Abstract
Population growth and climate change demand the constant development of new crop varieties that can produce higher yields, and better organoleptic and nutritional value under adverse biotic, and abiotic conditions. In this sense, traditional breeding and genetic transformation have been used for decades. Nevertheless, the first approach is time consuming endeavor, and is unable to keep up with increasing food demands. On the other hand, genetic transformation is often limited by consumer acceptance. Recent genome editing technologies, such as clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein (CRISPR-Cas9) system allows precise, specific, and low cost edition in a targeted genome region. The wide variety of applications for this technology includes increased yields and nutritional value, stress tolerance, and herbicide resistance. Crops of tropical origin have nutritional and economic importance; therefore, this review will analyze the advances and applications of CRISPR in crops of tropical origin to obtain varieties better adapted to current environmental conditions and market requirements.
Key message
Genome editing technologies, such as CRISPR, allows precise and specific modification of genetic information for the improvement of crops of tropical origin, including rice, maize, tomato, coffee, cacao, and citrus, to produce varieties with resistance or tolerance to biotic and abiotic factors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
According to the Food and Agriculture Organization of the United Nations (FAO), the impact of climate change on crop yield will depend on factors such as temperature, CO2 increases, and precipitation changes (FAO 2016). In this sense, food production systems rely on highly selected cultivars adapted to specific environments and growth conditions. Because of climate change, these cultivars may be grown under conditions for which they are not adapted, making them more vulnerable to different biotic (bacteria, fungi, insects, and viruses), and abiotic stresses (salinity, drought, temperature) (Govindaraj et al. 2015).
The progress achieved in the last decades in biotechnology and molecular biology has increased the available tools for improvement of agricultural crops. Some of these tools include techniques for the introduction of transgenes, directed mutagenesis, and accelerated reproduction techniques, among others (Lusser et al. 2012). Genetic improvement using Agrobacterium tumefaciens or particle gun bombardment has some limitations, such the inability to control the random integration of the transgene in the genome affecting the level, and stability of gene expression (Kohli et al. 2006). More specific, in the case of particle gun bombardment, the transgene copy number can be very high which depends on the amount of DNA delivered into cells. Also, some studies showed that it can cause extreme genome damage in the form of chromosome truncations, large deletions, and partial trisomy (Liu et al. 2019a, b). On the other hand, Agrobacterium-mediated transformation system has host-range limitations and it cannot be used to directly deliver molecules like proteins and RNA (RNP) unlike particle gun bombardment (Mello-Farias and Chaves 2008; Mohammed et al. 2019; Liu et al. 2019a, b).
In recent years, new precision plant biotechnologies (NPBT) such as zinc finger domains (ZFN), effector nucleases similar to transcriptional activators (TALEN), and short palindromic repeats grouped regularly interspaced associated with the Cas9 endonuclease (CRISPR/Cas9) have been developed and allow precise, and specific edition in a targeted genome region. The editing of a genome consists of producing directed, permanent, and inheritable mutations in a specific place of the genome, mediated by DNA repair systems in the cell, with the lowest probability of committing unwanted errors (off-targets) and leaving no foreign DNA sequences.
The newest and most widely used genome editing technology for the study of the function of genes and for the development of mutant lines with enhanced tolerance to biotic and abiotic stresses, herbicide resistance or improved yield is CRISPR/Cas (Petolino 2015; Cao et al. 2016; Khan et al. 2016; Lassoued et al. 2018a, b). This technology differs from ZFN and TALEN in terms of the DNA-binding system. Cas9 can be targeted to a specific genomic sequence by an easily engineered 20 base pair (bp) RNA guide sequence that binds to its DNA target by base-pairing. The wide variety of applications for this technology includes simple non-homologous end joining, homologous recombination, gene replacement, and base editing, among others. The applications and challenges of CRISPR/Cas9 in tropical crops has been recently reviewed (Haque et al. 2018). However, in this review, we will present an in-depth study of genome editing technologies research that has been reported in crops of tropical origin emphasizing in the pre-requisites, limitations, and advantages. Literature from 2013 to mid-2019 was searched using the PubMed database with the keywords “NAME OF THE CROP” and “CRISPR”.
Importance for breeding crops of tropical origin
One of the challenges that crops breeders face is the generation of varieties resistant to the adverse environmental conditions produced by climate change (Pereira 2016). Crops are susceptible to a large set of pathogens and pest, including fungi, bacteria, nematodes, insects, and viruses that cause important economic losses, especially in tropical climates. It is estimated that biotic stress cause losses worldwide up to 35% of the total food production (Bainsla and Meena 2016). As an example, losses in rice due to insects have been estimated in 40% (Oerke and Dehne 2004); and in sorghum the losses been valued over $1079 billion. On the other hand, abiotic stress (drought, salinity, cold, and heat) represent the primarily cause of crop losses worldwide, and yield losses are estimated around 50% of crop production (Ashraf et al. 2008).
This adverse biotic and abiotic factors influence negatively the survival, production and yields of staple food crops threatening food security (Meena et al. 2016). To counteract these losses, plant breeding programs focus the efforts on developing crops with improved yield, field performance and nutritional quality, as well as resistance to biotic and abiotic stresses (Ferrão et al. 2017). For decades, breeding strategies include selection, hybridization, mutation induction using chemical and physical agents, and somaclonal variation. However, advances for breeding crops against biotic and abiotic stresses using conventional strategies has been slow, principally due to the long time necessary to achieve the desired phenotype, and it is limited by the lack of resistance genes in the germoplasms (Bainsla and Meena 2016; Parmar et al. 2017).
Moreover, several crops of tropical origin are an important source of nutrients and carbohydrates for humans. Therefore, efforts have been focused to the enrichment of nutrients and the elimination of toxic elements. Several strategies including conventional and transgenic approaches have been used for the bio fortification of staple crops (Ricachenevsky et al. 2019). As an example, efforts have been made to increase the content of pro-vitamin A in banana (Paul and Qi 2016).
In this sense, modern biotechnology tools, such as tissue culture and genetic engineering, offer an alternative to conventional breeding in order to generate new cultivars with enhanced agronomic and nutritional characteristics (Parmar et al. 2017). In the last decades, transgenic crops, including those of tropical origin, have been developed and genetic modification has been performed to confer resistance against insects, bacteria and fungi diseases, and viruses; as well as tolerance against drought, high temperatures, salinity, and herbicides (Parmar et al. 2017). More recently, the availability of genome editing technologies, genome sequences, efficient tissue culture, and transformation methodologies could remarkably facilitate the breeding of crops of tropical origin.
Overview of CRISPR/Cas9 technology
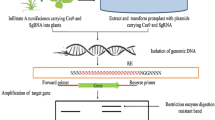
CRISPR/Cas9 originates from the immune response of bacteria and archaea against foreign DNA from viruses and plasmids. When bacteria survive initial infection by a bacteriophage, segments of the phage genome are inserted into the bacterial genome. Then, when the bacteria is infected a second time, CRISPR RNA is synthesized and guides the Cas endonuclease to the phage DNA for its degradation (Johnson et al. 2015). Several CRISPR systems which differ in the diversity of their protein components have been identified in bacteria. In general, the three main CRISPR systems are type I, II, and III, which use the endonucleases Cas3, Cas9, and Cas10, respectively (Chylinski et al. 2014). More information on the systems can be found in the work by Ishino et al. (2018). The CRISPR/Cas technology combined the two RNAs naturally involved in the CRISPR system (tracrRNA and crRNA) in a single artificial chimeric molecule called guide RNA or single guide RNA (sgRNA) (Fig. 1a). The sgRNA is designed in silico to guide the Cas9 to specific sites in the genome of the organism that need to be modified or improved. This results in a double strand break in the DNA (deoxyribonucleic acid). The DNA breakage triggers the Non-Homologous End Joining (NHEJ) repair system (Fig. 1b) which leads to insertions or deletions of small sequences (InDels) adjacent to the excision site (Porteus 2016; Scheben et al. 2016). NHEJ mediated gene inactivation is the simplest way of directed modification, and it is generally used to knockout genes that have a negative influence on a trait of interest in the plant (Bortesi and Fischer 2015). Homology directed repair (HDR) can also be generated when a DNA template with high sequence homology is introduced into the cleaved zones (Sander and Joung 2014). HDR is the repair mechanism used to insert a sequence or repair a mutation in a specific part of a gene (Kamburova et al. 2017; Malzahn et al. 2017).

Source Own elaboration based on Tang and Tang (2017)
Mechanism of genome editing through CRISPR/Cas9 in plants. (A) The cassette expressing Cas9 is driven by the 35S promoter and the guiding RNA is usually driven by the U6 promoter. (B) The CRISPR/Cas9 system is introduced into plant cells by protoplast transformation, particle bombardment or Agrobacterium transformation. Once in the cells, the sgRNA directs the Cas9 to the target site of the genome. Cas9 recognizes the PAM sequence and performs the double DNA chain break. Through the NHEJ repair system, deletions or insertions of bases (InDel) are generated in the target site, on the other hand, by means of the HDR repair system, precise corrections can be made in the DNA or directed sequences can be inserted. (C) Finally, gene editing could be detected by restriction enzymes or by sequencing.
A requirement for the CRISPR endonucleases to adhere to the target site of the DNA, aside from the pairing of the sgRNA to the target sequence, is the presence of a PAM (Protospacer adjacent motif). The PAM is a sequence of 2 to 6 nucleotides, generally found after the joining site of the leader sequence in the DNA. This sequence may vary depending on the type of Cas protein being used. The most commonly used is Cas9 from S. pyogenes, which recognizes the PAM sequence 5′NGG3′ (Endo et al. 2018). More information on the CRISPR/Cas mechanism and structures can be found in the work by Jiang and Doudna (2017).
In plants, the first step involved for using CRISPR, consist in the design and construction of the sgRNA for the specific gene or site where the modification will take place (Fig. 1a). The next step is the introduction of the CRISPR/Cas9 components into the plant cells using particle bombardment or Agrobacterium-mediated transformation (Fig. 1b). Finally, plants are regenerate from the genetically edited cells, tissues, or organs, and the mutations are verified through restriction analysis, PCR (polymerase chain reaction) and sequencing (Fig. 1c) (Kangquan et al. 2017).
The CRISPR/Cas9 system offers some advantages, such as simplicity, accessibility, versatility, and low cost, in comparison with other editing technologies, such as ZNFs (zinc/finger nucleases), and TALENs (transcription activator-like effector nuclease). For example, ZNFs and TALENs technologies recognize their target site by proteins so designing these proteins are far more complicated and expensive that designed a single guide RNA for CRISPR (Farooq et al. 2018). Also CRISPR/Cas9 make possible to target several different genes by multiples sgRNA in a single CRISPR array (Li et al. 2017). Another advantage of the CRISPR system is the ability to excise methylated DNA, which is particularly appropriate for monocotyledons with a high genomic GC content, such as rice (Bortesi and Fischer 2015).
The main drawback of CRISPR/Cas9 is the possibility to generate off-target mutations, which are the product of erroneous pairings of the sgRNA on unwanted sites in the DNA. Studies in Arabidopsis (Zhang et al. 2018a, b), citrus (Jia et al. 2017), rice (Baysal et al. 2016) and tomato (Li et al. 2017) genomes mutated by CRISPR/Cas9 have demonstrated that this system is very specific in plants, and off-target mutations do not happen often. Nevertheless, there are some cases in rice where off-target mutations were detected (Xie and Yang 2013). Therefore, the sequencing of the complete mutant genome is the most reliable way to detect off-target mutations. A less expensive method would be sequencing only the potential off-target regions predicted in silico (Wolt et al. 2016; Puchta 2017). Although problems caused by off-target mutations are not as severe in plants as in humans and animals (Kangquan et al. 2017), there are strategies to minimize their occurrence, like the use of alternatives to SpCas9. For example, SaCas9 is usually more precise due to a longer PAM sequence, and Cas12a uses a different molecular mechanism for DNA targeting, recognition and cutting (Schindele et al. 2018). There are also reports that insertion of the CRISPR system as an RNP (Ribonucleoprotein) reduces off-target mutations (Hahn and Nekrasov 2018). Another limitation is that for the proper recognition of the target site by the sgRNA a PAM sequence is required, this decreases the flexibility of possible targets sites, and also affects the design of sgRNA (Zhou et al. 2018b).
Applications of CRISPR/Cas9 in crops of tropical origin
CRISPR/Cas9 was used for the first time in the model plants Arabidopsis thaliana (Li et al. 2013), Nicotiana benthamiana (Li et al. 2013), and Oryza sativa (Shan et al. 2013) in 2013. Since then, this technology has been widely used as a genetic editing method for several plants (Zhang et al. 2017a, b; Belhaj et al. 2015). A review of 52 studies between 2014 and 2017 revealed that this technology has been mainly focused on rice, tomato, citrus, tobacco, maize, and wheat for tolerance to abiotic (drought, salinity), and biotic (bacteria, fungus, and viruses) factors, yield improvement, and bio-fortification (Ricroch et al. 2017) (Fig. 2). Nevertheless, in the last few years, applications of this technology have been extended to other crops such as cacao, maize, sorghum, and sweet orange (Table 1). Since 2013, the number of publications where CRISPR/Cas9 has been used in crops of tropical origin has increased especially in rice, tomatoes, and maize according to the studies present in the PubMed database (Fig. 3).
Applications of the CRISPR/Cas9 system for crop improvement. Based on the 52 studies reported between 2014 and 2017 by Ricroch et al. (2017) and 15 studies published in 2018–2019 which are mentioned in this review
The first approaches using CRISPR/Cas9 in crops of tropical origin aimed to evaluate the efficiency and to experiment with this technology. Therefore, many studies have used marker genes such as phytoene desaturase (PDS) as proof-of-concept for genome editing. Phytoene desaturase gene encodes an enzyme that limits carotenoid synthesis and converts phytoene into colored ζ-carotene in a two-step desaturation reaction. Successful knock out of this gene leads to albino phenotypes and allow the screening of mutants (Wang et al. 2009). This has been applied in banana (Hu et al. 2017a, b; Kaur et al. 2018), cassava (Odipio et al. 2017), citrus (Jia and Wang 2014; Zhang et al. 2017b; LeBlanc et al. 2018), coffee (Breitler et al. 2018), maize (Svitashev et al. 2016), rice (Miao et al. 2013; Liang et al. 2018), sorghum (Jiang et al. 2013), tomato (Pan et al. 2016), and watermelon (Tian et al. 2017) (Table 1). Recently, the green fluorescent protein (EGFP) has been used as an indicator for the screening of tobacco and grapes mutants generated by CRISPR/Cas9 as a result of the activation of the non-functional EGFP protein (Ren et al. 2019). This method could be applied for the identification of large populations of mutants of crops of tropical origin derived by new breeding technologies without the necessity of isolating DNA or generating mutants with an albino phenotype.
Banana
Musa spp. is one of the most important commercial commodities in tropical and subtropical developing countries. It is cultivated over 130 countries, and is a source of carbohydrates for millions of people worldwide (FAOSTAT 2017). Conventional banana breeding is limited by factors, such as sterility, poliploidy, and asexual propagation (Rout et al. 2008). Therefore, number of biotechnological tools, including tissue culture, and genetic engineering protocols have been developed to complement traditional breeding (Rout et al. 2008). Moreover, the sequencing of the banana genome (D’Hont et al. 2012) provides access and information about each gene making it possible to apply genome editing technologies for improving it (Table 2).
Among the first studies of CRISPR/Cas9, Hu et al. (2017a, b) modified the gene MaPDS in embryogenic cell suspensions of Boxi cultivar via A. tumefaciens. As a result, approximately 55% of the genetically modified lines displayed an albino and/or chimeric phenotype. Similarly, Kaur et al. (2018) edited the MaPDS gene in the Rasthali cultivar transforming cells suspensions via A. tumefaciens. In this case, most of the resulting mutants displayed a complete albino phenotype and did not survive the rooting phase. Also, some mutants displayed chimeric phenotypes (green and albino). Moreover, Naim et al. (2018) edited the PDS gene in Cavendish banana cv. Williams and genotyping of 19 independent events showed a 100% PDS mutation rate like insertions or deletions. These three studies demonstrated the efficiency of CRISPR/Cas9 for genome editing of endogenous genes in banana and opens the possibility to apply this system for the targeting of genes for the improvement of important agronomic characteristics. In this sense, CRISPR/Cas9 was used to confer resistant to Banana Streak Virus (eBSV) by editing the virus sequences in the B genome in a plantain with AAB genome called “Gonja Manjaya”. For this, three sgRNAs were designed to target the most conserved parts of the BSV strain Obino l’Ewai (BSOLV). The mutation efficiency was 85% at the target sites. After 1 month, six of eight edited lines remained asymptomatic and two lines showed moderate symptoms compared to wild type control plants (Tripathi et al. 2019). This study shows that targeted mutagenesis by CRISPR/Cas could be used for the rapid generation of disease-resistant plants. A similar approach is expected in the near future in banana against Fusarium oxysporum Tropical race 4 or TR4 (“Panama disease”). This is an aggressive and fast-spreading disease that has already affected Cavendish banana in Africa, Asia, and Central America, endangering this variety which is the most important worldwide (Pérez et al. 2014). For example, genes like MaAPS1 and MaAPL could be edited to generate resistance not only to abiotic stress (cold, and salinity), but also to TR4 (Haque et al. 2018).
Moreover, semi dwarf plants were obtained using CRISPR/Cas9 by editing the MaGA20ox2 gene in Musa acuminate (AAA group) (Shao et al. 2019). The Ga20ox2 (GA20 oxidase) is an important enzyme in the biosynthesis of gibberellin. It has been proven that mutation of this gene (383-base-pair deletion) has caused a semi-dwarf phenotype in rice (Sasaki et al. 2002). In the banana study, five GA20ox2 homologous genes were identified in the cultivar “Gros Michel”, and two sgRNAs were designed targeting exon 2 of each gene. At the end, seven mutant lines with semi-dwarf phenotype were obtained showing short insertions in the target regions (Shao et al. 2019). This study is important because generating dwarf or semi-dwarf fruit plants could have agronomical advantages like an easier maintenance, and harvesting. However, it has to be considered that inducing this phenotype does not affect yield or generate adverse effects in plants.
Cacao
Theobroma cacao L. is a diploid tree endemic to tropical jungles in South America (Argout et al. 2011). The seeds are used as a raw material for the chocolate industry, which is estimated at 90 billion dollars per year and it is grown by approximately 6 million small holders around the world (Anga 2014; Wickramasuriya and Dunwell 2018). Recent advances in tissue culture, and genetic transformation applications have allowed cacao improvement using modern biotechnological tools (Wickramasuriya and Dunwell 2018) (Table 2). A previous study using microRNA targeting the TcNPR3 (Non-Expressor of Pathogenesis-Related 3) showed that knockout of this gene induce resistance to infection with Phytophthora capsici (Shi et al. 2013). In this sense, CRISPR/Cas9 was used to knockout the TcNPR3 gene in this crop. The system was first tested in leaves and confirmed the resistance to P. tropicalis and an elevated expression of downstream defense genes. Afterward, stable transformation and edition of somatic embryos was performed to analyze the resistance in the whole plant (Fister et al. 2018). The availability of the draft genome of Theobroma cacao (Argout et al. 2011; Motamayor et al. 2002) could make possible in a near the application of genome editing technologies for the modification of genes associated with biotic, and abiotic stress, and improvement of bean quality.
Cassava
Manihot esculenta is a tropical crop of high economic importance and nutritional value as a source of carbohydrates for human consumption. Moreover, it is a staple food for 800 million people around the world and it is cultivated mainly by small farmers (Fondong and Rey 2018). Therefore, genetic improvement of this crop is very important. Nevertheless, conventional breeding and genetic engineering are very challenging (Fondong and Rey 2018). In this sense, genome editing technologies offer an opportunity to complement and accelerate conventional and modern (tissue culture and genetic transformation) breeding (Table 2) (Chavarriaga-Aguirre et al. 2016; Fondong and Rey 2018).
As a proof-of-concept, MePDS-1 and MePDS-2 genes were edited by delivering the CRISPR/Cas9 components via A. tumefaciens in friable embryogenic callus of cultivar 60444 and TME 204. The altered phenotype (albino or partially albino) was observed in 97.1% and 98.9% of the lines obtained for MePDS-1 and MePDS-2, respectively. Sequence analysis showed that 100% of the examined lines carried mutations on the target site, and found out that plants with complete albinism had homozygous mutations while the chimeric plants had heterozygous mutations (Odipio et al. 2017). This study and the availability of the cassava draft genome (Prochnik et al. 2012) open the possibility to use the CRISPR/Cas9 technology to confer virus resistance, and drought tolerance, increase the protein and amylose content, and reduce the content of hydrogen cyanide (Fondong and Rey 2018). For example, CRISPR/Cas9 has already been used to generate cassava plants resistant to Potyviridae family viruses that cause Cassava Brown Streak Disease (CBSD) (Gomez et al. 2018). For this, the target genes were two isoforms of the IF4E gene (nCBP1 and nCBP-2). Plants with both isoforms mutated showed a lower incidence of symptoms or delayed symptom onset, and a lower incidence of root necrosis (Gomez et al. 2018).
Additionally, glyphosate resistance was achieved by editing the endogenous promoter and first two exons of the gene EPSPS in embryogenic calli of TME 419 cassava cultivars using Agrobacterium tumefaciens. Homologous recombination (HR) or non-homologous end‐joining repair mechanisms were employed for correcting the 3.2‐kb deletion cause in the locus. As a result, six edited lines showed glyphosate resistance. This study demonstrated that the highest recovery of edited events was obtained using the HR mechanisms and opens the possibility for further development of glyphosate traits in other crops (Hummel et al. 2018).
On the other hand, AC2 and AC3 viral genes were also targeted using CRISPR/Cas9 in order to confer resistance to geminivirus (Mehta et al. 2019). In the past, resistance to geminivirus was achieved targeting AC1 viral gene through RNAi (Vanderschuren et al. 2009). Nevertheless, the results in the CRISPR/Cas9 study showed that during the inoculation in the greenhouse an effective resistance was not achieved. Moreover, it was shown that some virus genomes develop a conserved mutation conferring resistance to the excision of CRISPR-Cas9 (Mehta et al. 2019). This research is important because it shows that viruses could evolve to escape from antiviral CRISPR transgenics plants.
Citrus
Citrus fruits are economically important, especially in tropical zones (Kumar et al. 2018). Between 2012 and 2013 an average of 119.164 thousand tons of citrus were produced worldwide (FAO 2015). Various protocols for organogenesis and somatic embryogenesis, as well as, transformation techniques, including Agrobacterium-mediated transformation, particle gun bombardment and PEG-mediated transformation has been successfully developed and applied for the breeding of some species of this genus (Singh and Rajam 2009). Also, advances in sequencing of sweet orange (Citrus sinensis) (Xu et al. 2012) allow the application of genome editing technologies for the improvement of this crop.
The first report of CRISPR/Cas9 in citrus was made in sweet orange (Citrus sinensis, Valencia cultivar) where the CsPDS gene was edited by infiltrating leaves with A. tumefaciens. Mutation rates of 3.2% to 3.9% were obtained, with no evidence of off-target sites. In this study, 11 types of InDels were obtained, among which deletions of 1 bp to 12 bp were predominant (Jia and Wang 2014).
Moreover, the mutation efficacy of CRISPR/Cas9 has been improved by editing the CsPDS gene using the YAO promoter (promoter of the active meristem of the apical apex) instead of CaMV35S for Cas9 expression. As a result, 65% of plants displayed an altered phenotype, of which 85% were albino (Zhang et al. 2017b). Another study showed that thermal stress (37 °C) promoted higher levels of CRISPR/Cas9 mutations in the CsPDS gene and most of the transformed plants with CRISPR plasmid and subjected to thermal stress showed the expected albino phenotype. This study concludes that temperatures closer to the optimal growth temperature of S. pyogenes (37 °C) favored higher Cas9 activity (LeBlanc et al. 2018). Despite the promising results of this study, thermal stress could only work in species or varieties with high tolerance to heat stress otherwise it can generate physiological problems, such as foliar senescence and abscission, inhibition of shoot and root growth, reduce of the photosynthesis rate (Nievola et al. 2017).
On the other hand, several studies demonstrated the feasibility to use CRISPR/Cas9 for inducing biotic resistance plants in the Citrus genus. In this sense, this system was used to modify the CsLOB1 gene, which is responsible for susceptibility to Citrus Canker disease produced by Xanthomonas axonopodis pv citri bacteria, in Duncan grapefruit (Citrus x paradise). As a result, several genetically modified lines were obtained with a mutation efficiency of 23.80% to 89.36%. In some of these lines, plants inoculated with the pathogen displayed delayed and milder symptoms of the disease than the wild type plants (Jia et al. 2017, 2019). In Citrus sinensis the promoter of CsLOB1 gene was also edited by using five sgRNAs to modify EBEPthA4, which is an effector binding element to the promoter of the CsLOB1 gene in both alleles and activates its expression. Mutation rates of 11.5% to 64.7% were obtained. In four mutant lines, resistance to citrus canker disease was higher than in wild type plants and two of these lines showed no symptoms of the disease (Peng et al. 2017). The PthA4 effector binding elements (EBEs) in the CsLOB1 promoter was also edited in Duncan grapefruit but edited transgenic plants developed canker symptoms similarly as wild type and the authors concluded that mutation in the promoters of both alleles of CsLOB1 is probably needed to generate citrus canker-resistant plants (Jia et al. 2015).
Coffee
Coffee (Coffea spp.) is a crop of great relevance worldwide for its high value as a beverage (Alemayehu 2017) and for being one of the most important agricultural products, occupying the second place in international trade after oil (Labouisse et al. 2008). This crop is mainly cultivated in tropical and subtropical regions of the world deriving directly or indirectly the income of more than 125 million people (Tran et al. 2016). Previously, modern biotechnology tools (such as organogenesis, somatic embryogenesis, and genetic engineering) have been used to confer insect and herbicide resistance, and reduce the caffeine content (Ribas et al. 2006).
In a first attempt, Breitler et al. (2018) designed an algorithm, called CRIP (Coffee gRNA Identification Program), to detect target sites in the diploid C. canephora genome for CRISPR/Cas9 editing. As a result, 8.145.748 possible sgRNAs were found for C. canephora genome. These authors edited the CcPDS gene in embryogenic calluses via A. tumefaciens mediated transformation. Sequencing revealed that 30% of the modified plants displayed a mutation; however, none of the plants presented a completely albino phenotype and some anomalies like leaf size (very small) and pigmentation (yellowish, chlorosis or anthocyanins) were also observed in mutated plants.
Previously, the genetic transformation of coffee has been employed as a tool for the validation of gene function, and for the production of transgenic crops with agronomical important characteristics (Mishra and Slater 2012). Nevertheless, the application of the genome editing technologies in this crop requires knowledge of the specific target genes that will be modified. In this sense, the recent release of the Arabica (C. arabica L.) (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Carabica_er), and robusta (Coffea canephora) (Denoeud et al. 2014) genomes opens the possibility to modify genes related with cup quality, resistance to biotic and abiotic factors, or production.
Cucumber
Cucumis sativus L. is one popular member of the Cucurbitaceae family with high economic value (Huang et al. 2009; Maurya et al. 2015). Biotechnological methods utilized in the genus Cucumis for genetic improvement include embryo rescue, in vitro pollination, protoplast isolation, culture and fusion, organogenesis, somatic embryogenesis, Agrobacterium and particle gun-mediated transformation, and electroporation (Navrátilová et al. 2011; Wang et al. 2015). Moreover, the genome of Cucumis sativus var. sativus L. has been sequenced (Huang et al. 2009), providing useful information for improving cultivars with biotic and abiotic resistance and horticultural traits (Wang et al. 2015). Although significant progress has been made for genetic transformation of cucumber, it is still necessary to increase regeneration and transformation efficiency and to explore other methods that allow genetic improvement without modifying desirable characteristics of the cucumber plants (Wang et al. 2015). In this sense, genome editing technologies could be considered as an alternative for genetic improvement of this crop. In a first attempt, CRISPR/Cas9 technology was used to generate virus resistant cucumber plants by editing the eIF4E gene (Chandrasekaran et al. 2016). This gene encodes an essential translation factor essential for the life cycle of the Potyviridae (Piron et al. 2010). As a result, the homozygous edited lines showed resistance to different viruses, including cucumber vein-yellowing virus (CVYV); whereas, the heterozygous mutants were susceptible just as the unedited control plants (Chandrasekaran et al. 2016). An attempt has been made to improve the efficiency of CRISPR/Cas9 in this crop by using endogenous U6 protomers for sgRNA expression. For this, the CsWIP1, CsVFB1, CsMLO8, and CsGAD1 genes were targeted for mutation (Hu et al. 2017b). As a result the transformation efficiency was 1% which was better than the previous study made by Chandrasekaran et al. (2016).
Maize
Zea mays is an economical important crop used as food and feed as well as for bioenergy production, and industrial materials (Guo et al. 2018). Advances in in vitro regeneration and genetic transformation protocols, as well as the availability of the genome sequence have allowed the application of modern biotechnology for the genetic improvement of this crop (Table 2).
In order to create plants with better nutritional values, the gene ZmIPK (phosphoinositide 3-kinase) was edited by CRISPR/Cas9 with the purpose of reducing the phytic acid concentration in seeds (Liang et al. 2014). Phytic acid is an anti-nutrient as it reduces the bioavailability of important micronutrients; also it is a compound that cannot be digested and can also cause environmental pollution (Perera et al. 2018). Two sgRNAs were designed to edit the ZmIPK gene and mutation efficacy of 16.4% and 19.1% was achieved, respectively. Unfortunately, this study does not report whether the edited plants had lower levels of PA in the seeds (Liang et al. 2014).
Svitashev et al. (2016) successfully introduced the CRISPR/Cas9 system as ribonucleoproteins (RNP) into maize tissue via particle bombardment for editing the genes liguleless1 (LIG), acetolactate synthase (ALS2), and two male fertility genes (MS26 and MS45). These genes were chosen in order to compare the results with a previous study made by the same authors where they used CRISPR plasmids instead of RNPs (Svitashev et al. 2015). As a result, these authors found fewer off-target mutations when RNPs were used instead of plasmids.
Also in maize, Char et al. (2016) edited two polymorphic Argonaute (Ago) genes, ZmAgo18a and ZmAgo18b, implicated in interference-RNA biogenesis in anthers. The gene for dihydroflavonol 4-reductase (a1) enzyme and its homolog (a4) were also modified at two different sites, following the same procedure used for the AGO genes. In order to do that, they designed two sgRNA for each gene. Mutations were observed in one or two alleles of the Ago genes in the edited lines at percentages higher than 70%. The mutation frequency of the a1 and a4 genes was higher than 15% (Char et al. 2016).
For abiotic stress tolerance, the CRISPR/Cas9 system was used to insert the constitutive GOS2 promoter in the 5′ UTR end of the ARGOS8 gene through HDR. For this, the CRISPR/Cas9 system and DNA repair template (GOS2 promoter) were introduced into immature maize embryos by particle gun bombardment. The overexpression of this gene reduced ethylene sensitivity and increased grain yield in drought conditions in comparison with wild type plants (Shi et al. 2017). This study is a clear example of how CRISPR/Cas9 can be used in other ways besides inducing gene knockout and mutations.
Regarding herbicide resistance, specific amino acid in the ALS2 gene (proline to serine at position 165) was changed using CRISPR/Cas9 to generate resistance to the herbicide chlorsulfuron. The difference with a similar study in rice (Sun et al. 2016) was that these authors introduced the CRISPR/Cas9 system as a ribonucleoprotein (RNP) together with a DNA template of 127 nt for HDR repair system. As a result, all edited plants with this amino acid change were chlorsulfuron-resistant (Svitashev et al. 2016).
A more recent application is the generation of a maize haploid inducer line carrying a CRISPR/Cas9 cassette for agronomic traits. In this study, the authors introduced a cassette for editing the ZmLG1 gene (liguleless1), which loss-of function mutants dramatically reduces leaf angle. Also, they introduced a cassette for the UB2 gene, which is involved in regulating tassel branch number. After getting the haploid modified lines with CRISPR/Cas9 cassettes, they were crossed with the B73 line. As a result, the next generation manifested mutant phenotypes and presented the expected editions (Wang et al. 2019a, b, c, d).
The CRISPR/Cas9 system has also been used for studying the fertility restoration of maize CMS-C (Type C Cytoplasmic male sterility) (Jaqueth et al. 2019), the role of ZmACD6 for Ustilago maydis resistance (Zhang et al. 2019), for the characterization of Phytochrome-Interacting Factor (PIF) genes Zmpif3, Zmpif4, and Zmpif5 (Wu et al. 2019), for the induction of haploid plants thought the mutation of the ZmDMP gene (Zhong et al. 2019), for the study of ZmbZIP22 transcription factor (Li et al. 2018b) and ZmCCT9 gene (Huang et al. 2018), among others.
Rice
Oryza sativa L. is one of the most important crops in tropical and subtropical humid regions around the world. This crop feeds approximately 50% of the world population (Wang et al. 2016; Kumar and Tuteja 2012). In the last decades, in vitro regeneration and genetic transformation protocols, as well as the development of genomic resources have allowed the application of modern biotechnology for the genetic improvement of this crop (Table 2).
Since first applications of the CRISPR/Cas9 technology in rice (Miao et al. 2013; Shan et al. 2013; Xie and Yang 2013), the number of studies in this crop keep growing at a fascinating rate (Fig. 3). Here we present only some applications of this genome editing technology and more detailed review can be consulted in Romero and Gatica-Arias (2019).
A concept proof research on the use of CRISPR/Cas9 in rice was reported by Miao et al. (2013). These authors used the modified GUS (β-Glucuronidase) reporter gene in a transgenic line to evaluate whether the CRISPR/Cas9 system was capable of generating a DNA double-strand break (DBS) in plant cells. As a result, expression of the GUS gene was achieved by excising repeated regions of the GUS gene either using the chimeric sgRNA or the two RNA molecules crRNA-tracrARN. Moreover, in the same study, the oxygenase gene in chlorophyll a (CAO1), whose mutants have a lighter green color, and the LAZY gene, which regulates shoot gravitropism were edited. Mutations in T1 generation were observed in 83.3% of the edited lines for CAO1 gene and 91.6% for LAZY gene (Miao et al. 2013).
In one of the first studies in rice, the OsMPK5 (mitogen-activated protein kinase) gene has been edited in protoplasts (Xie and Yang 2013). This gene codes for a kinase protein activated by the stress response (Jagodzik et al. 2018). Using three sgRNAs for different sites in the gene, the mutations average was 3%-8% on the target sites. Off-target mutations were detected on a site in chromosome 12 with an incidence of 1.6% (Xie and Yang 2013).
The CRISPR/Cas9 system has been used to generate new rice varieties with tolerance to abiotic stress (Cordones et al. 2017). For example, CRISPR/Cas9 system has been used as a strategy to develop new varieties with reduced cesium (Cs+) accumulation. Cesium is a toxic metal present in polluted soils that affects rice production and that has become a problem, especially in places like Fukushima in Japan, where soils are contaminated with radioactive cesium (Nihei et al. 2015). In this sense, Cordones et al. (2017) inactivated the Cs+-permeable K+ transporter (OsHAK1) using two sgRNAs targeting exons 1 and 2 of this gene and obtained a mutation rate of 83%. As a result, the plants carried homo- or bi-allellic frameshift mutations causing the inactivation of the OsHAK1 gene. Also, Cs+ uptake in roots of the edited lines was 35 times lower than in wild type plants (Cordones et al. 2017).
There are other rice genes which are expected to be modified in the future using CRISPR/Cas to generate new varieties resistant to abiotic stress. For example, overexpression of the gene OsPRX2, which encodes 2-Cys peroxiredoxins, contributes to stomatal closure and increases tolerance to potassium deficiency (Mao et al. 2018). Thus, this gene could be overexpressed by CRISPR/Cas9 system using a similar approach to the study in maize made by Shi et al. (2017).
On the other hand, in the case of biotic stress, CRISPR/Cas9 technology has been used to obtain resistance to rice blast disease (Magnaporthe oryzae) by editing the OsERF922 gene, which acts as a negative regulator of resistance to this disease. The frequency of mutations on the target site in edited T0 was 42%. Afterwards, the inserted transgenes (genes from the CRISPR/Cas9 plasmid) were eliminated through segregation, but the induced mutations remained in the T1 and T2 generations. At the end, lesions caused by the pathogen significantly decreased in the edited lines in comparison to the control plants at the seedling and bunch stages (Wang et al. 2016).
In the case of herbicide resistance, the ALS (acetolactate synthase) gene was edited to achieve plants resistant to chlorsulfuron and bispyribac sodium (Sun et al. 2016). In order to do this, CRISPR/Cas9 plasmid was designed with two sgRNAs under the control of the U3 promoter and a donor template for HDR. The donor sequence consisted of a 476 bp fragment which, after insertion, would substitute two amino acids (W548 to L, and S627 to I) in the coding protein. In all of the analyzed lines, the W548 amino acid had been substituted with a L. The plants were sprayed with bispyribac sodium for 36 days and, as result, control non-edited plants died while the edited plants grew normally (Sun et al. 2016).
To enhance nutritional value, the CRISPR/Cas9 system was used to mutate the SBEI gene and its homolog SBEIIb, which encode the starch-branching enzyme (Sun et al. 2017). The first and third exons were edited for SBEI gene and SBEIIb gene, respectively. As a result, 32 and 21 mutants were obtained from 40 and 30 transgenic plants for the SBEI gene and SBEIIb gene, respectively. As a result, the SBEII mutants had a higher proportion of long unbranched amylopectin chains and an increased content of amylose (25%) and resistant starch (9.8%) (Sun et al. 2017). This is important because a cereal with higher amylose content is a good source of resistant starch, which is beneficial to human health (Higgins 2004). Also, the CRISPR/Cas9 technology has been used to disrupt one of the three fatty acid desaturase 2 genes, FAD2-1, in order to generate plants with high oleic and low linoleic content in seeds. The content of oleic acid increased to more than twice in homozygous mutants and increase 10% in heterozygous mutants compared to non-edited control plants (Abe et al. 2018). This study showed that targeting only the genes with dominant or higher expression could be enough to change agronomic traits.
In several studies, the CRISPR/Cas9 technology has been used to generate knockout mutants in order to study genes and their molecular roles. For example, each rice SR (Serine/arginine-rich proteins) locus has been target to produce single knockouts (Butt et al. 2019); OsPIN1 mutants have also been created to study the role of this gene (Li et al. 2019a, b). Some other mutants generated by CRISPR/Cas9 in rice include: OsDGD2β mutants (Basnet et al. 2019), OsPPa6 mutants (Wang et al. 2019b), PE-1 mutants (YuChun et al. 2019), OsLHT1 mutants (Wang et al. 2019c), OsPRP1 mutants (Nawaz et al. 2019), OsPKS2 mutants (Zou et al. 2018), OsPT4 mutants (Ye et al. 2017) (Fig. 3). A recent study demonstrated that using a soybean heat-shock protein gene promoter for Cas9, it was possible to successfully develop a heat-shock -inducible CRISPR/Cas9 system in rice. This system offers the advantage of increasing the mutation rate and off-target effects were low or not found (Nandy et al. 2019). In this sense, this system culd be applied for the improvement of the CRISPR/Cas9 platforms for targeting genes in other crop of topical origin.
Sorghum
Sorghum bicolor L. is the fifth cereal more important in the world providing food for millions of people in subtropical and semi-arid regions in Africa and Asia (Hariprasanna and Rakshit 2016; Che et al. 2018). Regarding the application of modern biotechnology for genetic improvement of this crop, several protocols for tissue culture, and genetic transformation have been developed (review by Girijashankar Swathisree 2009) (Table 1). Additionally, the genome of Sorghum bicolor has been sequenced (Paterson et al. 2009), and it represents a valuable resource for genetic improvement of this crop using modern biotechnological tools, such genome editing technologies.
In a first incursion, Jiang et al. (2013) verified the activity of the CRISPR system within the cells of immature sorghum embryos with a plasmid that encoded the Cas9 gene, one green fluorescent protein (GFP) gene, one sgRNA, and one out of frame red fluorescent protein gene (DsRED2). The sgRNA/Cas9 complex was designed to excise the DsRED2 gene and achieved an active gene via NHEJ repair. In their results, five of eighteen groups of transformed cells with positive GFP expression included sectors with DsRED2 expression (Jiang et al. 2013).
Five years later, the CRISPR/Cas9 system was used to knock out the endogenous histone H3 gene (b-CENH3) in immature sorghum embryos. It is known that this gene is involved in the regulation of chromosome segregation; therefore, its knockout can induce haploids. These researchers obtained lower regeneration percentages (8 to 16%) and the result was attributed to the possible lethality of the biallelic knockout of the Sb-CENH3 gene. In general, the inactivation efficacy in edited immature embryos was between 1% and 5%, and biallelic knockout was not observed (Che et al. 2018).
On the other hand, the CRISPR/Cas9 system has been used to increase the nutritional value of this crop by increasing the digestibility and protein quality (Li et al. 2018c). Kafirins (sorghum prolamins), especially a-Kafirin, are the most abundant storage proteins in the endosperm. Nevertheless, these proteins form bodies with poor digestibility (Oria et al. 2000). In order to increase the protein quality, the k1C gene families, which encode most of kafirins, were targeted. As a result, the T1 and T2 generations showed lower α-kafirin accumulation compared to non-edited control plants, also protein digestibility in the edited lines increased 1.3- to 1.5-fold (Li et al. 2018c).
Recently, Char et al. (2019) used the CRISPR/Cas9 system for targeting the genes SbFT and SbGA2ox5. As a result, the SbFT mutant plants exhibit significant difference in flowering time. Also, Liu et al. (2019b) edited the genes cinnamyl alcohol dehydrogenase (CAD) and phytoene desaturase (PDS) though the particle bombardment of the CRISPR/Cas9 components. In order to establish an efficient genome editing system in sorghum, these authors mentioned that it is important to have an efficient transformation and delivery system for the effective expression of the sgRNA and Cas proteins.
Tomato
Solanum lycopersicum is a vegetable crop of high economic importance and widely cultivated around the world, including tropical and subtropical regions (Hanson and Yang 2016). Conventional breeding of tomato is challenging due to the narrow genetic diversity existing in the germoplasm of cultivated tomatoes, the crossing incompatibility among cultivated and wild species, and the long time required for the introgression (Chaudhary et al. 2019). In the last years, efforts have been made to develop efficient tools for plant tissue culture, plant transformation, and sequence the genome as a complement for the conventional breeding programs (Table 2). The CRISPR/Cas9 system has been employed in S. lycopersicum and its wild relative S. pimpinellifolium. In this sense, Pan et al. (2016) edited the SPIDS (phytoene desaturase) and SIPIF4 (the PIF4 factor that interacts with phytochrome) genes with two independent sgRNAs for each gene. The mutation efficiency was 54.54% and 57.14% for the two sgRNAs in the SIPDS gene, and the mutants showed different types of albinism. In the case of the SIPIF4 gene, the mutation efficacy for the two sgRNAs was 84.00% and 89.47%, respectively. This study demonstrated the ability to mutate genes with high efficacy in tomato. The researchers concluded that high percentages of guanine and cytosine in the sgRNAs led to higher efficacy of editing.
Dahan-Meir et al. (2018) used CRISPR/Cas9 system in the Micro-Tom variety to edit the carotenoid isomerase (CRTISO) and phytoene synthase 1 (PSY1) genes of the carotenoid biosynthetic pathway. These genes were chosen as targets because of the easily detectable phenotype in the mutants. The crtiso mutant had orange fruits, yellowish young leaves, and pale petals, while the psy1 mutant displayed only yellow fruits. Mutation efficacies of 90.4% and 56.4% were obtained in the CRTISO and PSY1 loci, respectively. These authors also used a tomato variety with defective CRTISO genes (with one deletion) and inserted a template of the gene without the mutation by HDR. The deletion was repaired in 25% of the genetically edited plants.
In another study, D’Ambrosio et al. (2018) confirmed the efficacy of CRISPR/Cas9 as a rapid method for generating allelic variants in tomato. For this, the authors edited the marker genes Psy1 (phytoene synthase 1) and CrtR-b2 (beta-carotene hydroxylase 2), which are key genes in the biosynthesis of carotenoids and their knockout generates visible phenotypes in fruit (silencing of Psy1 leads to yellow coloring), and flowers (silencing of CrtR-b2 results in white petals). Two sgRNAs were designed for exon 1 of each gene and the plasmids were introduced in tomato cotyledons by A. tumefaciens. The mutated phenotype was observed in 69% of the modified plants.
Generating tomato plants tolerant to abiotic stress, such as heat stress, is an important objective in the breeding programs since higher temperatures cause physiological and biochemical damages. For example, heat stress in tomato decrease shoot weight, accumulate soluble phenols, and generate lowest peroxidase and polyphenol oxidase activity (Rivero et al. 2001). In this case, the CRISPR/Cas9 system was used to edit the SlMAPK3 gene in tomato (Wang et al. 2017). It has been previously demonstrated that mitogen-activated protein kinases (MAPKs) are genes involved in plant response to environmental stresses (Jalmi and Sinha 2015). In this study, sgRNA were designed to target two different sites of third exon with a mutation efficiency of 41.82%. As a result, editing types were almost small deletions, insertions, or substitutions. The authors of this study concluded that slmapk3 mutants had a lower tolerance to drought stress compared to non-edited plants (Wang et al. 2017). However, a more recent study of the same mutants lines also showed that knocking out SlMAPK3 gene enhances tolerance to heat stress in tomato plants because mutant lines showed less severe wilting and less membrane damage, lower reactive oxygen species (ROS) contents, and higher levels of antioxidant enzymes compared to non-edited plants (Yu et al. 2019). As a conclusion, both studies showed that these mutant lines were more susceptible to drought stress but more tolerant to heat stress than non-edited plants.
Regarding biotic stress, Tashkandi et al. (2018) used CRISPR/Cas9 to target the Tomato Yellow Leaf Curl Virus (TYLCV) genome to produce virus resistant plants. The authors generated tomato plants expressing sgRNA targeting the CP (viral coat protein) or RP (replicase) sequences of TYLCV. Six lines were obtained for each target (CP or RP) expressing the Cas9 gene. This study is important because target genes for editing were those of the virus and not those of the plant. Nevertheless, the problem of this approach is that next generations must keep the transgenes in order to maintain the resistance.
Tomato is also highly vulnerable to Phelipanche aegyptiaca, a weedy root parasitic, which causes many economic problems. There are no effective methods to control it (Hershenhorn et al. 2009). It has been proven that strigolactones in host roots stimulate seed germination of parasitic plants, such as Phelipanche aegyptiaca. This phytohormones are derived from carotenoids via a pathway involving the carotenoid cleavage dioxygenases 7 (CCD7), and CCD8 (Aly et al. 2014). The CRISPR/Cas9 system was used to edit the CCD8 gene in order to generate mutant lines resistant to this parasite. For that, a sgRNAs were designed for targeting exon 2 of this gene, and several mutated lines were obtained with no off-target mutation. The mutated lines in CDD8 gene presented insertions and deletions in the target site and also many morphological changes like dwarfing, excessive shoot branching, and adventitious root formation compared to non-edited plants. Also mutants showed a reduction in parasite infection compared to non-edited plants (Kumar et al. 2019).
Regarding the nutritional value of tomato, the γ-aminobutyric acid (GABA) is a health-promoting functional compound that helps people with nervousness, depression, and insomnia (Liu et al. 2015). In this crop, GABA has been manipulated through gene editing to modify six targets in five key genes for GABA metabolism (GABA-TP1, GABA-TP2, GABA-TP3, CAT9 and SSADH) using the pYLCRISPR/Cas9 multiplex system (Li et al. 2017). The edited efficiencies of the six target sites were 50.0%, 56.8%, 0.0%, 46.6%, 6.8%, and 9.1%, respectively. As a result, heterozygous mutations were the most predominant and homozygous mutations were least abundant. Plants with mutations in four genes at the same time were obtained and not off-target changes were found. Almost all edited plants had higher GABA accumulation in leaves compared to wild type control plants. Specifically, the edited lines with four knocked-out genes at the same time had the highest GABA levels. This study demonstrated the possibility to efficiently edit multiple genes at the same time in a metabolic pathway by creating multi-site knockout mutations (Li et al. 2017).
The CRISPR/Cas9 gene editing technology has also been implemented to enhance agronomic traits, such as parthenocarpy (seedless fruit without prior fertilization) in tomato. In order to achieve this, a knock down of SlIAA9 gene, which is important for control of parthenocarpy, was performed targeting exon 2 in the commercial cultivars Micro-Tom and Ailsa Craig using three different sgRNAs. The mutation rates in the Micro-Tom were 11.1% for sgRNA1, and 40.0–46.2% for sgRNA2, and sgRNA3, respectively. Parthenocarpic fruits were observed in the mutants, but not in wild type plants. Furthermore, fruit morphology of mutant plants was very similar to wild type (Ueta et al. 2017). Induced parthenocarpy is important because it confers many agricultural and industrial benefits. For example, pollination and fertilization are affected by the environment, so parthenocarpy could help to stabilize crop production in unstable environments. Also, seedless fruits facilitate many industrial processes of the fruits (Pandolfini 2009).
Additionally, the CRISPR/Cas9 system has been used in many studies to determine the functions and molecular roles of genes and regulatory elements in tomato. Some recent examples are: GID1 mutants (Illouz-Eliaz et al. 2019), SlMYB21 mutants (Schubert et al. 2019), LOL1 mutants (Borovsky et al. 2019), AP2a mutants (Wang et al. 2019d), SlNPR1 mutants (Li et al. 2019b), and SlMET1 mutants (Yang et al. 2019).
Genome editing has also been used for de novo domestication of plants, such as wild Solanum pimpinellifolium (currant tomato). For this, Zsögön et al. (2018) designed six sgRNAs to target six important loci that control important traits for yield and productivity, like plant growth habit (SELFPRUNING), fruit shape (OVATE) and size (FASCIATED FRUIT WEIGHT 2.2), fruit number (MULTIFLORA), and nutritional quality (LYCOPENE BETA CYCLASE). Four of the six targeted loci were edited successfully in 50 lines from T1. The edited loci were SELF-PRUNING (SP), OVATE (O), FRUIT WEIGHT 2.2 (FW2.2), and LYCOPENE BETA CYCLASE (CycB). These authors demonstrated the feasibility of using CRISPR/Cas9 system for rapid domestication of wild plants that could be extended to other crops of tropical origin.
Watermelon
Citrullus lanatus is an economic important tropical fruit with an estimated worldwide production of 29.6 million tonnes. Moreover, watermelon contains several compounds (vitamins, minerals, and other antioxidants) which play an important role in human health (Reetu and Maharishi 2017). Regarding biotechnological tools, protocols have been developed for somatic embryogenesis (Compton and Gray 1993), biolistic-mediated transformation (Suratman et al. 2010), electroporation of zygotic embryos and nodular buds (Hema et al. 2004); organogenesis (Krug et al. 2005), and Agrobacterium-mediated transformation (Cho et al. 2008). Moreover, the draft genome of watermelon has been released (Guo et al. 2013). These advances open the possibility of applying new genome editing technologies might be impact positively in the watermelon genetic improvement. In a first approach, phytoene desaturase gene (CIPDS) in protoplasts and calluses of this crop were knocked out using the CRISPR/Cas9 system (Tian et al. 2017). In transformed protoplasts with two CRISPR plasmids editing efficacy was 52.0% and 40.7%, respectively. From the calluses transformed with A. tumefaciens with a third plasmid, 16 transformed lines showing albino or quimeric phenotypes and not off-target mutations were found (Tian et al. 2017). Later in this crop, CRISPR/Cas9 was used to generate resistant plants to herbicides by editing ALS gene with a base editing efficiency of 23% at T0 generation and not off-target mutations were found. At the end, some edited plants were not affected after treated with tribenuron while non-edited control plants were severely damaged at 14 days after treatment (Tian et al. 2018).
Yam
The Dioscorea genus include about 600 species but only D. rotundata and D. alata are an economic important crops proving around 90% of the world consumption. It has been used in traditional medicine, and is an important source of carbohydrates, vitamins, and minerals (Aighewi et al. 2015). Breeding programs of yam face some challenging due to the long breeding cycle, polyploidy, heterozygous genetic background, and clonal propagation. In the genus Dioscorea protocols for organogenesis (Anike et al. 2012), somatic embryogenesis (Shu et al. 2005), protoplast culture (Tör et al. 1998), Agrobacterium tumefaciens-mediated transformation (Shi et al. 2012); particle gun mediated transformation (Tör et al. 1993) have been reported. Moreover, the genome of Dioscorea zingiberensis has been recently analyzed (Zhou et al. 2018a, b). Therefore, genetic engineering and genome editing technologies are valuable tools for improvement of this crop (Tripathi 2018). Diosgenin, a compound isolated from the rhizomes, is a precursor of many steroid hormones used as anti-inflammatory, anti-allergic, and antioxidant drugs (Jesus et al. 2016; Zhang et al. 2018b). In a first incursion of the CRISPR/Cas9 technology in Dioscorea zingiberensis, the farnesylpyrophosphate synthase (Dzfps) gene, which encodes an essential enzyme involved in the biosynthesis of secondary metabolites like diosgenin, has been edited. As a result, 9 out of 15 genetically edited plants showed mutations on the target site, and the expression of the Dzfps gene and the content of squalene were significantly less than in the control plants (Feng et al. 2018). This study is important because editing genes involved in diosgenin biosynthesis could be an alternative to increase the production of this metabolite, especially when germoplasm resources with high diosgenin content are needed. The study revealed that CRISPR-Cas9 can be an effective approach for genome modification in yam allowing the development of cultivars with increased pharmacological compounds (such as sapogenin, alkaloids, diterpenes, and steroidals), resistance to diseases, pests, and herbicides (Tripathi 2018).
Design of sgRNA for genome editing of plant cells of crops of tropical origin
At the time of writing, multiple software’s have been developed for predicting and designing CRISPR sgRNA constructs to target specific plant genomic loci. Nevertheless, all these tools are different and have some limitations. For example, some of them are user friendly and are available via web servers while others are not. Moreover, the genomes accessible in some tools are limited, and very few tools have been subjected to peer-review (Brazelton et al. 2015). Off-target effect determination is a challenging task and could be minimized at the time of choosing the appropriate designed sgRNA. Therefore, tools that search the rest of the genome for off-target prediction and allow using alternate PAM sequences should be selected. In the case of the studies analyzed in this review several different computational tools were used for the design of the sgRNA (Table 1).
Various systems that may be used for delivery of CRISPR into plant cells of crops of tropical origin
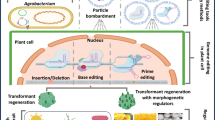
Several methods are available for delivering CRISPR reagents to plant cells (Fig. 4). CRISPR components can be expressed in the plant cell genome using heterologous DNA or RNA transgenes, or integrated directly into the nuclei as ribonucleotide protein complexes. The delivery systems of the CRISPR components depend on several factors, such as plant species, research purposes, experience, and available equipment. Moreover, the commercialization of non-GM genome edited crops can be favored by using specific delivery systems that circumvent restrictive regulatory burdens (Lowder et al. 2016).

Source Own elaboration based on Lowder et al. (2016)
Methods for the delivery of CRISPR components into plant cells and tissues. a Floral dip transformation with transgenic T-DNA carrying Agrobacteria. b Transient inoculation of plant leaf tissue or calli with Agrobacteria harboring Cas9 and gRNA T-DNA. c Viral vector delivery causes a transiently transformed plant (at left) to develop systemic infection upon viral capsid replication. d Transient gun bombardment of plant leaf tissue using a cassette containing Cas9 and gRNA or e delivery of gRNAs only to plant leaf tissues. f Delivery of ribonucleoprotein (RNP) complex to protoplasts using PEG transformation or g RNA delivery directly to protoplasts.
Agrobacterium mediated T-DNA delivery
Specific Agrobacterium transformation methods have been developed and optimized for different plant species, including crops of tropical origin (Wang 2015). The Agrobacterium transformation method uses pathogenic bacteria that have the ability to infect plant cells and transfer exogenous DNA horizontally or directly to the genome of the host plant (Mohammed et al. 2019).
Not surprisingly, Agrobacterium mediated T-DNA transformation method, followed by plant tissue culture dependent regeneration of stable mutants, and has been used for generating genome edited crops of tropical origin (Table 1). Nevertheless, the CRISPR systems have the advantage that desirable results can often be accomplished using transient expression in plant cells. Hence, it is not necessary the integration of the CRISPR directly into plant genomes.
In this sense, the delivery of CRISPR expression DNA cassettes into plant cells has been carried out using in planta transformation methods, such as floral dip in the model plant Arabidopsis (Feng et al. 2013; Jiang et al. 2013; Li et al. 2013) (Fig. 4a), and infiltration of tobacco somatic cells (Gao et al. 2015) (Fig. 4b). In planta methodologies allows that the transformation strategies could be used directly and quickly by being independent of in vitro tissue culture techniques. This strategy could be applied to recalcitrant crops of tropical origin.
Viral delivery
Plant RNA and DNA viruses offer a great potential for efficient delivery of CRISPR components into plant cells (Fig. 4c). Beet necrotic yellow vein virus-based vectors were employed to deliver NbPDS (N. benthamiana phytoene desaturase gene) guide RNAs for genome editing in tobacco transgenic plants expressing Cas9, leading to the generation of photo bleached phenotype in infected leaves (Jiang et al. 2019). Viral systems provide great opportunity for genome editing of crops of tropical origin as plants can be transiently infected moderately rapidly and viral replication of CRISPR components can spread to systemic infection of whole plants. Moreover, such systems offer a more facile and efficient delivery option compared to the laborious and highly technical process of Agrobacterium-mediated transformation and biolistic transformation. Nevertheless, the application of such systems is limited by the low editing efficiency (Lowder et al. 2016).
Plasmid delivery
Instead of being delivered by Agrobacterium, CRISPR components can be transferred into plant cells by expression plasmids using polyethylene glycol–calcium (PEG–Ca2+) transfection of protoplasts or biolistic particle delivery using a gene particle gun (Fig. 4d, e). The first approach has been mainly used for rapid testing of CRISPR activity in plant cells and such assays have been performed in watermelon, rice, and maize (Table 1). Whereas the biolistics transformation system consists of firing micro particles at high speeds, usually of gold or tungsten, coated with the DNA fragments to the plant cells. This technique has been successfully applied to rice, and maize (Table 1). Both approaches are robust enough and generally can be applied to deliver CRISPR-Cas9 reagents to almost any crops of tropical origin.
Ribonucleotide protein complex delivery
Previously, ZFNs (zinc-finger nucleases) and TALENs (transcription activator-like effector nucleases) were successfully delivered as proteins into cells to mediate genome editing (Lowder et al. 2016). This approach consists in the delivery of preassembled Cas protein-gRNA ribonucleoproteins (RNPs) into protoplasts derived from somatic tissues by PEG–Ca2+-mediated transfection or biolistic particle delivery (Fig. 4f). This technique has been successfully applied to plant protoplasts of rice (Woo et al. 2015) or into embryo cells by biolistic bombardment in maize (Svitashev et al. 2016). Nevertheless, the isolation and culture of protoplasts is a challenging task in most plant species and the frequency of obtaining genome-edited plants through biolistic bombardment is relatively low. Therefore, Toda et al. (2019) developed a system for the direct delivery of Cas9–gRNA RNPs into plant zygotes of rice produced by in vitro fertilization of isolated gametes (Toda et al. 2019). The RNPs delivery may offer advantages for certain applications such as avoiding potential GMOs regulatory laws or genome editing of asexually propagated crops, such as bananas (Lowder et al. 2016). Therefore, this plant-genome-editing system has enormous potential for the improvement of other important crop of tropical origin.
RNA delivery
This strategy consists in the transfer of RNA encoding genome editing components directly into the plant cells (Fig. 4g). This system has been used for the successfully delivery of mRNA transcripts of CRISPR-Cas9 and gRNA into wheat calli, although mutation frequencies were very low-1.1% (Zhang et al. 2016). Nevertheless, the RNA delivery could induce genome editing without transgene insertion into host genomes (Lowder et al. 2016). An additional benefit of this system could be the reduction of the probability of off-targeting that could negatively impact plant function or growth, since RNPs and mRNA degrade quickly after mutagenesis (Luo et al. 2015).
Screening of mutants
Various tools, including PCR (Polymerase chain reaction), sequencing PCR amplicons, next generation sequencing (NGS), whole genome sequencing (WGS), Southern blotting, DNA microarray, ELISA, genotypic and phenotypic screening, have been established and could be used to corroborate mutagenesis in genome edited plants (Grohmann et al. 2019). One of the most common, simply, cheaper, sensitive, and specific method for detection of mutations is PCR. This method requires information about the target DNA sequence and the design of complementary primers for the amplification of changes induced by genome editing. PCR-based methods have been used for screening of mutants generated by CRISPR in crops of tropical origin (Table 1). Short sequence changes, such as substitutions or InDels of few nucleotides, could be detected by real-time PCR or digital PCR (Grohmann et al. 2019).
Sanger sequencing of PCR amplicons and NGS are suitable methods for the detection of changes in the targeted gene, even if the modifications are small (Grohmann et al. 2019). Nevertheless, sequencing errors and bioinformatic analysis problems should be taken into account in order to avoid false positives results. These two methods have been used for the detection and corroboration of mutations in generated by CRISPR in crops of tropical origin (Table 1).
Moreover, WGS has been used for GMOs (Genetically Modified Organisms) detection, and might be adapted for genome-edited plants. This tool does not require prior information on a specific genetic modification and can be used as a detection approach for detection of off-target alterations. Nevertheless, efforts and costs are significantly increased compared to NGS. Also, it requires a high quality reference genome, derived from the parental plant, and bioinformatics capacity for the processing of millions of small DNA sequence reads. Furthermore, another limitation for the application of WGS is the size of the genome in question and the presence of repetitive sequences in the genome (Grohmann et al. 2019). This applies for a variety of crop plants of tropical origin, e.g., the genome of the arabica coffee (Coffea arabica L.) and maize.
DNA hybridization based methods, such as Southern blotting and DNA microarray, have been used for the detection of GMOs; nevertheless, the application for the detection of genome-edited plants is inappropriate since it requires a large amount of genetic material and the sensitivity is low compared to DNA-amplification and sequencing methods. Moreover, these methods are commonly used for the detection of longer altered nucleotide sequences and/or integrated foreign DNA, therefore, the detection of small or single nucleotide modifications are challenging (Grohmann et al. 2019).
Although, protein based-methods, such as ELISA, are available and have been used the characterization of GMO, the corroboration of changes induced by genome editing should be confirmed by subsequent DNA analyses (Grohmann et al. 2019). Similarly, metabolite-based methods allow the detection and identification of a wide variety of substances in a plant metabolite profile. Nevertheless, identified difference in the metabolite profile is no proof of a genetic modification since it is highly dynamic and fluctuating in response to developmental and environmental conditions (Grohmann et al. 2019).
Novel CRISPR technologies that may be used in breeding of crops of tropical origin
CRISPR/Cas12a system
This system, previously known as CRISPR/Cpf1, allows double strand breaks with efficiencies similar to those of CRISPR/Cas9. The nuclease Cas12a requires a small crRNA for its activity and the maximum efficiency and specificity is determined by 22 nt spacer (Schindele et al. 2018). Moreover, this nuclease identifies a T-rich PAMs located upstream of the guide and generated staggered ends (Schindele et al. 2018). In plants, LbCas12a from Lachnospiraceae bacterium and FnCas12a from Francisella novicida, promoted mutagenesis by genome editing in rice and tobacco (Endo et al. 2016) and citrus (Jia et al. 2019). Also, Cas12a could be used as a specific DNA binding protein and downregulate gene expression as demonstrated in Arabidopsis (Tang et al. 2017). Therefore, Cas12a nucleases could be a potential option for targeting genes in crops of tropical origin, specifically those genes with low GC content.
CRISPR/Cas13 system
This system, previously called C2c2, recognizes and cleaves specific single strand RNA in eukaryotic cells, offering a wide range of applications in bacteria, mammals, and plants (Ali et al. 2018; Schindele et al. 2018; Wolter and Puchta 2018). It offers great potential for agriculture and could be used for post-transcriptional repression (Abudayyeh et al. 2017), for combating RNA viruses as demonstrated in Nicotiana benthamiana (Aman et al. 2018), and as RNA binding protein for the discrimination of specific nucleic acids as reported for Zika virus (ZIKV) and the related flavivirus Dengue (DENV) (Gootenberg et al. 2017).
Base editing
Base editing is a recently new tool that allows the conversion of nucleotides without inducing double-stranded DNA breaks or using donor templates. In this sense, it is rapidly adopted for changing a C-G base pair into T-A, or A-T into G-C (Marx 2018). As proof-of-concept, the system CRISPR-Cas9 nickase-cytidine deaminase was successfully employed in protoplasts and regenerated rice and maize plants for targeting the conversion of cytosine to thymine within the protospacer with efficiency up to 40% (Zong et al. 2017). In tomato (Solanum lycopersicum), the CRISPR-Cas9 and activation-induced cytidine deaminase was used to develop marker free plants (Zenpei et al. 2017). Moreover, herbicide-tolerant rice plants have been developed through a substitution of T to C in a single amino acid (C2186R) of the gene Acetyl-coenzyme A carboxylase (ACC) (Li et al. 2018a, b, c).
Modification of gene expression and epigenome
The CRISPR/Cas9 system could be used in synthetic biology for engineering gene circuits and regulate ectopic gene expression by combining dCAS9 with different activator/inhibitor domains of transcription factors. In this sense, dCAS9 could be employed to edit epigenetic marks in the genome (Bortesi and Fischer 2015).
Challenges for genome editing in crops of tropical origin
There is still much work to do in this area of plant biotechnology and plant genome editing still faces some challenges. For example, for effective genome editing in crops of tropical origin, the genome of the target plant must first be sequenced. Moreover, plant transformation methods (via Agrobacterium, biolistic bombardment or protoplast transformation) is a major limiting factor for adopting CRISPR-Cas9 technology to many crops of tropical origin, especially those recalcitrant plants. Additionally, an efficient plant tissue culture regeneration system must be established. For example, regeneration of plants from protoplasts and intact somatic cells can be challenging and time consuming. Improving plant regeneration protocols and delivery approaches will become an important research priority to open up CRISPR delivery to these plants.
Regulatory aspects
Genome editing technologies can introduce advantageous traits for the improvement of crops of tropical origin that could be commercially available very soon. Some gene-edited crops such as white button mushrooms, wheat, soybeans and waxy corn have overcome USDA regulation. However, there is still a need to clarify their regulatory status, particularly with regard to global regulations on Genetically Modified Organisms (GMOs) (Kinderlerer 2008; McHughen and Smyth 2008). In many countries, the use of GMOs is regulated (Rosado and Craig 2017). Nevertheless, with respect to genome editing technologies, governments should consider their regulatory status and establish appropriate regulations, if necessary, without representing an obstacle to the commercialization of products derived from NPBTs (Seyran and Craig 2018). However, countries differ in how they regulate these technologies, which fall between genetic engineering and traditional techniques (Sprink et al. 2016; Eckerstorfer et al. 2019; Eriksson et al. 2019; Smyth 2019). In some jurisdictions, some genome editing technologies have been considered as simply as a variation of existing conventional plant breeding, while other countries have not determined what to do or how to proceed to regulate them (Lassoued et al. 2018b). So far, countries such as Argentina, Australia, Brazil, Chile, Colombia, the United States, and Israel have regulatory approaches that allow the cultivation and use of the products obtained by gene editing. The premise is that the final products do not contain genes from other organisms, unlike the transgenic ones, and the genetic changes produced by the human being, precisely resemble those genetic changes that occur spontaneously in nature (Smyth 2019).
Conclusions
Many studies have demonstrated the efficacy of the genome editing technologies for gene editing in highly important tropical crops, such as rice, maize, tomato, and citrus. On the other hand, the use of this technology in other tropical crops with similar commercial value is just beginning. This is the case for coffee, yams, bananas, and many other species in which the technology has not been applied. Without a doubt, due to the low cost, precision, and rapidity, the CRISPR system could accelerate the genetic improvement of tropical crops of economic importance and as a consequence contribute to global food security. In this sense, this new breeding technology could improve a variety of crop traits, including yield, nutritional value, and stress tolerance, and pest and herbicide resistance.
Abbreviations
- bp:
-
Base-pair
- CRISPR:
-
Clustered regularly interspaced short palindromic repeats
- Cas9:
-
CRISPR associated protein 9
- DBS:
-
DNA double-strand break
- DNA:
-
Deoxyribonucleic acid
- HDR:
-
Homology directed repair
- Indel:
-
Insertion or deletion of base(s)
- NHEJ:
-
Non-homologous end joining
- PAM:
-
Protospacer adjacent motif
- PCR:
-
Polymerase chain reaction
- RNA:
-
Ribonucleic acid
- RNP:
-
Ribonucleoprotein
- sgRNA:
-
Single guide RNA
References
Abe K, Araki E, Suzuki Y et al (2018) Production of high oleic/low linoleic rice by genome editing. Plant Physiol Biochem. https://doi.org/10.1016/j.plaphy.2018.04.033
Abudayyeh OO, Gootenberg JS, Essletzbichler P et al (2017) RNA targeting with CRISPR-Cas13. Nature 550:280–284
Aighewi BA, Asiedu R, Maroya N, Balogun M (2015) Improved propagation methods to raise the productivity of yam (Dioscorea rotundata Poir.). Food Security 7(4):823–834
Alemayehu D (2017) Review on genetic diversity of coffee (Coffea arabica L) in Ethiopia. Int J For Hortic 3:18–27
Ali Z, Mahas A, Mahfouz M (2018) CRISPR/Cas13 as a tool for RNA interference. Trends Plant Sci 23:374–378. https://doi.org/10.1016/j.tplants.2018.03.003
Aly R, Dubey N, Yahyaa M et al (2014) Gene silencing of CCD7and CCD8 in Phelipanche aegyptiaca by tobacco rattle virus system retarded the parasite development on the host. Plant Signaling & Behavior 9:e29376. https://doi.org/10.4161/psb.29376
Aman R, Ali Z, Butt H et al (2018) RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol 19:1
Anga J (2014) The world cocoa economy: current status, challenges and prospects. In: Multi-year expert meeting on commodities and development. ICCO, London
Anike FN, Konan K, Olivier K et al (2012) Efficient shoot organogenesis in petioles of yam (Dioscorea spp.). Plant Cell, Tissue Organ Cult 111:303–313. https://doi.org/10.1007/s11240-012-0195-9
Argout X, Salse J, Aury JM et al (2011) The genome of Theobroma cacao. Nat Genet 43:101–108
Ashraf M, Athar HR, Harris PJC, Kwon TR (2008) Some prospective strategies for improving crop salt tolerance. Adv Agron 97:45–110
Bainsla NK, Meena HP (2016) Breeding for resistance to biotic stresses in plants. In: Yadav P, Kumar S, Jain V (eds) Recent advances in plant stress physiology. Daya Publishing House, New Delhi, p 379
Basnet R, Hussain N, Shu Q (2019) OsDGD2β is the sole digalactosyldiacylglycerol synthase gene highly expressed in anther, and its mutation confers male sterility in rice. Rice (N Y) 12:66. https://doi.org/10.1186/s12284-019-0320-z
Battraw M, Hall TC (1991) Stable transformation of Sorghum bicolor protoplasts with chimeric neomycin phosphotransferase II and b-glucuronidase genes. Theor Appl Genet 82:161–168
Baysal C, Bortesi L, Zhu C et al (2016) CRISPR/Cas9 activity in the rice OsBEIIb gene does not induce off-target effects in the closely related paralog OsBEIIa. Mol Breed 36:108
Belhaj K, Chaparro-Garcia A, Kamoun S et al (2015) Editing plant genomes with CRISPR/Cas9. Curr Opin Biotechnol 32:76–84
Borovsky Y, Monsonego N, Mohan V et al (2019) The zinc-finger transcription factor CcLOL1 controls chloroplast development and immature pepper fruit color in Capsicum chinense and its function is conserved in tomato. Plant J 99:41–55. https://doi.org/10.1111/tpj.14305
Bortesi L, Fischer R (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv 33:41–52
Brazelton VA, Zarecor S, Wright DA et al (2015) A quick guide to CRISPR sgRNA design tools. GM Crops Food 6:266–276. https://doi.org/10.1080/21645698.2015.1137690
Breitler JC, Dechamp E, Campa C et al (2018) CRISPR/Cas9-mediated efficient targeted mutagenesis has the potential to accelerate the domestication of Coffea canephora. Plant Cell, Tissue Organ Cult. https://doi.org/10.1007/s11240-018-1429-2
Butt H, Piatek A, Li L et al (2019) Multiplex CRISPR mutagenesis of the serine/arginine-rich (SR) gene family in rice. Genes 10:596. https://doi.org/10.3390/genes10080596
Cao HX, Wenqin W, Hien TTL et al (2016) The power of CRISPR-CAS9-induced genome editing to speed up plant breeding. Int J Genomics. https://doi.org/10.1155/2016/5078796
Chandrasekaran J, Brumin M, Wolf D et al (2016) Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol Plant Pathol 17:1140–1153
Char SN, Neelakandan AK, Nahampun H et al (2016) An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol J. https://doi.org/10.1111/pbi.12611
Char SN, Wei J, Mu Q et al (2019) An Agrobacterium -delivered CRISPR/Cas9 system for targeted mutagenesis in sorghum. Plant Biotechnol J. https://doi.org/10.1111/pbi.13229
Chaudhary J, Alisha A, Bhatt V et al (2019) Mutation breeding in tomato: advances, applicability and challenges. Plants 8:128. https://doi.org/10.3390/plants8050128
Chavarriaga-Aguirre P, Brand A, Medina A et al (2016) The potential of using biotechnology to improve cassava: a review. In Vitro Cell Dev Biol Plant 52:461–478
Che P, Anand A, Wu E et al (2018) Developing a flexible, high-efficiency Agrobacterium-mediated sorghum transformation system with broad application. Plant Biotechnol 16:1388–1395. https://doi.org/10.1111/pbi.12879
Cho MA, Moon CY, Liu JR et al (2008) Agrobacterium-mediated transformation in Citrullus lanatus. Biol Plant 52:365–369. https://doi.org/10.1007/s10535-008-0076-6
Chylinski K, Makarova KS, Charpentier E et al (2014) Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res 42:6091–6105. https://doi.org/10.1093/nar/gku241
Compton M, Gray DJ (1993) Somatic embryogenesis and plant regeneration from immature cotyledons of watermelon. Plant Cell Rep 12:61–65. https://doi.org/10.1007/bf00241935
Cordones MN, Mohamed S et al (2017) Production of low-Cs + rice plants by inactivation of the K + transporter OsHAK1 with the CRISPR-Cas system. Plant J 92:43–56. https://doi.org/10.1111/tpj.13632
D’Ambrosio C, Stigliani AL, Giorio G (2018) CRISPR/Cas9 editing of carotenoid genes in tomato. Transgenic Res 27:367–378
D’Hont A, Denoeud F, Aury J-M et al (2012) The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488:213–217. https://doi.org/10.1038/nature11241
Dahan-Meir T, Filler-Hayut S, Melamed-Bessudo C et al (2018) Efficient in planta targeting in tomato using geminiviral replicons and the CRISPRCas9 system. Plant J 95:5–16
Denoeud F, Carretero-Paulet L, Dereeper A et al (2014) The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345:1181–1184
Do PT, Lee H, Mookkan M et al (2016) Rapid and efficient Agrobacterium-mediated transformation of sorghum (Sorghum bicolor) employing standard binary vectors and bar gene as a selectable marker. Plant Cell Rep 35:2065. https://doi.org/10.1007/s00299-016-2019-6
Eckerstorfer MF, Engelhard M, Heissenberger A et al (2019) Plants developed by new genetic modification techniques—comparison of existing regulatory frameworks in the EU and Non-EU countries. Front Bioeng Biotechnol 7:26. https://doi.org/10.3389/fbioe.2019.00026
Emons AMC, Kieft H (1995) Somatic embryogenesis in maize (Zea mays L.). In: Balaji YPS (ed) Biotechnology in agriculture and forestry. Springer, Berlin, pp 24–39. https://doi.org/10.1007/978-3-642-78643-3_3
Endo M, Mikami M, Endo A et al (2018) Genome editing in plants by engineered CRISPR–Cas9 recognizing NG PAM. Nat Plants. https://doi.org/10.1038/s41477-018-0321-8
Eriksson D, Kershen D, Nepomuceno A et al (2019) A comparison of the EU regulatory approach to directed mutagenesis with that of other jurisdictions, consequences for international trade and potential steps forward. New Phytol 222:1673–1684
FAO (2015) Citrus fruit statistics 2015. http://www.fao.org/3/a-i5558e.pdf. Accessed 29 Sept 2018
FAO (2016) Climate change and food security: risks and responses. http://www.fao.org/3/a-i5188e.pdf. Accessed 29 Sept 2018
FAOSTAT (2017) Food and Agriculture Organization of the United Nations. On-line and Multilingual Database. http://faostat.fao.org/. Accessed 15 Aug 2018
Farooq R, Hussain K, Nazir S et al (2018) CRISPR/Cas9; A robust technology for producing genetically engineered plants. Cell Mol Biol 64:31–38. https://doi.org/10.14715/cmb/2018.64.14.6
Feng Z, Zhang B, Ding W et al (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23:1229–1232. https://doi.org/10.1038/cr.2013.114
Feng S, Song W, Fu R et al (2018) Application of the CRISPR/Cas9 system in Dioscorea zingiberensis. Plant Cell, Tissue Organ Cult. https://doi.org/10.1007/s11240-018-1450-5
Ferrão LFV, Ortiz R, Garcia AAF (2017) Genomic selection: state of the art. In: Campos H, Caligari PDS (eds) Genetic improvement of tropical crops. Springer, Cham, pp 19–57. https://doi.org/10.1007/978-3-319-59819-2_2
Fister AS, Landherr L, Maximova SN et al (2018) Transient expression of CRISPR/Cas9 machinery targeting TcNPR3 enhances defense response in Theobroma cacao. Front Plant Sci 9:268. https://doi.org/10.3389/fpls.2018.00268
Fondong VN and Rey C (2018) Recent biotechnological advances in the improvement of cassava. https://www.intechopen.com/books/cassava/recent-biotechnological-advances-in-the-improvement-of-cassava. Accessed 31 Aug 2019
Gao J, Wang G, Ma S et al (2015) CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol Biol 87:99–110. https://doi.org/10.1007/s11103-014-0263-0
Girijashankar V, Sharma KK, Balakrishna P et al (2007) Direct somatic embryogenesis and organogenesis pathway of plant regeneration can seldom occur simultaneously within the same explant of sorghum. SAT eJournal 3(1):1–3
Girijashankar V, Swathisree V (2009) Genetic transformation of Sorghum bicolor. Physiol Mol Biol Plants 15(4):287–302. https://doi.org/10.1007/s12298-009-0033-7
Gomez MA, Lin ZD, Moll T et al (2018) Simultaneous CRISPR/Cas9-mediated editing of cassava elF4E isoforms nCBP-1 and nCBP-2 confers elevated resistance to cassava brown streak disease. bioRxiv. https://doi.org/10.1101/209874
Gootenberg JS, Abudayyeh OO, Lee JW et al (2017) Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356:438–442
Gotsch N (1997) Cocoa biotechnology: status, constraints and future prospects. Biotechnol Adv 15(2):333–352
Govindaraj M, Vetriventhan M, Srinivasan M (2015) Importance of genetic diversity assessment in crop plants and its recent advances: an overview of its analytical perspectives. Genet Res Int 2015:1–14. https://doi.org/10.1155/2015/431487
Grohmann L, Keilwagen J, Duensing N et al (2019) Detection and identification of genome editing in plants: challenges and opportunities. Front Plant Sci 10:236. https://doi.org/10.3389/fpls.2019.00236
Guo S, Zhang J, Sun H et al (2013) The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet 45:51–58. https://doi.org/10.1038/ng.2470
Guo X, Duan X, Wu Y et al (2018) Genetic engineering of maize (Zea mays L.) with improved grain nutrients. J Agric Food Chem. https://doi.org/10.1021/acs.jafc.7b05390
Hahn F, Nekrasov V (2018) CRISPR/Cas precision: do we need to worry about off-targeting in plants? Plant Cell Rep. https://doi.org/10.1007/s00299-018-2355-9
Hanson P, Yang R (2016) Genetic improvement of tomato (Solanum lycopersicum L.) for phytonutrient content at AVRDC-the World Vegetable Center. Ekin J 2:1–10
Haque E, Taniguchi H, Hassan MM et al (2018) Application of CRISPR/Cas9 genome editing technology for the improvement of crops cultivated in tropical climates: recent progress, prospects, and challenges. Front Plant Sci. https://doi.org/10.3389/fpls.2018.00617
Hariprasanna K, Rakshit S (2016) Economic importance of sorghum. Sorghum Genome. https://doi.org/10.1007/978-3-319-47789-3_1
Hayashimoto A, Li Z, Murai N (1990) A polyethylene glycol-mediated protoplast transformation system for production of fertile transgenic rice plants. Plant Physiol 93:857–863. https://doi.org/10.1104/pp.93.3.857
Hema MV, Prasad T, Vani A (2004) Transient expression and stable integration of chimeric Gus gene in watermelon following electroporation. J Hortic Sci Biotechnol 79:364–369
Hershenhorn J, Eizenberg H, Dor E et al (2009) Phelipanche aegyptiacamanagement in tomato. Weed Res 49:34–47. https://doi.org/10.1111/j.1365-3180.2009.00739.x
Hiei Y, Ohta S, Komari T et al (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282. https://doi.org/10.1046/j.1365-313x.1994.6020271.x
Higgins J (2004) Resistant starch: metabolic effects and potential health benefits. J AOAC Int 87:761–768
Hu C, Deng G, Sun X et al (2017a) Establishment of an efficient CRISPR/Cas9-mediated gene editing system in banana. Sci Agric Sin 50:1294–1301
Hu B, Li D, Liu X et al (2017b) Engineering non-transgenic gynoecious cucumber using an improved transformation protocol and optimized CRISPR/Cas9 system. Mol Plant 10:1575–1578. https://doi.org/10.1016/j.molp.2017.09.005
Hua K, Tao X, Yuan F et al (2018) Precise A·T to G·C base editing in the rice genome. Mol Plant 11:627–630. https://doi.org/10.1016/j.molp.2018.02.007
Huang S, Li R, Zhang Z et al (2009) The genome of the cucumber, Cucumis sativus L. Nat Genet 41:1275–1281. https://doi.org/10.1038/ng.475
Huang C, Sun H, Xu D et al (2018) ZmCCT9 enhances maize adaptation to higher latitudes. Proc Natl Acad Sci USA 115:334–341. https://doi.org/10.1073/pnas.1718058115
Hummel AW, Chauhan RD, Cermak T et al (2018) Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnol J 16:1275–1282
Illouz-Eliaz N, Ramon U, Shohat H et al (2019) Multiple gibberellin receptors contribute to phenotypic stability under changing environments. Plant Cell 31:1506–1519. https://doi.org/10.1105/tpc.19.00235
Ishino Y, Krupovic M, Forterre P (2018) History of CRISPR-Cas from encounter with a mysterious repeated sequence to genome editing technology. J Bacteriol. https://doi.org/10.1128/jb.00580-17
Jagodzik P, Tajdel-Zielinska M, Ciesla A et al (2018) Mitogen-activated protein kinase cascades in plant hormone signaling. Front Plant Sci. https://doi.org/10.3389/fpls.2018.01387
Jalmi S, Sinha A (2015) ROS mediated MAPK signaling in abiotic and biotic stress-striking similarities and differences. Front Plant Sci. https://doi.org/10.3389/fpls.2015.00769
Jaqueth JS, Hou Z, Zheng P et al (2019) Fertility restoration of maize CMS-C altered by a single amino acid substitution within the Rf4 bHLH transcription factor. Plant J. https://doi.org/10.1111/tpj.14521
Jesus M, Martins A, Gallardo E et al (2016) Diosgenin: recent highlights on pharmacology and analytical methodology. J Anal Methods Chem 016:1–16. https://doi.org/10.1155/2016/4156293
Jia H, Wang N (2014) Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE 9:1–6
Jia H, Orbovic V, Jones JB et al (2015) Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4:dCsLOB1.3 infection. Plant Biotechnol J 14:1291–1301. https://doi.org/10.1111/pbi.12495
Jia H, Zhang Y, Orbović V et al (2017) Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol J 15:817–823. https://doi.org/10.1111/pbi.12677
Jia H, Zou X, Orbovic V et al (2019) Genome editing in citrus tree with CRISPR/Cas9. Plant Genome Ed CRISPR Syst 1917:235–241. https://doi.org/10.1007/978-1-4939-8991-1_17
Jiang F, Doudna JA (2017) CRISPR–Cas9 structures and mechanisms. Annu Rev Biophys 46:505–529
Jiang W, Zhou H, Bi H et al (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41:e188. https://doi.org/10.1093/nar/gkt780
Jiang N, Zhang C, Liu JY et al (2019) Development of beet necrotic yellow vein virus-based vectors for multiple-gene expression and guide RNA delivery in plant genome editing. Plant Biotechnol J 17:1302–1315
Johnson RA, Gurevich V, Filler S et al (2015) Comparative assessments of CRISPR-Cas nucleases’ cleavage efficiency in planta. Plant Mol Biol 87:143–156
Kamburova VS, Nikitina EV, Shermatov SE et al (2017) Genome editing in plants: an overview of tools and applications. Int J Agron 2017:1–15
Kangquan Y, Caixia G, Jin-Long Q (2017) Progress and prospects in plant genome editing. Nat Plants. https://doi.org/10.1038/nplants.2017.107
Kaur N, Alok A, Shivani et al (2018) CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. rasthali genome. Funct Integr Genomics 18:89–99. https://doi.org/10.1007/s10142-017-0577-5
Khan Z, Khan SH, Mubarik MS et al (2016) Use of TALEs and TALEN technology for genetic improvement of plants. Plant Mol Biol Rep 35:1–19
Kinderlerer J (2008) The Cartagena protocol on biosafety. Collect Biosaf Rev 4:12–65
Kohli A, Melendi PG, Abranches R et al (2006) The quest to understand the basis and mechanisms that control expression of introduced transgenes in crop plants. Plant Signal Behav 1:185–195
Krug MG, Stipp LC, Rodriguez AP et al (2005) In vitro organogenesis in watermelon cotyledons. Pesqui Agropecu Bras 40:861–865. https://doi.org/10.1590/s0100-204x2005000900004
Kumar R, Tuteja N (2012) Development of Agrobacterium-mediated transformation technology for mature seed-derived callus tissues of indica rice cultivar IR64. GM Crops Food Biotechnol Agric Food Chain 3:123–128
Kumar K, Gill MI, Gosal SS (2018) Somatic embryogenesis, in vitro selection and plantlet regeneration for citrus improvement. Biotechnol Crop Improv 1:373–406
Kumar V, Abu J, Marzouk S et al (2019) CRISPR/Cas9-mediated mutagenesis of CAROTENOID CLEAVAGE DIOXYGENASE 8 in tomato provides resistance against the parasitic weed Phelipanche aegyptiaca. Sci Rep 9:11438. https://doi.org/10.1038/s41598-019-47893-z
Labouisse JP, Bellachew B, Kotecha S et al (2008) Current status of coffee (Coffea arabica L.) genetic resources in Ethiopia: implications for conservarion. Genet Resour Crop Evol 55:1079–1093. https://doi.org/10.1007/s10722-008-9361-7
Lassoued R, Hesseln H, Phillips PWB et al (2018a) Top plant breeding techniques for improving food security: an expert Delphi survey of the opportunities and challenges. Int J Agric Resour Gov Ecol 14:321–337
Lassoued R, Smyth SJ, Phillips PWB et al (2018b) Regulatory uncertainty around new breeding techniques. Front Plant Sci 9:1291. https://doi.org/10.3389/fpls.2018.01291
LeBlanc C, Zhang F, Mendez J et al (2018) Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J 93:377–386. https://doi.org/10.1111/tpj.13782
Lee H, Zhang ZJ (2013) Agrobacterium-mediated transformation of maize (Zea mays) immature embryos. Cereal Genomics. https://doi.org/10.1007/978-1-62703-715-0_22
Li L, Qu R, de Kochko A et al (1993) An improved rice transformation system using the biolistic method. Plant Cell Rep 12:250–255. https://doi.org/10.1007/bf00237129
Li JF, Norville JE, Aach J, McCormack M et al (2013) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31:688–691. https://doi.org/10.1038/nbt.2654
Li R, Li R, Li X et al (2017) Multiplexed CRISPR/Cas9-mediated metabolic engineering of gamma-aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnol J 16:415–427. https://doi.org/10.1111/pbi.12781
Li C, Zong Y, Wang Y, Jin S et al (2018a) Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol 19:59
Li C, Yue Y, Chen H et al (2018b) ZmbZIP22 is a transcription factor that regulates 27-kD γ-zein gene transcription during maize endosperm development. Plant Cell 30:2402–2424
Li A, Jia S, Yobi A et al (2018c) Editing of an alpha-kafirin gene family increases digestibility and protein quality in sorghum. Plant Physiol 77:1425–1438. https://doi.org/10.1104/pp.18.00200
Li Y, Zhu J, Wu L et al (2019a) Functional divergence of PIN1 paralogous genes in rice. Plant Cell Physiol. https://doi.org/10.1093/pcp/pcz159
Li R, Liu C, Zhao R et al (2019b) CRISPR/Cas9-Mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol. https://doi.org/10.1186/s12870-018-1627-4
Liang Z, Zhang K, Chen K et al (2014) Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J Genet Genomics 41:63–68
Liang Z, Chen K, Yan Y et al (2018) Genotyping genome-edited mutations in plants using CRISPR ribonucleoprotein complexes. Plant Biotechnol J. https://doi.org/10.1111/pbi.12938
Liu L, Liu C, Wang Y et al (2015) Herbal medicine for anxiety, depression and insomnia. Curr Neuropharmacol 13:481–493. https://doi.org/10.2174/1570159x13041508311227
Liu J, Gao P, Sun X et al (2017) Efficient regeneration and genetic transformation platform applicable to five Musa varieties. Electron J Biotechnol 25:33–38
Liu J, Nannas N, Fu F et al (2019a) Genome-scale sequence disruption following biolistic transformation in rice and maize. Plant Cell. https://doi.org/10.1105/tpc.18.00613
Liu G, Li J, Godwin ID (2019b) Genome editing by CRISPR/Cas9 in Sorghum through biolistic bombardment. Sorghum. https://doi.org/10.1007/978-1-4939-9039-9_12
Lowder L, Malzahn A, Qi Y (2016) Rapid evolution of manifold CRISPR systems for plant genome editing. Front Plant Sci 7:1683. https://doi.org/10.3389/fpls.2016.01683
Luo S, Li J, Stoddard TJ et al (2015) Non-transgenic plant genome editing using purified sequence-specific nucleases. Mol Plant 8:1425–1427. https://doi.org/10.1016/j.molp.2015.05.012
Lusser M, Parisi C, Plan D et al (2012) Deployment of new biotechnologies in plant breeding. Nat Biotechnol 30:231–239. https://doi.org/10.1038/nbt.2142
Lyznik L, Kamo K, Grimes H et al (1989) Stable transformation of maize: the impact of feeder cells on protoplast growth and transformation efficiency. Plant Cell Rep 8(5):292–295. https://doi.org/10.1007/bf00274133
Ma J, Liu T, Qiu D (2015) Optimization of Agrobacterium-mediated transformation conditions for tomato (Solanum lycopersicum L.). Plant Omics J 8:529–536
Malzahn A, Lowder L, Qi Y (2017) Plant genome editing with TALEN and CRISPR. Cell Biosci 7:21. https://doi.org/10.1186/s13578-017-0148-4
Mao X, Zheng Y, Xiao K et al (2018) OsPRX2 contributes to stomatal closure and improves potassium deficiency tolerance in rice. Biochem Biophys Res Commun 495:461–467. https://doi.org/10.1016/j.bbrc.2017.11.045
Marx V (2018) Base editing a CRISPR way. Nat Methods 15:767–770
Maurya GP, Vivek P, Singh GP, Meena Kumar (2015) An economic analysis of cucumber cultivation in sultanpur district of Uttar Pradesh (India). Int J Agric Sci Res 5:23–28
McHughen A, Smyth S (2008) US regulatory system for genetically modified [genetically modified organism (GMO), rDNA or transgenic crop cultivars. Plant Biotechnol J 6:2–12
Meena HP, Bainsla NK, Yadav DK (2016) Breeding for abiotic stress tolerance in crop plants. In: Yadav P, Kumar S, Jain V (eds) Recent advances in plant stress physiology. Daya Publishing House, New Delhi, pp 329–379
Mehta D, Stürchler A, Anjanappa R et al (2019) Linking CRISPR-Cas9 interference in cassava to the evolution of editing-resistant geminiviruses. Genome Biol 20:80. https://doi.org/10.1186/s13059-019-1678-3
Mello-Farias PC, Chaves AL (2008) Advances in Agrobacterium-mediated plant transformation with enphasys on soybean. Sci Agric 65:95–106. https://doi.org/10.1590/s0103-90162008000100014
Miao J, Guo D, Zhang J et al (2013) Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res 23:1233–1236. https://doi.org/10.1038/cr.2013.123
Mikami M, Toki S, Endo M (2015) Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol Biol 88:561–572. https://doi.org/10.1007/s11103-015-0342-x
Mishra MK, Slater A (2012) Recent advances in the genetic transformation of coffee. Biotechnol Res Int 2012:580857. https://doi.org/10.1155/2012/580857
Mohammed S, Samad AA, Rahmat Z (2019) Agrobacterium-mediated transformation of rice: constraints and possible solutions. Rice Sci 26:133–146. https://doi.org/10.1016/j.rsci.2019.04.001
Motamayor JC, Risterucci AM, Lopez PA et al (2002) Cacao domestication I: the origin of the cacao cultivated by the Mayas. Heredity 89:380–386
Mushke R, Yarra R, Bulle M (2016) Efficient in vitro direct shoot organogenesis from seedling derived split node explants of maize (Zea mays L.). J Genet Eng Biotechnol 14:49–53. https://doi.org/10.1016/j.jgeb.2016.03.001
Naim F, Dugdale B, Kleidon J et al (2018) Gene editing the phytoene desaturase alleles of Cavendish banana using CRISPR/Cas9. Transgenic Res. https://doi.org/10.1007/s11248-018-0083-0
Nandy S, Pathak B, Zhao S et al (2019) Heat-shock-inducible CRISPR/Cas9 system generates heritable mutations in rice. Plant Direct 3:e00145. https://doi.org/10.1002/pld3.145
Nannas NJ, Dawe RK (2015) Genetic and genomic toolbox of Zea mays. Genetics 199:655–669. https://doi.org/10.1534/genetics.114.165183
Navrátilová B, Skálová D, Ondřej V et al (2011) Biotechnological methods utilized in Cucumis research—a review. Hortic Sci (Prague) 38:150–158
Nawaz G, Han Y, Usman B et al (2019) Knockout of OsPRP1, a gene encoding proline-rich protein, confers enhanced cold sensitivity in rice (Oryza sativa L.) at the seedling stage. 3 Biotech 9:254. https://doi.org/10.1007/s13205-019-1787-4
Newman PO, Krishnaraj S, Saxena PK (1996) Regeneration of tomato (Lycopersicon esculentum Mill.): somatic embryogenesis and shoot organogenesis from hypocotyl explaints induced with 6-benzyladenine. Int J Plant Sci 157:554–560. https://doi.org/10.1086/297375
Nhut DT, Le BV, Van Thanh KT (2000) Somatic embryogenesis and direct shoot regeneration of rice (Oryza sativa L.) using thin cell layer culture of apical meristematic tissue. J Plant Physiol 157:559–565. https://doi.org/10.1016/s0176-1617(00)80112-1
Nievola C, Carvalho C, Carvalho V et al (2017) Rapid responses of plants to temperature changes. Temperature 4:371–405. https://doi.org/10.1080/23328940.2017.1377812
Nihei N, Tanoi K, Nakanishi TM (2015) Inspections of radiocesium concentration levels in rice from Fukushima Prefecture after the Fukushima Dai-ichi Nuclear Power Plant accident. Scientific Reports 5:8653. https://doi.org/10.1038/srep08653
Nirwan RS, Kothari SL (2004) High frequency shoot organogenesis in Sorghum bicolor (L) Moench. J Plant Biochem Biotechnol 13:149. https://doi.org/10.1007/BF03263212
Odipio J, Alieai T, Ingelbrecht I et al (2017) Efficient CRISPR/Cas9 genome editing of phyteone desaturase in cassava. Front Plant Sci 8:1780. https://doi.org/10.3389/fpls.2017.01780
Oerke EC, Dehne HW (2004) Safeguarding production—losses in major crops and the role of crop protection. Crop Prot 23:275–285
Oria MP, Hamaker BR, Axtell JD et al (2000) A highly digestible sorghum mutant cultivar exhibits a unique folded structure of endosperm protein bodies. Proc Natl Acad Sci USA 97:5065–5070. https://doi.org/10.1073/pnas.080076297
Pan C, Ye L, Quin L et al (2016) CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci Rep 6:24765
Pandolfini T (2009) Seedless fruit production by hormonal regulation of fruit set. Nutrients 1:168–177. https://doi.org/10.3390/nu1020168
Parmar N, Singh KH, Sharma D et al (2017) Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: a comprehensive review. 3 Biotech 7:239
Paterson AH, Bowers JE, Bruggmann R et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556. https://doi.org/10.1038/nature07723
Paul JW, Qi Y (2016) CRISPR/Cas9 for plant genome editing: accomplishments, problems and prospects. Plant Cell Rep 35:1417–1427. https://doi.org/10.1007/s00299-016-1985-z
Peng A, Chen S, Lei T et al (2017) Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol J. https://doi.org/10.1111/pbi.12733
Pereira A (2016) Plant abiotic stress challenges from the changing environment. Front Plant Sci 7:1123. https://doi.org/10.3389/fpls.2016.01123
Perera I, Seneweera S, Hirotsu N (2018) Manipulating the phytic acid content of rice grain toward improving micronutrient bioavailability. Rice 11(1):4. https://doi.org/10.1186/s12284-018-0200-y
Pérez L, Dita M, Martínez E (2014) Prevention and diagnostic of Fusarium Wilt (Panama disease) of banana caused by Fusarium oxysporum f. sp. cubense Tropical Race 4 (TR4). http://www.fao.org/fileadmin/templates/banana/documents/Docs_Resources_2015/TR4/13ManualFusarium.pdf. Accessed 1 Sept 2019
Pérez L, Soto E, Farré G et al (2019) CRISPR/Cas9 mutations in the rice Waxy/GBSSI gene induce allele-specific and zygosity-dependent feedback effects on endosperm starch biosynthesis. Plant Cell Rep. https://doi.org/10.1007/s00299-019-02388-z
Petolino JF (2015) Genome editing in plants via designed zinc finger nucleases. In Vitro Cell Dev Biol Plant 51:1–8
Piron F, Nicolaï M, Minoïa S et al (2010) An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PLoS ONE 5(6):e11313. https://doi.org/10.1371/journal.pone.0011313
Porteus M (2016) Genome editing: a new approach to human therapeutics. Annu Rev Pharmacol Toxicol 56:163–190
Prochnik S, Marri PR, Desany B et al (2012) The cassava genome: current progress, future directions. Trop Plant Biol 5:88–94. https://doi.org/10.1007/s12042-011-9088-z
Puchta H (2017) Applying CRISPR/Cas for genome engineering in plants: the best is yet to come. Curr Opin Plant Biol 36:1–8. https://doi.org/10.1016/j.pbi.2016.11.011
Ray S, Lahiri S, Halder M et al (2015) An efficient method of isolation and transformation of protoplasts from tomato leaf mesophyll tissue using the binary vector pCambia 1302. Int Adv Res J Sci Eng Technol 2:146–150
Razali R, Bougouffa S, Morton MJL et al (2018) The genome sequence of the wild tomato Solanum pimpinellifolium provides insights into salinity tolerance. Front Plant Sci 9:1402. https://doi.org/10.3389/fpls.2018.01402
Reetu V, Maharishi T (2017) Watermelon: a valuable horticultural crop with nutritional benefits. Popular Kheti 5. www.popularkheti.com. Accessed 30 Aug 2019
Ren C, Guo Y, Gathunga EK et al (2019) Recovery of the non-functional EGFP-assisted identification of mutants generated by CRISPR/Cas9. Plant Cell Rep. https://doi.org/10.1007/s00299-019-02465-3
Ribas A, Pereira LFP, Vieira LGE (2006) Genetic transformation of coffee. Braz J Plant Physiol 18:83–94
Ricachenevsky FK, Vasconcelos MW, Shou H, Johnson AAT, Sperotto RA (2019) Improving the nutritional content and quality of crops: promises, achievements, and future challenges. Front Plant Sci. https://doi.org/10.3389/fpls.2019.00738
Ricroch A, Clairand P, Harwood W (2017) Use of CRISPR systems in plant genome editing: toward new opportunities in agriculture. Emerg Top Life Sci 1:169–182. https://doi.org/10.1042/ETLS20170085
Rivero R, Ruiz J, Garcı́a P et al (2001) Resistance to cold and heat stress: accumulation of phenolic compounds in tomato and watermelon plants. Plant Sci 160:315–321. https://doi.org/10.1016/s0168-9452(00)00395-2
Romero M, Gatica-Arias A (2019) CRISPR/Cas9: development and application in rice breeding. Rice Sci 26:265–281
Rosado A, Craig W (2017) Biosafety regulatory systems overseeing the use of genetically modified organisms in the Latin America and Caribbean region. AgBioForum 20:120–132
Rout GR, Samantaray S, Premananda D (2008) Biotechnology of the banana: a review of recent progress. Plant Biol 2:512–524. https://doi.org/10.1055/s-2000-7470
Ruma D, Dhaliwal MS, Kaur A et al (2009) Transformation of tomato using biolistic gun for transient expression of the β-glucuronidase gene. Indian J Biotechnol 8:363–369
Sander JD, Joung JK (2014) CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32:347–355
Sasaki A, Ashikari M, Ueguchi-Tanaka M et al (2002) A mutant gibberellin-synthesis gene in rice. Nature 416:701–702. https://doi.org/10.1038/416701a
Scheben A, Yuan Y, Edwards D (2016) Advances in genomics for adopting crops to climate change. Curr Plant Biol 6:2–10
Schindele P, Wolter F, Puchta H (2018) Transforming plant biology and breeding with CRISPR/Cas9, Cas12 and Cas13. FEBS Lett 592:1954–1967
Schubert R, Dobritzsch S, Gruber C et al (2019) Tomato MYB21 acts in ovules to mediate jasmonate-regulated fertility. Plant Cell. https://doi.org/10.1105/tpc.18.00978
Seyran E, Craig W (2018) New breeding techniques and their possible regulation. AgBioForum 21(1):1–12
Shan QW, Wang YP, Li J et al (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31:686–688
Shao X, Wu S, Dou T et al (2019) Using CRISPR/Cas9 genome editing system to create Ma GA 20ox2 gene modified semi-dwarf banana. Plant Biotechnol J. https://doi.org/10.1111/pbi.13216
Shi L, Fan JQ, Hu CG et al (2012) Improved production of transgenic Dioscorea zingiberensis (Dioscoreaceae) by Agrobacterium tumefaciens-mediated transformation. Genet Mol Res 11:244–253. https://doi.org/10.4238/2012
Shi Z, Zhang Y, Maximova SN et al (2013) TcNPR3 from Theobroma cacao functions as a repressor of the pathogen defense response. BMC Plant Biol 13:204. https://doi.org/10.1186/1471-2229-13-204
Shi J, Gao H, Wang H et al (2017) ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J 15:207–216. https://doi.org/10.1111/pbi.12603
Shu Y, Ying-Cai Y, Hong-Hui L (2005) Plant regeneration through somatic embryogenesis from callus cultures of Dioscorea zingiberensis. Plant Cell, Tissue Organ Cult 80:157–161. https://doi.org/10.1007/s11240-004-9543-8
Singh S, Rajam MV (2009) Citrus biotechnology: achievements, limitations and future directions. Physiol Mol Biol Plants 15:3. https://doi.org/10.1007/s12298-009-0001-2
Smyth SJ (2019) Global status of the regulation of genome editing technologies. https://www.cabi.org/cabreviews/review/20193130669. Accessed 30 Aug 2019
Sprink T, Eriksson D, Schiemann J, Hartung F (2016) Regulatory hurdles for genome editing: process- vs. product-based approaches in different regulatory contexts. Plant Cell Rep 35:1493–1506. https://doi.org/10.1007/s00299-016-1990-2
Sun Y, Zhang X, Wu C et al (2016) Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mol Plant 9:628–631
Sun Y, Jiao G, Liu Z et al (2017) Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front Plant Sci 8:298. https://doi.org/10.3389/fpls.2017.00298
Suratman F, Huyop F, Wagiran A et al (2010) Biolistic transformation of Citrullus vulgaris Schrad (Watermelon). Biotechnology 9:119–130
Svitashev S, Young JK, Schwartz C et al (2015) Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol 169:931–945. https://doi.org/10.1104/pp.15.00793
Svitashev S, Schwartz C, Lenderts B (2016) Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat Commun 7:13274. https://doi.org/10.1038/ncomms13274
Tadesse Y, Sági L, Swennen R et al (2003) Optimization of transformation conditions and production of transgenic sorghum (Sorghum bicolor) via microparticle bombardment. Plant Cell, Tissue Organ Cult 75:1. https://doi.org/10.1023/A:1024664817800
Tang W, Tang AY (2017) Applications and roles of the CRISPR system in genome editing of plants. J For Res 28:15–28. https://doi.org/10.1007/s11676-016-0281-7
Tang X, Lowder LG, Zhang T et al (2017) A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants 3:17018
Tashkandi M, Ali Z, Aljedaani F et al (2018) Engineering resistance against tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signal Behav 13:e1525996. https://doi.org/10.1080/15592324.2018.1525996
The 100 tomato Genome Sequencing Consortium, Aflitos S, Schijlen E et al (2014) Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant J 80:136–148. https://doi.org/10.1111/tpj.12616
Tian S, Jiang L, Gao Q et al (2017) Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep 36:399–406. https://doi.org/10.1007/s00299-016-2089-5
Tian S, Jiang L, Cui X et al (2018) Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep 37:1353–1356. https://doi.org/10.1007/s00299-018-2299-0
Toda E, Koiso N, Takebayashi A et al (2019) An efficient DNA- and selectable-marker-free genome-editing system using zygotes in rice. Nat Plants 5:363–368
Tör M, Ainsworth C, Mantell S (1993) Stable transformation of the food yam Dioscorea alata L. by particle bombardment. Plant Cell Rep 12:468–473. https://doi.org/10.1007/bf00234714
Tör M, Twyford CT, Funes I, Boccon-Gibod J et al (1998) Isolation and culture of protoplasts from immature leaves and embryogenic cell suspensions of Dioscorea yams: tools for transient gene expression studies. Plant Cell, Tissue Organ Cult 53:113–126. https://doi.org/10.1023/a:1006028406641
Tran H, Lee LS, Furtado A et al (2016) Advances in genomics for the improvement of quality in coffee. J Sci Food Agric 96:3300–3312
Tripathi L (2018) Prospects of genetic engineering and gene editing in yam improvement. https://pag.confex.com›xxvii›oid›Recording3992›paper37167_1. Accessed 31 Aug 2019
Tripathi J, Ntui V, Ron M et al (2019) CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun Biol. https://doi.org/10.1038/s42003-019-0288-7
Ueta R, Abe C, Watanabe T et al (2017) Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci Rep 7:507
Vanderschuren H, Alder A, Zhang P et al (2009) Dose-dependent RNAi-mediated geminivirus resistance in the tropical root crop cassava. Plant Mol Biol 70:265–272. https://doi.org/10.1007/s11103-009-9472-3
Wang K (2015) Agrobacterium protocols. Methods Mol Biol. https://doi.org/10.1007/978-1-4939-1695-5
Wang M, Wang G, Ji J et al (2009) The effect of pds gene silencing on chloroplast pigment composition, thylakoid membrane structure and photosynthesis efficiency in tobacco plants. Plant Sci 177:222–226. https://doi.org/10.1016/j.plantsci.2009.04.006
Wang S, Seong SK, Ye X, He C, Suk YK, Pil SC (2015) Current status of genetic transformation technology developed in cucumber (Cucumis sativus L.) WANG. J Integr Agric 14:469–482
Wang F, Wang C, Liu P et al (2016) Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OSERF922. PLoS ONE 11:e0154027. https://doi.org/10.1371/journal.pone.0154027
Wang L, Chen L, Li R et al (2017) Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J Agric Food Chem 65:8674–8682. https://doi.org/10.1021/acs.jafc.7b02745
Wang B, Zhu L, Zhao B, Zhao Y et al (2019a) Development of a haploid-inducer mediated genome editing (IMGE) system for accelerating maize breeding. Mol Plant. https://doi.org/10.1016/j.molp.2019.03.006
Wang B, Xie G, Liu Z et al (2019b) Mutagenesis reveals that the OsPPa6 gene is required for enhancing the alkaline tolerance in rice. Front Plant Sci. https://doi.org/10.3389/fpls.2019.00759
Wang X, Yang G, Shi M et al (2019c) Disruption of an amino acid transporter LHT1 leads to growth inhibition and low yields in rice. BMC Plant Biol. https://doi.org/10.1186/s12870-019-1885-9
Wang R, Tavano EC, Lammers M et al (2019d) Re-evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9-mutagenesis. Sci Rep. https://doi.org/10.1038/s41598-018-38170-6
Wickramasuriya AM, Dunwell JM (2018) Cacao biotechnology: current status and future prospects. Plant Biotechnol J 16:4–17
Wolt JD, Wang K, Sashital D et al (2016) Achieving plant CRISPR targeting that limits off-target effects. Plant Genome 9:1–8. https://doi.org/10.3835/plantgenome2016.05.0047
Wolter F, Puchta H (2018) The CRISPR/Cas revolution reaches the RNA world: Cas13, a new Swiss Army knife for plant biologists. Plant J 94:767–775
Woo JW, Kim J, Kwon SI et al (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33:1162–1164. https://doi.org/10.1038/nbt.3389
Wright M, Dawson J, Dunder E et al (2001) Efficient biolistic transformation of maize (Zea mays L.) and wheat (Triticum aestivum L.) using the phosphomannose isomerase gene, pmi, as the selectable marker. Plant Cell Rep 20:429–436. https://doi.org/10.1007/s002990100318
Wu G, Zhao Y, Shen R et al (2019) Characterization of maize phytochrome-interacting factors in light signaling and photomorphogenesis. Plant Physiol. https://doi.org/10.1104/pp.19.00239
Xie K, Yang Y (2013) RNA-guided genome editing in plants using a CRISPR-Cas system. Mol Plant 6:1975–1983. https://doi.org/10.1093/mp/ssT119
Xu Q, Chen LL, Ruan X et al (2012) The draft genome of sweet orange (Citrus sinensis). Nat Genet 45:59–66. https://doi.org/10.1038/ng.2472
Xu R, Li H, Qin R et al (2014) Gene targeting using the Agrobacterium tumefaciens-mediated CRISPR-Cas system in rice. Rice. https://doi.org/10.1186/s12284-014-0005-6
Yang Y, Tang K, Datsenka TU et al (2019) Critical function of DNA methyltransferase 1 in tomato development and regulation of the DNA methylome and transcriptome. J Integr Plant Biol. https://doi.org/10.1111/jipb.12778
Ye Y, Li P, Xu T et al (2017) OsPT4 contributes to arsenate uptake and transport in rice. Front Plant Sci. https://doi.org/10.3389/fpls.2017.02197
Yin X, Biswal AK, Dionora J et al (2017) CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep 36:745–757. https://doi.org/10.1007/s00299-017-2118-z
Yoshida KT, Fujii S, Sakata M et al (1994) Control of organogenesis and embryogenesis in rice calli. Ikushugaku Zasshi 44:355–360. https://doi.org/10.1270/jsbbs1951.44.355
Yu J, Wang J, Lin W et al (2005) The genomes of Oryza sativa: a history of duplications. PLoS Biol 3:e38. https://doi.org/10.1371/journal.pbio.0030038
Yu W, Wang L, Zhao R et al (2019) Knockout of SlMAPK3 enhances tolerance to heat stress involving ROS homeostasis in tomato plants. BMC Plant Biol. https://doi.org/10.1186/s12870-019-1939-z
YuChun R, Xu N, Li S et al (2019) PE-1, encoding heme oxygenase 1, impacts heading date and chloroplast development in rice (Oryza sativa. L). J Agric Food Chem. https://doi.org/10.1021/acs.jafc.9b01676
Zenpei S, Sachiko K, Mariko T et al (2017) Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol 35:441–443
Zhang Y, Liang Z, Zong Y et al (2016) Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun 7:12617. https://doi.org/10.1038/ncomms12617
Zhang H, Zhang J, Lang Z et al (2017a) Genome editing—principles and applications for functional genomics research and crop improvement. Crit Rev Plant Sci 36:291–309
Zhang F, LeBlanc C, Irish VF et al (2017b) Rapid and efficient CRISPR/Cas9 gene editing in Citrus using the YAO promoter. Plant Cell Rep 36:1883–1887. https://doi.org/10.1007/s00299-017-2202-4
Zhang Q, Xing HL, Wang ZP et al (2018a) Potential high-frequency off-target mutagenesis induced by CRISPR/Cas9 in Arabidopsis and its prevention. Plant Mol Biol 96:445–456
Zhang X, Jin M, Tadesse N et al (2018b) Dioscorea zingiberensis C. H. Wright: an overview on its traditional use, phytochemistry, pharmacology, clinical applications, quality control, and toxicity. J Ethnopharmacol 220:283–293. https://doi.org/10.1016/j.jep.2018.03.017
Zhang Z, Guo J, Zhao Y et al (2019) Identification and characterization of maize ACD6-like gene reveal ZmACD6 as the maize orthologue conferring resistance to Ustilago maydis. Plant Signal Behav. https://doi.org/10.1080/15592324.2019.1651604
Zhong Y, Liu C, Qi X et al (2019) Mutation of ZmDMP enhances haploid induction in maize. Nat Plants 5:575–580. https://doi.org/10.1038/s41477-019-0443-7
Zhou W, Li B, Li L et al (2018a) Genome survey sequencing of Dioscorea zingiberensis. Genome 61:567–574. https://doi.org/10.1139/gen-2018-0011
Zhou L, Peng R, Zhang R et al (2018b) The applications of CRISPR/Cas system in molecular detection. J Cell Mol Med 22:5807–5815. https://doi.org/10.1111/jcmm.13925
Zong Y, Wang Y, Li C et al (2017) Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol 5:438–440. https://doi.org/10.1038/nbt.3811
Zou T, Liu M, Xiao Q et al (2018) OsPKS2 is required for rice male fertility by participating in pollen wall formation. Plant Cell Rep 37:759–773. https://doi.org/10.1007/s00299-018-2265-x
Zsögön A, Čermák T, Naves ER et al (2018) De novo domestication of wild tomato using genome editing. Nat Biotechnol. https://doi.org/10.1038/nbt.4272
Acknowledgements
This work was financed by “Espacio de Estudios Avanzados de la Universidad de Costa Rica” (UCREA Project No. 801-B7-294). The authors would like to thank Prof. Dr. Stefan Schillberg (Fraunhofer IME, Aachen, Germany) for his helpful comments on this review.
Author information
Authors and Affiliations
Contributions
R.R.-V. researched and wrote the manuscript; A.G.-A. conceived the manuscript, wrote, and edited the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest. All authors read and approved the final review.
Additional information
Communicated by Degao Liu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rojas-Vásquez, R., Gatica-Arias, A. Use of genome editing technologies for genetic improvement of crops of tropical origin. Plant Cell Tiss Organ Cult 140, 215–244 (2020). https://doi.org/10.1007/s11240-019-01707-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-019-01707-2