition between every site impossible. Considering that nirS gene is more widely used to resolve the diversity and community structure of denitrifiers in different ecological sites, especially those with similar ecological environments to East Lake [17, 20, 27], we selected nirS gene as a functional marker gene to characterize the diversity and community composition of denitrifier in East Lake sediments.
In our study, both nirS gene RFLP and sequencing analyses grouped the NirS-type communities in East Lake into three clusters. Cluster I was the dominant cluster (65.7 % of total) in three moderately eutrophic sub-lakes. This cluster was related to the reference sequences from greenhouse soil, river, estuary, or even coastal sediments, indicating that this cluster may have good adaptability and flexibility to make them evolve, adapt, and settle into a broad range of ecological environments. CCA analyses further showed cluster I weakly related to the nutrient concentration (Fig. 6), and it was not selected by increasingly eutrophic environments. Cluster II was the dominant group in Miao Lake. Cluster III also mainly existed in Miao Lake and was even absent from Guo Zheng Lake. In contrast to other moderately eutrophic sub-lakes, Miao Lake was seriously polluted due to sewage injection. As shown by nutrient analysis (Table 1), the nutrient content of this site was significantly higher than other sub-lakes of East Lake, river sediment, or even constructed wetland sediment. Some reference strains of these two clusters come from rice paddy soil and estuary sediment. The nutrient contents of these environments are generally higher than the reference sites in cluster I such as greenhouse soil and coastal sediments. Some reference strains such as A. faecalis and Thauera sp. are generally have adapted to heavily polluted or fecal environments. This reflected that characteristics of cluster II and III are eutrophic or even hypertrophic environment preference. CCA analyses further testified that cluster II and III are positively related to the concentration of soluble NO2–N, NH4–N, TN, TP, TOC, and COD (Fig. 6). This could further prove that clusters II and III are more preferred in heavily polluted environments.
The fast expanding information of nirS sequences has provided a comprehensive measure of the diversity of the community from environmental samples and also facilitated the comparative analysis of NirS-type communities from different ecological regions. By comparing our sequence data with the reference sequences from other environments, several sub-clusters were found to specifically exist in this ecological environment and without counterpart in other environments. When we compare NirS-type community from East Lake with other ecological environments, PCoA analysis revealed that most nirS sequences from the three sub-lakes Tuan, Tang Ling, and Guo Zheng were divergent from the communities from river sediment and estuary, but had their closest matches with the clones originating from constructed wetland sediment. For the significant environmental difference among the freshwater lake, the open ocean, and the polluted river, NirS-type communities from these sites were distinctly divergent. Identified reference strains from eastern South Pacific are those oceanic bacteria such as Ralstonia eutropha, Roseobacter denitrificans, and Pseudomonas fluorescens [6], and the bacteria from Pearl River sediment are Comamonadaceae, Dechloromonas aromatica, Pseudomonas stutzeri, etc. [17]. However, as revealed by PCoA analysis, the communities in East Lake are similar with those in constructed wetland sediment in Spain. A more detailed view of the community structure found that the constructed wetland community could also be divided into three groups, and the ratio of every group was also similar with those in East Lake [27]. According to the description by Ruiz-Rueda et al. [27], this constructed wetland was set up as a tertiary treatment of a wastewater treatment plant located in the vicinity of a highly developed tourist resort in Spain. Routine physical and chemical monitoring of this site revealed it to be a eutrophic wetland. The nutrient environmental similarity of this wetland sediment with three sub-lakes of East Lake may explain the similarity of denitrifier communities between them. On the other hand, the communities from these two sites were divergent from the forest soil, river, and estuary sediment (Fig. 5). Although the NirS-type communities in the river, estuary, and lake sediment originally should have a terrestrial origin, the environment–species interaction made the functional denitrifying bacterial communities conformable to external environmental factors. Thus, the “traits” or “function” of bacterial communities determine their existence and richness. This phenomenon was further evidenced by the communities in Pearl River sediment [17] and Pearl River Estuary sediment. Although these communities may have the same origin, the environmental divergence between river and estuary makes them phylogenetically divergent (Fig. 5).
In summary, NirS-type denitrifier communities from three moderate eutrophication sub-lakes are similar with the community in constructed wetland sediment and divergent from those in forest soil, river, estuary sediment, and open ocean. These finding suggest that similar environmental conditions are inhabited by similar denitrifier communities despite large geographic distances. Communities from the hypertrophic sub-lake Miao Lake are divergent from all the reference communities and cluster uniquely. Our results support the argument that environmental factors (e.g., nutrient content) regulate the composition and distribution of the functional bacterial groups. These implicitly suggest that to maximize denitrification in eutrophic environments, selecting communities from these environments is required.
Reference
Abell GC, Revill AT, Smith C, Bissett AP, Volkman JK, Robert SS (2010) Archaeal ammonia oxidizers and nirS-type denitrifiers dominate sediment nitrifying and denitrifying populations in a subtropical macrotidal estuary. ISME J 4:286–300
Berk BC, Ferguson SJ, Moir JW, Richardson DJ (1995) Enzyme and associated electron transport systems that catalyze the respiratory reduction of nitrogen oxides and oxyanions. Biochim Biophys Acta 1232:97–173
Braker G, Fesefeldt A, Witzel KP (1998) Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl Environ Microbiol 64:3769–3775
Brezonik PL, Lee GF (1968) Denitrification as a nitrogen sink in lake Mendota. Wis Environ Sci Tech 2:120–125
Bruesewitz DA, Hamilton DP, Schipper LA (2011) Denitrification potential in lake sediment increases across a gradient of catchment agriculture. Ecosystem 14:341–352
Castro-González M, Braker G, Farías L, Ulloa O (2005) Communities of nirS-type denitrifiers in the water column of the oxygen minimum zone in the eastern South Pacific. Environ Microbiol 7:1298–1306
Cawse PA (1967) The determination of nitrate in soil solutions by ultraviolet spectrophotometry. Analyst 92:311–315
Chen H (1989) Impact of aquaculture on the ecosystem the Donghu lake, Wuhan. Acta Hydrobiol Sin 13:359–368
Dang H, Wang C, Li J, Li T, Tian F, Jin W, Ding Y, Zhang Z (2009) Diversity and distribution of sediment nirS-encoding bacterial assemblages in response to environmental gradients in the eutrophied Jiaozhou Bay, China. Microb Ecol 58:161–169
Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJ, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJ, Janssen-Megens EM, Francoijs KJ, Stunnenberg H, Weissenbach J, Jetten MS, Strous M (2010) Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548
Focht DD, Chang AC (1975) Nitrification and denitrification processes related to waste water treatment. Adv Appl Microbiol 19:153–186
Gayle B, Boardman G, Sherrard J, Benoit R (1989) Biological denitrification of water. J Environ Eng 115:930–943
Glockner AB, Jüngst A, Zumft WG (1993) Copper-containing nitrite reductase from Pseudomonas aureofaciens is functional in a mutationally cytochrome cd 1-free background (NirS) of Pseudomonas stutzeri. Arch Microbiol 160:18–26
Goering JJ, Dugdale VA (1966) Estimates of the rates of denitrification in a subarctic lake. Limnol Oceanogr 11:113–117
Hasegawa T, Okino T (2004) Seasonal variation of denitrification rate in Lake Suwa sediment. Limnol 5:33–39
Hochstein LI, Tomlinson GA (1988) The enzymes associated with denitrification. Annu Rev Microbiol 42:231–261
Huang S, Chen C, Yang X, Wu Q, Zhang R (2011) Distribution of typical denitrifying functional genes and diversity of the nirS-encoding bacterial community related to environmental characteristics of river sediments. Biogeosciences 8:3041–3051
Jones CM, Hallin S (2010) Ecological and evolutionary factors underlying global and local assembly of denitrifier communities. ISME J 4:633–641
Kandeler E, Deiglmayr K, Tscherko D, Bru D, Philippot L (2006) Abundance of narG, nirS, nirK, and nosZ genes of denitrifying bacteria during primary successions of a glacier foreland. Appl Environ Microbiol 72:5957–5962
Katsuyama C, Kondo N, Suwa Y, Yamagishi T, Itoh M, Ohte N, Kimura H, Nagaosa K, Kato K (2008) Denitrification activity and relevant bacteria revealed by nitrite reductase gene fragments in soil of temperate mixed forest. Microbes Environ 23:337–345
Knowles R (1982) Denitrification. Microbiol Rev 46:43–70
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235
Lozupone CA, Hamady M, Kelley ST, Knight R (2007) Quantitative and qualitative diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73:1576–1585
Nims RW, Darbyshire JF, Saavedra JE, Christodoulou D, Hanbauer I, Cox GW, Grisham MB, Laval F, Cook JA, Krishna MC, Wink DA (1995) Colorimetric methods for the determination of nitric oxide concentration in neutral aqueous solutions. Methods 7:48–54
Plumb RH Jr. Procedures for handling and chemical analysis of sediment and water samples. Environmental Protection Agency/Crops of Engineers Technical Committee on Criteria for Dredged and Fill Material 1981, 5, AD/A103 788
Qiu D, Wu Z, Liu B, Deng J, Fu G, He F (2001) The restoration of aquatic macrophytes for improving water quality in a hypertrophic shallow lake in Hubei Province, China. Ecol Eng 18:147–156
Ruiz-Rueda O, Trias R, Garcia-Gil LJ, Bañeras L (2007) Diversity of the nitrite reductase gene nirS in the sediment of a free-water surface constructed wetland. Int Microbiol 10:253–260
Rysgaard S, Risgaard-Petersen N, Nielsen LP, Revsbech NP (1993) Nitrification and denitrification in lake and estuarine sediments measured by the 15N dilution technique and isotope pairing. Appl Environ Microbiol 59:2093–2098
Schumacher BA, Methods for the determination of total organic carbon (TOC) in soils and sediments. United States Environmental Protection Agency 2002, NCEA-C-1282
ter Braak CJF (1986) Canonical correspondence analysis: a new eigenvector method for multivariate direct gradient analysis. Ecology 67:1167–1179
Throbäck IN, Enwall K, Jarvis A, Hallin S (2004) Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol Ecol 49:401–417
Yentsch CS, Menzel DW (1963) A method for the determination of phytoplankton chlorophyll and phaeophytin by fluorescence. Deep-Sea Res Oceanogr Abs 10:221–231
Zumft WG (1997) Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev: MMBR 61:533–616
Acknowledgments
We are grateful to B. Zhu and X. Chen for their help in sample collection. This work was financially supported by Key Project of Chinese Ministry of Education (No. 212118) and Human Recourse Foundation of Wuhan Polytechnic University. Dr. JK Yang is an incumbent of the Chutian Scholar Program position.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yang, JK., Cheng, ZB., Li, J. et al. Community Composition of NirS-Type Denitrifier in a Shallow Eutrophic Lake. Microb Ecol 66, 796–805 (2013). https://doi.org/10.1007/s00248-013-0265-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-013-0265-5