Abstract
Key message
Induced mutations in the waxy locus in rice endosperm did not abolish GBSS activity completely. Compensatory mechanisms in endosperm and leaves caused a major reprogramming of the starch biosynthetic machinery.
Abstract
The mutation of genes in the starch biosynthesis pathway has a profound effect on starch quality and quantity and is an important target for plant breeders. Mutations in endosperm starch biosynthetic genes may impact starch metabolism in vegetative tissues such as leaves in unexpected ways due to the complex feedback mechanisms regulating the pathway. Surprisingly this aspect of global starch metabolism has received little attention. We used CRISPR/Cas9 to introduce mutations affecting the Waxy (Wx) locus encoding granule-bound starch synthase I (GBSSI) in rice endosperm. Our specific objective was to develop a mechanistic understanding of how the endogenous starch biosynthetic machinery might be affected at the transcriptional level following the targeted knock out of GBSSI in the endosperm. We found that the mutations reduced but did not abolish GBSS activity in seeds due to partial compensation caused by the upregulation of GBSSII. The GBSS activity in the mutants was 61–71% of wild-type levels, similarly to two irradiation mutants, but the amylose content declined to 8–12% in heterozygous seeds and to as low as 5% in homozygous seeds, accompanied by abnormal cellular organization in the aleurone layer and amorphous starch grain structures. Expression of many other starch biosynthetic genes was modulated in seeds and leaves. This modulation of gene expression resulted in changes in AGPase and sucrose synthase activity that explained the corresponding levels of starch and soluble sugars.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Starch is the major component of rice endosperm and comprises a mixture of the polysaccharides amylose and amylopectin. Amylose is a linear chain of α(1,4)-linked glucose monomers whereas amylopectin contains additional α(1,6)-linked branches every 24–30 residues (Martin and Smith 1995). Starch from different plant species varies in its physicochemical properties due to the ratio of amylose and amylopectin and differences in chain length and/or amylopectin branching density (Jobling 2004). Differences in enzyme activities may thus induce changes in starch composition.
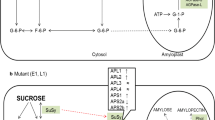
Starch synthesis in plants begins with the conversion of glucose 1-phosphate to ADP-glucose, catalyzed by ADP-glucose pyrophosphorylase (AGPase). This is followed by the polymerization of ADP-glucose to form amylose and amylopectin via the coordinated activity of AGPase and starch synthases (SSs), which form the α(1,4)-linked glycosidic bonds of both molecules, and starch-branching enzymes (SBEs), which form the α(1,6)-linked glycosidic bonds in amylopectin. Starch synthases catalyze the transfer of the glucosyl moiety of the soluble precursor ADP-glucose to the reducing end of a pre-existing α(1,4)-linked glucan primer (Tetlow et al. 2004) whereas SBEs cleave internal amylose α(1,4) glycosidic bonds and transfer the reducing ends to C6 hydroxyls to form α(1,6) linkages (Jiang et al. 2013). The latter can be removed by starch debranching enzyme isoamylase, hence the amylopectin content of starch is sensitive to the balance between branching and debranching activities. The structure of amylopectin is also influenced by the two different SBE isoforms, with SBEI showing higher affinity towards amylose than amylopectin and a preference for longer glucan chains and SBEII showing the opposite properties (Tetlow et al. 2004). Disproportionating enzyme (DPE) and starch phosphorylase (PHO) also play a role in the initiation and elongation of starch polymers (Satoh et al. 2008). The pathway is summarized in Fig. 1.
The coordination of different starch biosynthesis genes in rice (modified from Thitisaksakul et al. 2012). SuSy sucrose synthase, G-1-P glucose-1-phosphate, G-6-P glucose-6-phosphate, ADP-Glu ADP-glucose, PPi inorganic diphosphate, F-6-P fructose-6-phosphate, ATP adenosine triphosphate, ADP adenosine diphosphate, UDP uridine diphosphate, PHO α-glucan phosphorylase, AGPase ADP-glucose pyrophosphorylase, ISAs isoamylase-type starch debranching enzymes, PUL pullulanase, SS starch synthases, GBSSI granule-bound starch synthase 1, SBEs starch-branching enzymes
There are two major groups of starch synthases (Fig. 1). The first group is the classical starch synthases and comprises four isoforms, some represented by multiple paralogs: SSI, SSIIa/b/c, SSIIIa/b and SSIVa/b (Nakamura 2002). These synthesize the linear chains of amylopectin and their distribution between granular and stromal fractions can vary between species, tissues and developmental stages (Ball and Morell 2003). The second group is the granule-bound starch synthases (GBSS) which are restricted to the granule matrix. There are two isoforms, GBSSI and GBSSII, the first expressed mainly in the endosperm and the second mainly in the leaves (Ohdan et al. 2005). GBSSI catalyzes the extension of long glucan chains primarily in amylose (Maddelein et al. 1994). The GBSSI protein is 609 amino acids in length, with a catalytic site spanning residues 78–609 composed of α-helices and β-sheets that form a substrate-binding cleft. An N-terminal transit peptide outside the catalytic center is required to import the protein into the starch granules (Momma and Fujimoto 2012).
In rice, GBSSI is encoded by the Waxy (Wx) gene, so named because of the waxy appearance of the amylose-free grain in Wx null mutants (Hirano 1993). As well as the Wx null allele, two other natural alleles are common in rice (Sano 1984). The Wxa allele is predominant in indica subspecies, and has strong GBSSI activity, and thus more amylose in the endosperm. Wxb has weaker GBSSI activity and the amount of amylose and amylopectin in the endosperm is more variable (Hirano and Sano 1998; Umemoto and Terashima 2002). Because of its impact on grain quality, Wx is an important target in rice quality improvement programs and in studies of starch biosynthesis and metabolism (Tran et al. 2011; Zhang et al. 2012). The Wx gene has been modified by conventional mutagenesis such as irradiation, chemical mutagenesis and T-DNA/transposon insertional mutagenesis but these approaches generate random lesions and large populations must be screened to isolate useful mutants. Such techniques have largely been supplanted by targeted mutagenesis using designer nucleases, particularly CRISPR/Cas9, because this can modify the target gene without altering agronomic traits (reviewed by Bortesi et al. 2016; Zhu et al. 2017).
Several previous studies have targeted GBSSI in rice using the CRISPR/Cas9 system (Ma et al. 2015; Zhang et al. 2018). However, these studies did not consider the broader impact on starch metabolism, reflecting feedback mechanisms that may be activated to restore homeostasis in the developing seed. In one report (Ma et al. 2015), wild-type Cas9 (Cas9WT) was used to target three different sites simultaneously of GBSSI gene, although only plants with one or two target site mutations were recovered. The amylose content in the T1 seeds was reduced from 14.6 to 2.6% (Ma et al. 2015). More recently, four Wx frameshift mutants were generated using CRISPR/Cas9 and the proportion of amylose in the seeds was reduced without affecting the overall starch content or agronomic properties such as seed number, yield and size (Zhang et al. 2018). The starch pathway is tightly regulated by the ratio of triose-phosphate to inorganic phosphate, and the disruption of this balance may lead to further changes in the expression of other genes involved in starch metabolism (Preiss 1982). In a Wx mutant generated by irradiation (GM077), the loss of GBSSI activity and lower starch and amylose levels induced the expression of GBSSII, AGPases, starch synthases and isoamylases (Zhang et al. 2012). Given that the mutation of endosperm-specific GBSSI is viewed as a good strategy to modulate amylose production in the endosperm without affecting amylose metabolism in vegetative tissues, impact on GBSSII expression (affecting starch biosynthesis in the leaves) and on enzymes related to amylopectin biosynthesis and starch degradation needs to be investigated in detail.
We hypothesized that at the protein level GBSSI enzyme activity might be lost in part or entirely depending on the nature of the mutation. In the former case we anticipated endosperm amylose levels to be reduced with no further impact on starch metabolism, as reported in other studies (Ma et al. 2015; Zhang et al. 2018). We, therefore, used the CRISPR/Cas9 system to generate truncated (nonfunctional) or partially active GBSSI mutants in rice endosperm and carried out a comprehensive analysis of starch and soluble sugar levels, grain morphology and analyzed the expression of a number of genes involved in starch metabolism. Our overarching objective was to establish a mechanistic basis to support further targeted interventions to generate rice grains with varying starch content and composition for various applications.
Materials and methods
Target sites and sgRNA design
Target sites for Cas9WT (single sgRNA) and Cas9 nickase (Cas9D10A, two sgRNAs targeting adjacent sites) were selected within the rice Wx coding sequence (GenBank EU735072.1) using E-CRISP (Heigwer et al. 2014) with the following parameters: only NGG PAM, only G as the 5′ base, off-target tolerates many mismatches, non-seed region ignored, introns ignored. These parameters were selected to minimize off-target cleavage. The catalytic efficiency of each sgRNAs was predicted using gRNA scorer (Chari et al. 2015). The following three targets sites were selected: TS1 = 5′-GTCGGCGATGCCGAAGC↓CGGTGG-3′ (D10A CRISPR/Cas9, targeted nucleotides 1337–1359), TS2 = 5′-GCTGCTCCGCCACGGGT↓TCCAGG-3′ (D10A CRISPR/Cas9, targeted nucleotides 1377–1399) and TS3 = 5′-CCGGCTTCGGCATCGCC↓GACAGG-3′ (WT CRISPR/Cas9, targeted nucleotides 5082–5104), where the arrows indicate the expected site of the DSB.
Vector construction
The Cas9WT vector pJIT163-2NSCas9 and the sgRNA vector pU3-gRNA were obtained from Dr. C. Gao, Chinese Academy of Sciences, Beijing, China (Shan et al. 2014). The nickase vector pJIT163-2NSCas9D10A was constructed in-house by mutating the cas9 gene in vector pJIT163-2NSCas9 to produce Cas9D10A and combining it with the maize ubiquitin-1 promoter and Cauliflower mosaic virus 35S terminator. The three sgRNAs described above were prepared as synthetic double-stranded oligonucleotides and introduced separately into pU3-gRNA at the AarI restriction site allowing all genomic sites with the form 5′-(20)-NGG-3′ to be targeted. The hpt selectable marker gene was provided on a separate vector as previously described (Christou et al. 1991).
Rice transformation and recovery of transgenic plants
Seven-day-old mature zygotic embryos (Oryza sativa ssp. Japonica cv. EYI) were transferred to osmotic medium (MS medium supplemented with 0.3 g/L casein hydrolysate, 0.5 g/L proline, 72.8 g/L mannitol and 30 g/L sucrose) 4 h before bombardment with 10 mg gold particles coated with the transformation vectors. The Cas9 vector (wild type or nickase), the corresponding sgRNA vector(s) and the selectable marker hpt were introduced at a 3:3:1 molar ratio (Cas9WT:sgRNA:hpt) or a 3:3:3:1 molar (Cas9D10A:sgRNA1:sgRNA2:hpt) as previously described (Sudhakar et al. 1998; Valdez et al. 1998). The embryos were returned to osmotic medium for 12 h before selection on MS medium supplemented with 0.3 g/L casein, 0.5 g/L proline, 30 g/L sucrose, 50 mg/L hygromycin and 2.5 mg/L 2,4-dichlorophenoxyacetic acid in the dark for 2–3 weeks. Callus was maintained on selective medium for 6 weeks with sub-culturing every 2 weeks as described (Farré et al. 2012). Transgenic plantlets were regenerated and hardened off in soil. For negative controls, we regenerated negative transformants (also bombarded with Cas9WT/Cas9D10A, hpt and the appropriate sgRNAs but which did not undergo mutation) at the same time as the mutated lines. We compared two Wx Oryza sativa ssp. Japonica mutants [KUR and Mushashimochi (Musa)] with our mutant lines.
Confirmation of the presence of cas9 and gRNA in regenerated rice plants
Genomic DNA was isolated from leaves of regenerated plants by phenol extraction and ethanol precipitation (Bassie et al. 2008; Kang and Yang 2004). The presence of the Cas9WT sequence was confirmed by PCR using primers 5′-GTC CGA TAA TGT GCC CAG CGA-3′ and 5′-GAA ATC CCT CCC CTT GTC CCA-3′. The presence of the Cas9D10A sequence was determined using primers 5′-GCA AAG AAC TTT CGA TAA CGG CAG CAT CCC TCA CC-3′ and 5′-CCT TCA CTT CCC GGA TCA GCT TGT CAT TCT CAT CGT-3′. The presence of the pU3-gRNA vectors was confirmed using the conserved primers 5′-TTG GGT AAC GCC AGG GTT TT-3′ and 5′-TGT GGA TAG CCG AGG TGG TA-3′.
Analysis of induced mutations
The Wx mutations induced by Cas9WT and Cas9D10A were detected by PCR using primers 5′-GGG TGC AAC GGC CAG GAT ATT-3′ and 5′-TGA AGA CGA CGA CGG TCA GC-3′. The PCR products were sequenced using an ABI 3730xl DNA analyzer by Stabvida (http://www.stabvida.com/es/). To confirm the mutations, PCR products generated using the primers listed above were purified using the Geneclean II Kit (MP Biomedicals), transferred to the pGEM-T Easy vector (Promega) and introduced into competent Escherichia coli cells. PCR fragments of ~ 760 bp were selected, purified and sequenced using an ABI 3730xl DNA analyzer by Stabvida. At least five clones per PCR product were sequenced using primer M13Fwd.
Protein structural modeling and phylogenetic analysis
The GBSSI sequences were translated into polypeptides (http://web.expasy.org/translate/) and automated homology modeling was carried out using Phyre2 (Kelley et al. 2015) with rice GBSSI catalytic domain (Protein Databank: 3vuf) as the template. The model of the mutant protein was superimposed on the wild-type version using DS Visualizer (http:/accelrys.com/products/collaborative-science/biovia-discovery-studio/visualization.html). The structure with the ADP and malto-oligosaccharide was docked to the protein using swissdock (http://www.swissdock.ch/docking). Sequence alignment and phylogenetic tree construction was carried out using the phylogeny.fr (Dereeper et al. 2008) server (http://www.phylogeny.fr/index.cgi) with default parameters.
Enzymatic activity assays
Leaves were cut into discs and immersed in 2% paraformaldehyde, 2% polyvinylpyrrolidone 40 (pH 7) for 2.5 h at 4 °C before washing three times in distilled water. AGPase and SuSy activity was then measured using a proprietary kit, according to the supplier’s instructions (CSIC 2016).
The GBSS activity in 10 pooled frozen seeds was determined according to Nakamura et al. (1989) and Jiang et al. (2004). The seeds were weighed and homogenized in 10 mL ice-cold 50 mM HEPES–NaOH (pH 7.5) containing 10 mM MgCl2, 2 mM EDTA, 50 mM 2-mercaptoethanol, 12.5% (v/v) glycerol and 5% (w/v) polyvinylpyrrolidone 40. Thirty (30) µL of the homogenate were added to 1.8 mL of the same buffer and centrifuged at 2000×g for 20 min at 4 °C. The pellet was resuspended in 2 mL of GBSS assay buffer (100 µL 14 mM ADP-glucose and 700 µL 50 mg/mL amylopectin). After incubation at 30 °C for 5 min, the reaction was initiated by adding 50 µL of 40 mM phosphoenolpyruvate, 50 mM MgCl2, and 1 IU pyruvate kinase, incubated at 30 °C for 30 min, and was stopped after 20 min by heating in a boiling water bath. A control sample was prepared by boiling the enzyme extract before starting the reaction, to determine the background signal. The ADP produced by GBSS was converted to ATP by the action of pyruvate kinase. The ATP was measured by adding 5 mL of luciferin reagent to 50 µL of the enzymatic reaction after the production of ATP and measuring the emission at 370–630 nm in a Berthold FB 12 luminometer.
Starch and soluble sugars
Flag leaf samples harvested at 7 pm were homogenized under liquid nitrogen and extracted in perchloric acid to measure the starch content, or in ethanol to measure the content of soluble sugars. The quantity of each carbohydrate was determined by spectrophotometry at 620 nm (Juliano 1971; Yoshida et al. 1976). To measure the amylose content, milled-rice grains were powdered with a faience pestle and mortar and the powder was transferred to a paper envelope and dried for 1 h at 135 °C. We discarded transparent seeds of the mutated lines in the T1 generation as they represented the wild-type phenotype. All remaining seeds were opaque. We pooled 20 such seeds and processed them to powder for all subsequent analyses. We transferred 100 ± 0.01 mg of dried powder to a conical flask and added 1 mL 95% ethanol and 9 mL 1 M NaOH. The suspension was boiled in a water bath for 10 min, cooled at room temperature for 10 min and then topped up to 100 mL with distilled water. A 5-mL aliquot of the solution was transferred to a 100-mL volumetric flask and mixed with 1 mL 1 M acetic acid, 2 mL 0.2% potassium iodide and 92 mL distilled water. Three amylose solutions (3%, 11.5% and 14%) were prepared for comparison. The starch content was determined by measuring the absorbance at 630 nm in a Unicam UV4-100 UV–Vis spectrophotometer after 30 min.
RNA extraction and real-time quantitative RT-PCR analysis
Total leaf/seed RNA was isolated using the RNeasy Plant Mini Kit (Qiagen) and DNA was digested with DNase I from the RNase-free DNase Set (Qiagen). Total RNA was quantified using a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific) and 2 µg total RNA was used as template for first strand cDNA synthesis with Quantitech reverse transcriptase (Qiagen) in a 20-µL total reaction volume, following the manufacturer’s recommendations. Real-time qRT-PCR was performed on a BioRad CFX96 system using 20-µL mixtures containing 5 ng cDNA, 1x iQ SYBR green supermix and 0.5 µM forward and reverse primers. The OsAPL1, OsAPL3, OsAPL4, OsAPS1, OsAPS2a/b, OsAPL2, OsSSI, OsSSIIa, OsSSIIb, OsSSIIc, OsSSIIIa, OsSSIIIb, OsSSIVa, OsSSIVb, OsGBSSI, OsGBSSII, OsBEI, OsBEIIa, OsBEIIb, OsISA1, OsISA2, OsISA3, OsPUL, OsDPE1, OsDPE2, OsPHOH and OsPHOL cDNAs were amplified using appropriate primers (Ohdan et al. 2005) as described by Tang et al. (2016). Serial dilutions of cDNA (80–0.0256 ng) were used to generate standard curves for each gene. PCR was performed in triplicate using 96-well optical reaction plates. Values represent the mean of three biological replicates ± SE. Amplification efficiencies were compared by plotting the ΔCt values of different primer combinations of serial dilutions against the log of starting template concentrations using the CFX96 software. The rice housekeeping OsUBQ5 (LOC_Os01g22490) gene was used as an internal control.
Seed phenotype and microscopy
The seed hull was removed to observe the external appearance of the grain in mutant lines using a magnifying lens. Thin sections (2 µm) of rice endosperm and leaves were prepared with a diamond knife using a Reichert Jung Ultramicrotome Ultracut E and were mounted on glass slides for analysis under a Zeiss Axioplan light microscope coupled to a Leica DC 200 digital camera. Tissue for sectioning was prepared by embedding in glycol methacrylate using the Technovit 7100 kit according to the manufacturer’s protocol (Kulzer, Hanau, Germany). Grains were cut through the center to expose the endosperm. Two drops of 1.0% Richardson Blue were placed on the endosperm surface and images were captured after 3–5 min. Rice seeds were fractured in half and mounted in stubs with carbon tape to keep them vertical. They were then processed for scanning electron microscopy (SEM) by dehydration at 60 °C for 24 h followed by carbon coating using an Edwards Auto 306 and gold sputtering using a Balzers SCD050 Sputter Coater. The samples were stored at 60 °C or in a vacuum chamber before analysis on a Jeol JSM-6300. Seed length, width and thickness were measure using Digimatic Caliper CD-6″CX. Seed volume was calculated multiplying length, width and thickness of seeds.
Statistical analysis
A general linear model was used to determine statistically significant differences in normalized expression of starch pathway genes. All the analyses were performed using the JMP Pro (JMP, SAS Institute Inc., Cary, NC, 2013). Five-factorial analysis of variance (ANOVA) with tissue, gene, genotype, gene_type and isoform as random factors was applied per normalized expression on log-transformed data. Other statistical analysis were determine by Student’s test.
Accession numbers
GenBank EU735072.1 (OsWaxy gene sequence)
UniProtKB/Swiss-Prot: P04713.1 (Waxy protein Oryza sativa subsp. indica)
UniProtKB/Swiss-Prot: Q0DEV5 (Waxy protein Oryza sativa subsp. japonica)
UniProtKB/Swiss-Prot: P09842 (Waxy protein Hordeum vulgare (barley))
UniProtKB/Swiss-Prot: P27736 (Waxy protein Triticum aestivum (wheat))
UniProtKB/Swiss-Prot: Q9MAQ0 (Waxy protein Arabidopsis thaliana)
UniProtKB/Swiss-Prot: A1YZE0 (Waxy protein Glycine max (soybean))
UniProtKB/Swiss-Prot: M9Q2A3 (Waxy protein Nicotiana tabacum (tobacco))
UniProtKB/Swiss-Prot: D2D315 (Waxy protein Gossypium hirsutum (cotton))
UniProtKB/Swiss-Prot: K4CPX6 (Waxy protein Solanum lycopersicum (tomato)).
Results
Recovery and characterization of GBSSI mutants
We analyzed six induced mutant lines in detail firstly in T1 seeds (Table 1). PCR analysis revealed that line 1 featured the substitution of a single nucleotide at site TS1 (Supplementary Figure S1a). The remaining five lines all featured mutations at sites TS2/TS3. Lines 2, 4 and 5 featured deletions of 55, 56 and 246 nucleotides, respectively, line 3 featured an insertion of 28 nucleotides, and line 6 featured a 2-nucleotide substitution (Supplementary Figure S1b–f). In T0 plants, all the lines contained heterozygous mutations. In T1 plants, all the lines contained heterozygous mutations except line 2, in which the mutation was homozygous. In T2 plants, all the lines contained homozygous mutations. As well as testing for on-target mutations, E-CRISP identified potential off-target cleavage sites at three loci based on the number of mismatches allowed in the target sequence and 2 bp upstream of the double-strand break (DSB). The three potential off-target sites were identified for TS1 in chromosomes 1, 5 and 7, but sequencing of these loci revealed no evidence of off-target mutations. No off-target sites were predicted for TS2 and TS3.
Structural comparisons and phylogenetic analysis of protein sequence
To investigate potential changes at the protein level, we translated the mutant sequences (Supplementary Figure S2) and generated 3D models using the SWISS-MODEL program (Supplementary Figure S3). The line 1 missense mutation resulted in the amino acid substitution Q33H, lines 2–4 featured indels and concurrent frameshifts that generated a severely truncated and nonfunctional protein, the indel in line 5 removed the N-terminal portion of the protein without affecting the catalytic site, and line 6 was a synonymous substitution with no effect on protein structure.
The wild-type protein structure contains binding clefts for ADP and malto-oligosaccharides. Although there was little overall structural change in the line 1 mutant, superimposing the mutant sequence over that of the wild-type protein revealed surface changes that constricted the malto-oligosaccharide pocket (indicated with black arrows) and prevented these substrates accessing the catalytic center (Fig. 2). Furthermore, amino acids 1–77 are essential for the import of GBSSI into starch granules so we compared the sequence of GBSSI enzymes from other plants to determine whether the Q33H substitution removed a functionally critical residue. We constructed a phylogenetic tree from the GBSSI sequences of the japonica and indica rice subspecies, as well as barley, wheat, tomato, tobacco, cotton, soybean and Arabidopsis thaliana, which revealed that Q33 is highly conserved among cereals but not in dicots (Fig. 3). These data suggest that Q33 may be required for the efficient import of GBSSI into starch granules in cereals and that the line 1 mutant may, therefore, suffer from the inefficient import of the enzyme into starch granules.
Structure of GBSSI in wild-type rice and mutant line 1. a Superimposition of wild-type GBSSI (pink) and the line 1 mutant (green). The principal structural differences are encircled by dashed lines. b Wild-type surface model with ADP and malto-oligosaccharide in the catalytic site indicated with a black arrow. c Line 1 mutant surface model with ADP and malto-oligosaccharide in the catalytic site indicated with a black arrow. d Superimposition of wild-type GBSSI (pink) and the line 1 mutant (green) with ADP (yellow) and malto-oligosaccharide (orange) in the catalytic site. (Color figure online)
Sequence analysis of the GBSSI protein. a Sequence alignment of GBSSI proteins from various monocot and dicots plants. Highly conserved residues are shaded in blue and moderately conserved residues in gray. The red box highlights residue 33, which is mutated in line 1. b Phylogenetic tree generated from the aligned sequences using the Phylogeny.fr server with default parameters. The UniProt accession numbers of the sequences are: Oryza sativa subsp. japonica (rice) Q0DEV5; Oryza sativa subsp. indica (rice) P04713; Hordeum vulgare (barley) P09842; Triticum aestivum (wheat) P27736; Arabidopsis thaliana Q9MAQ0, Glycine max (soybean) A1YZE0; Nicotiana tabacum (tobacco) M9Q2A3; Gossypium hirsutum (cotton) D2D315; and Solanum lycopersicum (tomato) K4CPX6. (Color figure online)
Changes in enzymatic activity of GBSS by the loss of GBSSI activity
GBSS activity was measured in the T2 seeds of the six mutant lines and compared to wild-type seeds as well as two Wx Oryza sativa ssp. Japonica mutants, namely KUR generated by exposure to neutrons (Yatou and Amano 1991) and Musashimochi generated by exposure to gamma rays (Itoh et al. 1997). Consistent with the observation that line 6 did not exhibit any changes at the protein level, there was no significant difference between line 6 and wild-type seeds, given that the mutation had no effect on the protein sequence or structure. In the other five mutant lines, the GBSS activity fell by 61–71% compared to wild-type activity, which was similar to the GBSS activity in Musa and slightly lower than the activity in KUR (Fig. 4a).
Enzyme activity in wild-type and mutant rice plants. a GBSS1 activity in T2 seeds of wild-type (WT) plants, the Wx mutant lines and the two Wx irradiation mutants, KUR and Musa. b AGPase activity in the T0 flag leaves of WT and mutant lines. c Sucrose synthase activity in the T0 flag leaves of WT and mutant lines. Values are means ± SDs (n = 3 biological replicates, 2 technical replicates for each biological replicate). The asterisk indicates a statistically significant difference between WT and mutant, as determined by Student’s t test (*P < 0.05, **P < 0.001)
We also investigated whether the loss of GBSS activity in the mutants affected the activity of AGPase and sucrose synthase in T2 seeds. Most of the remaining lines, as well as the KUR and Musa mutants, showed a 10–30% drop in AGPase activity, but in lines 1 and 3 the activity of AGPase increased by 30–40%; line 6 showed similar AGPase/Susy activity compared to the wild-type. Although the trends were clear, these differences were not statistically significant. Conversely, almost all mutant lines, as well as KUR and Musa, showed a > 50% increase in soluble sucrose synthase activity, with a statistically significant > 100% increase in line 3. Line 5 showed a modest increase of 28%, and line 1 showed a 50% decrease compared to wild-type plants, but these differences were only statistically significant in line 3 (Fig. 4b, c).
As reported earlier there is a correlation between AGPase and SuSy activities, where Susy provides an alternative route for the synthesis of ADP-glucose which might be induced by GBSSI mutations (Pérez et al. 2018). The ratio of SuSy/AGPase activities indicated a correlation between the two. In the wild-type both enzymes had an activity of ca: 1 mmol NADPH or sucrose/min/mg FW. Higher levels of AGPase activity reduced levels of SuSy activity such as in line 1 and lower levels of AGPase activity resulted in higher levels of SuSy activity (Supplementary Figure S4).
Deregulation of starch-related family gene expression induced by GBSSI mutations
Next, we measured the expression of the rice Wx gene in T0 flag leaves (Supplementary Figure S5) and T1 (Supplementary Figure S5) and T2 (Fig. 5) endosperm and compared the expression of other genes involved in starch biosynthesis (OsAPL1, OsAPL3, OsAPL4, OsAPS1, OsAPS2a/b, OsAPL2, OsSSI, OsSSIIa, OsSSIIb, OsSSIIc, OsSSIIIa, OsSSIIIb, OsSSIVa, OsSSIVb, OsGBSSII, OsBEI, OsBEIIa, OsBEIIb, OsISA1, OsISA2, OsISA3, OsPUL, OsDPE1, OsDPE2, OsPHOH and OsPHOL). We created a heat map of the expression profiles based on percentiles to visualize the most significant changes. The intense red color represents extreme (≥ 10-fold) downregulation compared to wild-type and the red gradient represents moderate (< 10-fold but ≥ 1.12-fold) downregulation compared to wild-type. The intense green color represents extreme (≥ tenfold) upregulation compared to wild-type and the green gradient represents moderate (< tenfold but ≥ twofold) upregulation compared to the wild-type. All other values close to onefold (no change) are colored yellow. In T0 leaves (Supplementary Figure S5), the relative expression levels of OsSSI, OsSSIIIa, OsBEI, OsPUL, OsPHOL, OsDPE1 and OsDPE2 increased significantly except in line 6 where all genes responded in a similar manner to wild-type plants as expected. OsAPS1, OsAPL3, OsSSIVb, OsPHOH and OsGBSSI were strongly downregulated. In contrast, OsAPS2a/b, OsBEI, OsAPL4, OsSSIIIa, OsPUL, OsGBSSII, OsPHOH and OsPHOL were strongly upregulated and OsAPL1, OsSSIIIb, OsSSIVb, OsBEIIa and OsGBSSI were strongly downregulated in the T1 seeds (Supplementary Figure S5). Genes with no changes in levels of expression in T2 with respect to wild-type were not included in the heat map. T2 seeds exhibited a similar expression profile with the, KUR and Musa mutants and also with T1 seeds (Fig. 5).
Heat map showing fold-changes in the expression of starch biosynthesis and degradation pathway genes in T2 seeds in wild-type and mutant rice plants. The red gradient shows increasing degrees of downregulation and the green gradient shows increasing degrees of upregulation, with yellow indicating no change in expression. The red gradient is expanded in the lower ranges because this is where most of the values lie while the green gradient is linear. The data for T0 leaves and T1 seeds are shown in Supplementary Figure S5. (Color figure online)
An analysis of variance for normalized expression on log-transformed data of T0 leaves and T1 seeds was carried out using Tissue (leave/seed); Genotype (Wild-type and separate mutant lines); Gene (Family of genes); Gene_Type (Function of genes) and isoform (different isoforms of the same gene) as independent factors. Gene_Type, Gene (Gene_Type) and Isoform (Gene_Type, Gene) were the most significant factors, although their interactions with tissue were also highly significant (Table 2). For the highest order interaction, Tissue*Isoform (Gene_Type, Gene) is the most significant interaction and the data was graphically represented, where Tissue and OsAPL1, OsAPL3, OsAPL4, OsAPS2a, OsDPE1, OsGBSSII, OsSSI, OsSSIIIa, OsSSIIIb and OsSSIVb isoforms were highly significant. Factor Gene (family) was statistically significant except on ISA and Gene_Type was statistically significant (Fig. 6).
Mean normalized expression correlating different plant tissues with the different starch pathway gene isoforms depending on gene family (Gene) and the enzymatic function (Gene_Type). Asterisks indicate a statistically significant difference, as determined by t test (*P < 0.05) on log-transformed data
Changes in starch, amylose and soluble sugar levels in GBSSI mutants
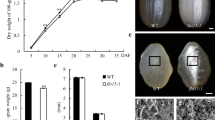
The starch content of the T2 seeds was similar to wild-type levels in all six mutant lines and in the KUR and Musa mutants (Fig. 7a). In contrast, the soluble sugar content in T2 seeds was significantly (23–57%) higher than wild-type levels in all the mutant lines except lines 1 and 6, the latter being effectively wild-type due to the synonymous nature of the mutation (Fig. 7b). Ignoring line 6, the amylose content of the T1 seeds in most mutant lines was 40–50% below wild-type levels, but > 75% lower in line 2, probably reflecting the homozygous nature of the mutation (Fig. 7c). Like the Musa mutant, the T1 seeds of line 2 contained less than 5% amylose which can be considered as a Wx phenotype, whereas the other lines (except line 6) contained 8–12% amylose. In T2 seeds, the mutations in lines 1–5 were homozygous and the amylose content was, therefore, lower than in T1 seeds. Lines 1, 2 and 4 (like Musa) can thus be considered as Wx lines because they contained less than 5% of amylose, whereas lines 3 and 5 (like KUR) can be considered as very-low-amylose lines because they contained 6–11.5% amylose (Fig. 7d).
Seed carbohydrate content in wild-type and mutant rice plants. a Total starch content of T2 seeds from wild-type (WT) plants, the Wx mutant lines and the two Wx irradiation mutants KUR and Musa. b Soluble sugar content of T2 seeds. c Amylose content of T1 seeds. d Amylose content of T2 seeds. Values are expressed as means ± SD (n = 3 biological replicates, 2 technical replicates for each biological replicate). Asterisks indicate a statistically significant difference, as determined by Student’s t test (*P < 0.05; **P < 0.01). Asterisks in black indicate a statistically significant difference between wild-type and mutant lines. Asterisks in blue indicate a statistically difference between KUR and the other lines. Asterisks in red indicate a statistically significant difference between Musa and the other lines. (Color figure online)
We measured no statistically significant changes in starch and soluble sugar levels. The ratio of soluble sugars/starch indicated a direct relationship as expected (Supplementary Figure S4).
Phenotype and microscopy changes produced by GBSSI mutations
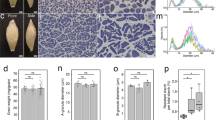
The KUR and Musa Wx lines have opaque seeds, characteristic of the Wx phenotype (Fig. 8). The T1 seeds of lines 1, 2 and 5 were completely opaque, whereas those of lines 3 and 4 were semi-opaque and those of line 6 were indistinguishable from wild-type seeds. Statistical analysis was done by measuring length, width and thickness of 20 seeds. Musashimochi and line 6 showed similar size as wild-type; line 5 only differed in seed volume; line 4 had differences in width, thickness and volume; while line 1, line 2 and line 3 showed significant changes in all parameters (Supplementary Figure S6). Richardson blue staining of fixed T1 mutant seeds showed changes with unstructured cells and disorganization in the aleurone layer in mutant lines (Supplementary Figure S7). More detailed morphological analysis by SEM in T1 seeds revealed that wild-type starch granules are angular with sharply defined edges whereas KUR/Musa seeds feature more rounded granules with softer, less-defined edges. In the T1 seeds of our mutant lines, the homozygous line 2 was similar to the KUR/Musa morphology whereas the other lines were more reminiscent of the wild-type morphology (Supplementary Figure S8).
Seed phenotypes of wild-type (WT) plants, the Wx mutant lines and the two Wx irradiation mutants KUR and Musa. Scale bar, 5 mm. a Wild-type vs KUR. b Wild-type vs Musa. c Wild-type vs line 1. d Wild-type vs line 2. e Wild-type vs line 3. f Wild-type vs line 4. g Wild-type vs line 5. h Wild-type vs line 6
Discussion
Starch accounts for ~ 90% of the dry weight of rice grains. It has two components: linear amylose with a small number of long branches, and amylopectin with a large number of short branches. Starch synthesis begins when AGPases catalyze the formation of short chains of ADP-glucose monomers (Li et al. 2017) and continues with the elongation of amylose and amylopectin by granule-bound starch synthase (GBSS) and soluble starch synthase (SS), respectively (Ohdan et al. 2005). In rice, there are two GBSS isoforms: GBSSI is mainly expressed in the endosperm and uses malto-oligosaccharides or short amylopectin chains as primers to synthesize amylose (Jeon et al. 2010; Momma and Fujimoto 2012) whereas GBSSII is mainly expressed in leaves and other vegetative tissues that accumulate transient starch (Tetlow 2011). GBSSI, encoded by the Waxy (Wx) locus, is therefore, the primary determinant of amylose levels in rice endosperm.
Waxy mutants have been generated by conventional mutagenesis using chemical mutagens, irradiation or T-DNA/transposons, including the lines KUR and Musashimochi (Musa) induced by neutron irradiation and gamma irradiation, respectively (Yatou and Amano 1991; Itoh et al. 1997). KUR has an amylose content of 7.5% and Musa has an amylose content of 1.3% compared to 20.8%, with no changes in the overall amount of starch compared to wild-type plants in either case. Similar phenotypes have been generated by the targeted mutation or knockdown of Wx in rice, confirming that GBSSI is the key determinant of amylose levels in the endosperm (Terada et al. 2000; Itoh et al. 2003; Fujita et al. 2006; Tran et al. 2011). More recently, CRISPR/Cas9 has been used to target Wx. For example, Ma et al. (2015) targeted three different Wx sites simultaneously. However, these studies have looked solely at the impact on starch and agronomic properties such as seed number and length (Li et al. 2017; Zhang et al. 2018) overlooking impact on other enzymes in the starch biosynthesis pathway.
Mutations in the Waxy gene at the DNA level and impact on mutant Waxy protein
We had previously used CRISPR/Cas9 to target starch biosynthetic genes active in the endosperm and have observed a profound impact on the wider starch biosynthetic pathway in vegetative tissues (Baysal et al. 2016; Pérez et al. 2018). To investigate whether similar effects would occur when we targeted Wx, we designed sgRNAs targeting three sites in exon 1 and obtained three different types of mutation: a missense mutation caused by the substitution Q33H resulting in a moderate structural change compared to the wild-type enzyme (line 1), a synonymous substitution caused by two nucleotide exchanges that did not change the sense of the corresponding codon (line 6), and larger indels causing frameshifts and complete loss of function due to early truncation (lines 2–4) or the abolition of protein import to the starch granules (line 5). All lines except line 2 were heterozygous in the T1 generation (line 2 was a homozygous mutant) and all six lines were homozygous mutants in the T2 generation.
Reduced levels of amylose are associated with Waxy gene mutations
The major consequence of mutating the Wx gene described earlier was the modification of the relative abundance of amylose and amylopectin without changing the overall starch content (Zhang et al. 2012, 2018). We compared the amylose content of our mutants with wild-type plants and the KUR and Musa irradiation mutants. Line 6 showed no difference to wild-type plants as expected because the GBSSI enzyme retained its normal activity. In the T1 generation, the heterozygous seeds of lines 1 and 3–5 had a lower amylose content than wild-type seeds but higher than both irradiation mutants, whereas the amylose content of the homozygous seeds of line 2 was between that of KUR and Musa. Consistently with the homozygous GBSSI mutation in the T2 generation, seeds of lines 1–5 all lay between the KUR and Musa mutants, with amylose levels of 4–9%. The mutation of other starch pathway genes increases the abundance of soluble sugars, e.g., as shown for AGPases (Rösti et al. 2007; Tang et al. 2016). The analysis of our T2 mutant lines likewise showed an increase in soluble sugars in lines 1, 2, 4, and 5 but not 3, which may reflect a metabolic bottleneck caused by the mutation or an increase in starch degradation due to feedback regulation, as discussed in more detail below. The KUR and Musa lines also showed higher levels of soluble sugars. Our data agree with earlier studies which reported a direct correlation between starch and soluble sugars (Preiss 1982). The phenotypes described above were concordant with the GBSS activity of the seeds. Our mutants and the two irradiation mutants showed only 42.4–69.16% of the GBSS activity compared to the wild-type lines.
Impact of mutation on grain phenotype
Mutations in starch biosynthesis genes are often recognized by their characteristic grain phenotype. APL2 and APS2 mutants have shrunken seeds (Kawagoe et al. 2005; Tang et al. 2016), mutations in SS or SBEI result in grains with a chalky appearance (Ryoo et al. 2007; Zhang et al. 2011), mutations in SBEIIb generate opaque grains (Sun et al. 2017), and mutations in ISA1 give rise to sugary grains (Wong et al. 2003) in contrast to ISA2 mutations with no phenotype because the enzyme has negligible activity (Li et al. 2017). The waxy phenotype that underlies the name of the GBSSI gene reflects the accumulation of amylopectin at the expense of amylose, and is characterized by grains that are white and opaque rather than translucent like wild-type grains (Zhang et al. 2012, 2018). In KUR and Musa, the grains are completely opaque due to the substantial loss of GBSS activity, and the T1 seeds of our lines 1, 2 and 5 were comparable, suggesting a similar degree of GBSS impairment. Wild-type and seeds from line 6 were fully translucent, whereas the remaining lines (3 and 4) were characterized by semi-opaque seeds indicating the wild-type GBSSI allele was more active in these lines or some compensatory mechanism was activated. Changes in the length, width, thickness and volume of grains in starch pathway gene mutants were reported earlier (Wang et al. 2013; Tang et al. 2016). Statistical analysis for these parameters in grains of lines 1, 2 and 3 showed significant changes with respect to wild-type; lines 4 and 5 showed moderate changes which were still significant.
The grain phenotypes we observed visually were also reflected by changes in cellular and subcellular organization. The neatly arranged cells of the aleurone layer in the wild-type seeds were disrupted in the mutants, suggesting that cell structure is at least partly dependent on normal starch synthesis. The reason for this became apparent at the subcellular level, where major differences in the shape and structure of starch granules were observed. The wild-type phenotype featured polygonal granules with sharp edges whereas our mutant lines and the Musa mutant showed rounded and amorphous granules, with greater deviation in the seeds with the lower amylose content. Similar granule structures have been reported in other Wx mutants (Liu et al. 2009; Zhang et al. 2018).
Association between mutations and tertiary protein structure
The mutations described above have a dramatic effect on the phenotype of rice grains by perturbing the structure of GBSSI and thus reducing its activity. Amino acids 1–77 of GBSSI correspond to the coiled coil region that interacts with a similar region of the protein PTST and allows both proteins to be imported into starch granules, whereupon the coiled coil region is proteolytically cleaved off (Seung et al. 2015). This means that mutations affecting the PTST protein can phenocopy the loss of GBSSI activity by preventing the import of the enzyme into starch granules (Seung et al. 2015). We targeted the first exon of Wx which corresponds to the coiled coil region. In lines 2–4, the resulting frameshift mutation caused the introduction of a nonsense codon and the heavily truncated product was nonfunctional. In contrast, the mutation in line 5 caused part of the coiled coil to be deleted without affecting the remainder of the protein, but still we observed the loss-of-function phenotype because the import of the enzyme was blocked. In line 1, the mutation was a more subtle amino acid exchange, replacing the polar and uncharged glutamine residue at position 33 with the positively charged histidine. Phylogenetic analysis revealed that the glutamine is highly conserved in cereals, but not in dicots, suggesting that it may play a role in the import of GBSSI into cereal starch granules and the mutation may, therefore, reduce the efficiency of import. This may be one explanation for the partial loss of activity we observed. However, we also found that the tertiary structure of the enzyme was affected. We modeled the tertiary structure of GBSSI in line 1 using the crystal structure of the wild-type enzyme as a template (Momma and Fujimoto 2012). The wild-type structure features α-helices and β-sheets that form two substrate-binding clefts, one for ADP-glucose and other for malto-oligosaccharide precursors. In line 1, the tertiary structure was modified at the N-terminus and C-terminus resulting in a surface change that constricted the malto-oligosaccharide pocket and changed the position of a hydrogen bond, indicating that the mutation may reduce the affinity of the enzyme for its substrate.
Deregulation of starch pathway genes generated by Waxy gene mutations
One of the key aspects of starch biosynthesis which is rarely investigated in mutational studies is the tight feedback regulation in the pathway, which manifests in the modulation of gene expression among other starch genes when one member is mutated. We, therefore, analyzed a panel of relevant genes in T0 leaves and T1/T2 seeds relative to wild-type plants and (for seeds only) the two irradiation mutants. Line 6 was essentially identical to wild-type plants in all respects and this line was used as a second control. In wild-type T0 leaves, the major AGPases are APL1, APL3 and APS2a and the major SEB is SBEI (Ohdan et al. 2005). In our mutant lines, APL1 and APL3 were strongly downregulated in the leaves, perhaps due to negative feedback caused by the accumulation of ADP-glucose, whereas SBEI was induced, perhaps because the surplus ADP-glucose is used to synthesize amylopectin. In terms of starch degradation the expression of ISA1 and ISA3 was downregulated whereas PUL was induced, and the DPE1/2 and PHOL genes were upregulated whereas PHOH, the major isoform expressed in leaves (Ohdan et al. 2005), was suppressed. The three debranching enzymes are normally expressed in the leaves but whereas ISA1 has a known role in the maintenance of amylopectin, ISA2 has no intrinsic activity unless associated with another ISA isoform and the role of PUL is unclear (Li et al. 2017), although it may compensate the loss of ISA activity (Jeon et al. 2010). We hypothesized that these changes reflect the capacity of PUL to substitute partially for the loss of ISA activity when the absence of GBSSI generates abnormal starch structures that are atypical ISA1 substrates. Similarly, PHOL is more important than PHOH for the maintenance of starch structure. Finally we considered the expression of soluble starch synthases in the T0 leaves. Although GBSSI is low expressed in leaves and loss of enzymatic activity in the endosperm should not have any effect, we nevertheless observed an increase in SSI and SSIIIa expression and a decrease in SSIVb expression compared to wild-type plants. SSI is strongly expressed in leaves and it forms a complex with SSIIIa and other proteins to synthesize short-chain glucans for amylopectin biosynthesis (Crofts et al. 2015). SSIV regulates the number of starch granules, and its modulation may reflect a response to the effect of abnormal starch on granule structures (Li et al. 2017).
A different set of starch-related genes was modulated in T1/T2 seeds compared to T0 leaves. In T1/T2 seeds, APS2a/b and APL4 were upregulated and APL1 was downregulated in our mutants and in the KUR and Musa lines. The rice apl1 mutant showed no change in AGPase activity in the endosperm or in the leaves, and the leaves were reported to contained < 5% of normal starch levels but normal levels of soluble sugars (Rösti et al. 2007). These data suggest that the APL1 subunit is necessary for starch synthesis in leaves but not in non-photosynthetic organs. The increase in APL4 and APS2a expression allows these proteins to form a functional heterotetrameric structure that is not normally found at significant levels in seeds. APS2b is the major AGPase small subunit in endosperm and its activity is needed to form ADP-glucose (Ohdan et al. 2005). We also observed the upregulation of SBEI and SBEIIB, whose products form a complex to facilitate endosperm starch synthesis (Tetlow et al. 2004), but the downregulation of SBEIIa needed to maintain the short-chain content of leaf starch, which does not appear to play a role in endosperm starch synthesis (Nakamura 2002). Concerning starch degradation PHO and PUL were upregulated in our mutants; PUL debranches pullulan and amylopectin thus its upregulation may help to deal with unusual starch structures by debranch these wear structures (Nakamura 2002). Finally, we found that the loss of GBSSI expression induced a compensatory increase in GBSSII expression, which is normally restricted to non-storage tissues. However, given the overall decrease in GBSS enzyme activity we observed in the mutant seeds, this compensation is clearly not enough to restore the normal phenotype. The loss of amylose in the mutant seeds also strongly induced the expression of SSIIIa, but inhibited SSIIIb and SSIVb. In wild-type plants SSIIIa is more strongly expressed in seeds whereas SSIIIb is expressed at the onset of grain formation and declines rapidly thereafter, and SSIVb regulates the number of starch granules (Ohdan et al. 2005). The expression profile in the mutant, therefore, mirrors but exaggerates the normal situation, with SSIVb expression not required because the granule number is already limited in the mutants. The model of variance analysis of normalized expression showed statistically significant differences. Gene_Type, Gene (Gene_Type) and Isoform (Gene_Type, Gene) were the most significant factors, but Genotype was also highly significant. Our statistical analysis further supported our conclusion that the nature of the induced mutations influenced changes in gene expression of other starch biosynthetic genes in a tissue dependent manner.
Impact of the loss of GBSSI enzyme activity on overall GBSS activity and other related enzymes
Given the differences between mutant and wild-type plants in terms of AGPase gene expression, we also measured the overall AGPase activity and the activity of sucrose synthase (SuSy) in T2 seeds, which provides an alternative pathway for starch synthesis in the absence of AGPase (Li et al. 2013). We found that the opposing changes in the expression levels of different AGPase subunits did not result in any statistically significant differences in overall AGPase activity between wild-type and mutant lines. However, there was an increase in SuSy activity in the mutants which was statistically significant at least in line 3. This may reflect the accumulation of precursors that cannot be converted into amylose by AGPase and the ability of SuSy to use ADP as substrate for the synthesis of ADP-glucose (Baroja-Fernández et al. 2012). The ratio of SuSy/AGPase exhibited an inverse relationship with activity levels of the two enzymes as was hypothesized earlier (Li et al. 2013).
Our collective results provide a basis to suggest that the GBSS activity levels in the mutant lines result from overexpression of GBSSII to compensate for the loss of GBSSI activity. GBSS activity in lines 2, 3 and 5 was lower than in lines 1 and 4 because there was a smaller compensatory increase in GBSSII expression to address the loss of GBSSI in the former lines. The increase in AGPase activity in lines 1 and 3 reflected the more profound increase in APL4 and APS2a/b expression and the less severe suppression of APL1 compared to the other mutants. AGPase activity in line 2 was particularly low due to the severe suppression of APL1 and relatively weak induction of APL4. In lines 4 and 5, the small subunit genes (APS2a/b) were upregulated, but without a corresponding increase in APL4 expression the quantity of the heterotetrameric enzyme could not increase. In lines 2–5, the accumulation of soluble sugars due to the loss of GBSSI activity resulted in an increase in sucrose synthase activity, but in line 1 the weak AGPase activity does not produce enough soluble sugars to induce sucrose synthase, hence the low activity in that line.
In summary, mutating the first exon of the rice Wx gene encoding GBSSI resulted in the expected partial loss of GBSS activity and the corresponding loss of amylose in the endosperm, but also caused the unexpected expression of other downstream starch pathway genes, partly to deal with abundant intermediates and unusual starch structures (amylopectin has more branches and takes up more space than amylose, so the replacement of amylose with amylopectin results in hyperbranched starch that occupies a greater volume than normal). The increase in GBSSII expression did not compensate for the loss of GBSSI, reflected in the amylose content of the mutant lines. Modifying the peptide signal needed for import into starch granules is sufficient to block GBSSI activity without affecting the catalytic center in any other way. Our results provide critical mechanistic insight into the complex feedback relationships among genes, enzymes, intermediates and end products in the starch biosynthesis and degradation pathways. This mechanistic understanding provides a basis for more targeted interventions to modulate starch biosynthesis in plants in a more precise manner to generate plants with altered starch content and composition for various applications.
Author contribution statement
LP and ES generated all the mutant lines, performed the experiments and wrote the article. GV performed protein modeling. LB helped with GBSSI activity and RT-PCR experiments. VM helped with optical microscopy and performed electron microscopy. GF helped with tissue culture, cloning and confirmation of the lines. JJ helped with real-time PCR analysis and preparation of the samples for starch and amylose experiments. AJS, MS and JAR performed AGPase and Susy activity assays. PM helped with starch content determination. CZ helped designing experiments. IR performed ANOVA statistical analysis. PC conceived the project, designed experiments and reviewed and edited drafts of the manuscript.
References
Ball SG, Morell MK (2003) From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Ann Rev Plant Biol 54:207–233
Baroja-Fernández E, Muñoz FJ, Li J, Bahaji A, Almagro G, Montero M, Etxeberria E, Hidalgo M, Sesma T, Pozueta-Romero J (2012) Sucrose synthase activity in the sus1/sus2/sus3/sus4 Arabidopsis mutant is sufficient to support normal cellulose and starch production. Proc Natl Acad Sci USA 109:321–326
Bassie L, Zhu C, Romagosa I, Christou P, Capell T (2008) Transgenic wheat plants expressing an oat arginine decarboxylase cDNA exhibit increases in polyamine content in vegetative tissue and seeds. Mol Breed 22:39–50
Baysal C, Bortesi L, Zhu C, Farré G, Schillberg S, Christou P (2016) CRISPR/Cas9 activity in the rice OsBEIIb gene does not induce off-target effects in the closely related paralog OsBEIIa. Mol Breed 36:108
Bortesi L, Zhu C, Zischewski J, Perez L, Bassié L, Nadi R, Forni G, Boyd-Lade S, Soto E, Jin X, Medina V, Villorbina G, Muñoz P, Farré G, Fischer R, Twyman R, Capell T, Christou P, Schillberg S (2016) Patterns of CRISPR/Cas9 activity in plants, animals and microbes. Plant Biotechnol J 14:2203–2216
Chari R, Mali P, Moosburner M, Church GM (2015) Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Methods 12:823
Christou P, Ford TL, Kofron M (1991) Production of transgenic rice (Oryza sativa L.) plants from agronomically important indica and japonica varieties via electric discharge particle acceleration of exogenous DNA into immature zygotic embryos. Nat Biotechnol 9:957–962
CISC (Consejo Superior de Investigaciones Científicas) (2016) Método para la determinación “in situ” de actividades enzimáticas relacionadas con el metabolismo del carbono en hojas. ES Patent no 7:915.111.623.106, 2016-05-06
Crofts N, Abe N, Oitome NF, Matsushima R, Hayashi M, Tetlow IJ, Emes MJ, Nakamura Y, Fujita N (2015) Amylopectin biosynthetic enzymes from developing rice seed form enzymatically active protein complexes. J Exp Bot 66:4469–4482
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Claverie JM, Gascuel O (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucl Acids Res 36:W465–W469
Farré G, Sudhakar D, Naqvi S, Sandmann G, Christou P, Capell T, Zhu C (2012) Transgenic rice grains expressing a heterologous ρ-hydroxyphenylpyruvate dioxygenase shift tocopherol synthesis from the γ to the α isoform without increasing absolute tocopherol levels. Transgenic Res 21:1093–1097
Fujita N, Yoshida M, Asakura N, Ohdan T, Miyao A, Hirochika H, Nakamura Y (2006) Function and characterization of starch synthase I using mutants in rice. Plant Physiol 140:1070–1084
Heigwer F, Kerr G, Boutros M (2014) E-CRISP: fast CRISPR target site identification. Nat Methods 11:122
Hirano HY (1993) Genetic variation and gene regulation at the wx locus in rice. Gamma Field Symp 24:63–79
Hirano HY, Sano Y (1998) Enhancement of Wx gene expression and the accumulation of amylose in response to cool temperatures during seed development in rice. Plant Cell Physiol 39:807–812
Itoh K, Nakajima M, Shimamoto K (1997) Silencing of waxy genes in rice containing Wx transgenes. Mol Gen Genet 255:351–358
Itoh K, Ozaki H, Okada K, Hori H, Takeda Y, Mitsui T (2003) Introduction of Wx transgene into rice wx mutants leads to both high-and low-amylose rice. Plant Cell Physiol 44:473–480
Jeon JS, Ryoo N, Hahn TR, Walia H, Nakamura Y (2010) Starch biosynthesis in cereal endosperm. Plant Physiol Biochem 48:383–392
Jiang D, Cao WX, Dai TB, Jing Q (2004) Diurnal changes in activities of related enzymes to starch synthesis in grains of winter wheat. Acta Bot Sin 46:51–57
Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucl Acids Res 41:e188–e188
Jobling S (2004) Improving starch for food and industrial applications. Curr Opin Plant Biol 7:210–218
Juliano BO (1971) A simplified assay for milled-rice amylose. Cereal Sci Today 16:334–336
Kang TJ, Yang MS (2004) Rapid and reliable extraction of genomic DNA from various wild-type and transgenic plants. BMC Biotechnol 4:20
Kawagoe Y, Kubo A, Satoh H, Takaiwa F, Nakamura Y (2005) Roles of isoamylase and ADP-glucose pyrophosphorylase in starch granule synthesis in rice endosperm. Plant J 42:164–174
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protov 10:845
Li J, Baroja-Fernández E, Bahaji A, Muñoz FJ, Ovecka M, Montero M (2013) Enhancing sucrose synthase activity results in an increased levels of starch and ADP-Glucose in maize (Zea mays L.) seed endosperms. Plant Cell Physiol 54:282–294
Li C, Powell PO, Gilbert RG (2017) Recent progress toward understanding the role of starch biosynthetic enzymes in the cereal endosperm. Amylase 1:59–74
Liu L, Ma X, Liu S, Zhu C, Jiang L, Wang Y, Shen Y, Ren Y, Dong H, Chen L, Liu X, Zhao Z, Zhai H, Wan J (2009) Identification and characterization of a novel Waxy allele from a Yunnan rice landrace. Plant Mol Biol 71:609–626
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Liu YG (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8:1274–1284
Maddelein ML, Libessart N, Bellanger F, Delrue B, D’Hulst C, Van den Koornhuyse N, Fontaine T, Wieruszeski JM, Decq A, Ball S (1994) Toward an understanding of the biogenesis of the starch granule. Determination of granule-bound and soluble starch synthase functions in amylopectin synthesis. J Biol Chem 269:25150–25157
Martin C, Smith AM (1995) Starch biosynthesis. Plant Cell 7:971–985
Momma M, Fujimoto Z (2012) Interdomain disulfide bridge in the rice granule bound starch synthase I catalytic domain as elucidated by X-ray structure analysis. Biosci Biotechnol Biochem 76:1591–1595
Nakamura Y (2002) Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: rice endosperm as a model tissue. Plant Cell Physiol 43:718–725
Nakamura Y, Yuki K, Park SY, Ohya T (1989) Carbohydrate metabolism in the developing endosperm of rice grains. Plant Cell Physiol 30:833–839
Ohdan T, Francisco Jr PB, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y (2005) Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J Exp Bot 56:3229–3244
Pérez L, Soto E, Villorbina G, Bassie L, Medina V, Muñoz P, Capell T, Zhu C, Christou P, Farré G (2018) CRISPR/Cas9-induced monoallelic mutations in the cytosolic AGPase large subunit gene APL2 induce the ectopic expression of APL2 and the corresponding small subunit gene APS2b in rice leaves. Transgenic Res 27:423–439
Preiss J (1982) Regulation of the biosynthesis and degradation of starch. Annu Rev Plant Physiol 33:431–454
Rösti S, Fahy B, Denyer K (2007) A mutant of rice lacking the leaf large subunit of ADP-glucose pyrophosphorylase has drastically reduced leaf starch content but grows normally. Funct Plant Biol 34:480–489
Ryoo N, Yu C, Park CS, Baik MY, Park IM, Cho MH, Bhoo SH, An G, Hahn TR, Jeon JS (2007) Knockout of a starch synthase gene OsSSIIIa/Flo5 causes white-core floury endosperm in rice (Oryza sativa L.). Plant Cell Rep 26:1083–1095
Sano Y (1984) Differential regulation of waxy gene expression in rice endosperm. Theor Appl Genet 68:467–473
Satoh H, Shibahara K, Tokunaga T, Nishi A, Tasaki M, Hwang SK, Okita TW, Kaneko N, Fujita N, Yoshida M, Hosaka Y, Sato A, Utsumi Y, Ohdan T, Nakamura Y (2008) Mutation of the plastidial α-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm. Plant Cell 20:1833–1849
Seung D, Soyk S, Coiro M, Maier BA, Eicke S, Zeeman SC (2015) PROTEIN TARGETING TO STARCH is required for localising GRANULE-BOUND STARCH SYNTHASE to starch granules and for normal amylose synthesis in Arabidopsis. PLOS Biol 13:e1002080
Shan Q, Wang Y, Li J, Gao C (2014) Genome editing in rice and wheat using the CRISPR/Cas system. Nature Protoc 9:2395
Sudhakar D, Bong BB, Tinjuangjun P, Maqbool SB, Valdez M, Jefferson R, Christou P (1998) An efficient rice transformation system utilizing mature seed-derived explants and a portable, inexpensive particle bombardment device. Transgenic Res 7:289–294
Sun Y, Jiao G, Liu Z, Zhang X, Li J, Guo X, Du J, Francis F, Zhao Y, Xia L (2017) Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front Plant Sci 8:298
Tang XJ, Peng C, Zhang J, Cai Y, You XM, Kong F, Yan HG, Wang GX, Wang L, Jin J, Chen WW, Chen XG, Ma J, Wang P, Jiang L, Zhang WW, Wan JM (2016) ADP-glucose pyrophosphorylase large subunit 2 is essential for storage substance accumulation and subunit interactions in rice endosperm. Plant Sci 249:70–83
Terada R, Nakajima M, Isshiki M, Okagaki RJ, Wessler SR, Shimamoto K (2000) Antisense waxy genes with highly active promoters effectively suppress waxy gene expression in transgenic rice. Plant Cell Physiol 41:881–888
Tetlow IJ (2011) Starch biosynthesis in developing seeds. Seed Sci Res 21:5–32
Tetlow IJ, Morell MK, Emes MJ (2004) Recent developments in understanding the regulation of starch metabolism in higher plants. J Exp Bot 55:2131–2145
Thitisaksakul M, Jiménez RC, Arias MC, Beckles DM (2012) Effects of environmental factors on cereal starch biosynthesis and composition. J Cereal Sci 56:67–80
Tran NA, Daygon VD, Resurreccion AP, Cuevas RP, Corpuz HM, Fitzgerald MA (2011) A single nucleotide polymorphism in the Waxy gene explains a significant component of gel consistency. Theor Appl Genet 123:519–525
Umemoto T, Terashima K (2002) Activity of granule-bound starch synthase is an important determinant of amylose content in rice endosperm. Funct Plant Biol 29:1121–1124
Valdez M, Cabrera-Ponce JL, Sudhakar D, Herrera-Estrella L, Christou P (1998) Transgenic Central American, West African and Asian elite rice varieties resulting from particle bombardment of foreign DNA into mature seed-derived explants utilizing three different bombardment devices. Ann Bot 82:795–801
Wang JC, Xu H, Zhu Y, Liu QQ, Cai XL (2013) OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J Exp Bot 64:3453–3466
Wong KS, Kubo A, Jane JL, Harada K, Satoh H, Nakamura Y (2003) Structures and properties of amylopectin and phytoglycogen in the endosperm of sugary-1 mutants of rice. J Cereal Sci 37:139–149
Yatou O, Amano E (1991) DNA structure of mutant genes in the waxy locus in rice. In: Plant mutation breeding for crop improvement, vol 2. IAEA, Vienna/FAO, Rome, pp 385–389
Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Determination of sugar and starch in plant tissue. In: Laboratory manual for physiological studies of rice, 3rd edn. IRRI, Los Baños, Philippines, pp 46–49
Zhang G, Cheng Z, Zhang X, Guo X, Su N, Jiang L, Mao L, Wan J (2011) Double repression of soluble starch synthase genes SSIIa and SSIIIa in rice (Oryza sativa L.) uncovers interactive effects on the physicochemical properties of starch. Genome 54:448–459
Zhang MZ, Fang JH, Yan X, Liu J, Bao JS, Fransson G, Andersson R, Jansson C, Aman P, Sun C (2012) Molecular insights into how a deficiency of amylose affects carbon allocation–carbohydrate and oil analyses and gene expression profiling in the seeds of a rice waxy mutant. BMC Plant Biol 12:230
Zhang J, Zhang H, Botella JR, Zhu JK (2018) Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. J Int Plant Biol 60:369–375
Zhu C, Bortesi L, Baysal C, Twyman RM, Fischer R, Capell T, Schillberg S, Christou P (2017) Characteristics of genome editing mutations in cereal crops. Trends Plant Sci 22:38–52
Acknowledgements
We thank Dr. Caixia Gao (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China) for providing pJIT163-2NLSCas9 containing the rice codon-optimized cas9, and the empty pU3-gRNA vector. We thank Professor Kimiko Itoh, Graduate School of Science and Technology (Niigata University, Japan) for providing the KUR and Musa Wx lines. This work was supported by funding from the Spanish Ministry of Economy and Competitiveness (MINECO) (BIO2014-54426), and a Juan de la Cierva fellowship to GF (IJCI-2014-19528). LP is the recipient of MINECO fellowship. ES is the recipient of a PhD fellowship from the University of Lleida (BIO2014-54441-P).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that no conflicts of interest exist.
Additional information
Communicated by Fabien Nogué.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pérez, L., Soto, E., Farré, G. et al. CRISPR/Cas9 mutations in the rice Waxy/GBSSI gene induce allele-specific and zygosity-dependent feedback effects on endosperm starch biosynthesis. Plant Cell Rep 38, 417–433 (2019). https://doi.org/10.1007/s00299-019-02388-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-019-02388-z