Abstract
Sharks compose one of the most diverse and abundant groups of consumers in the ocean. Consumption and digestion are essential processes for obtaining nutrients and energy necessary to meet a broad and variable range of metabolic demands. Despite years of studying prey capture behavior and feeding habits of sharks, there has been little exploration into the nutritional physiology of these animals. To fully understand the physiology of the digestive tract, it is critical to consider multiple facets, including the evolution of the system, feeding mechanisms, digestive morphology, digestive strategies, digestive biochemistry, and gastrointestinal microbiomes. In each of these categories, we make comparisons to what is currently known about teleost nutritional physiology, as well as what methodology is used, and describe how similar techniques can be used in shark research. We also identify knowledge gaps and provide suggestions to continue the progression of the field, ending with a summary of new directions that should be addressed in future studies regarding the nutritional physiology of sharks.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sharks make up one of the most abundant and diverse groups of consumers in the ocean (Fig. 1, Compagno 2008). They may play an important ecological role in energy fluxes in marine environments and in impacting the biodiversity of lower trophic levels that we depend on as a food and economic resource (e.g., Wetherbee et al. 1990; Cortés et al. 2008). However, beyond prey capture methods and dietary analyses, the nutritional physiology of sharks is woefully understudied. They consume a broad range of diet types (smaller sharks, marine mammals, teleosts, crustaceans, zooplankton, etc.) but are generally known to be largely carnivorous, consuming prey items high in protein and lipids (Wetherbee et al. 1990; Cortés et al. 2008; Bucking 2016). We will regularly refer to the phylogeny (Fig. 1) in order to show how the diverse families of sharks are related. Indeed, matching physiological concepts with genetic underpinnings and evolutionary background is crucial to understanding the patterns and processes involved in the evolution of the digestive strategies that sharks possess.

Illustrations by R. Aidan Martin (2003)
Phylogeny of sharks to the family level based on the tree from Vélez-Zuazo & Agnarsson 2011. Light gray lines show which families belong to certain orders. Illustrations of specific shark species discussed in the text are shown (not to scale). The letters above each illustration correspond to the family in the phylogeny to which that species belongs. A) Squalus acanthias (spiny dogfish), B) Carcharodon carcharias (white shark), C) Cetorhinus maximus (basking shark), D) Rhincodon typus (whale shark), E) Ginglymostoma cirratum (nurse shark), F) Cephaloscyllium ventriosum (swell shark), G) Triakis semifasciata (leopard shark), H) Negaprion brevirostris (lemon shark), I) Sphyrna tiburo (bonnethead shark).
The broader field of nutritional physiology has a foundation based largely on economic theory: the digestive tract is energetically expensive to maintain (Cant et al. 1996), and thus, from basic economic principles, the Adaptive Modulation Hypothesis (AMH; Karasov 1992; Karasov and Martinez del Rio 2007) suggests that gut function should match with what is consumed in terms of quantity and biochemical composition (Martine and Fuhrman 1995; Karasov and Douglas 2013). Shark evolution presumably follows AMH and sharks should, therefore, have guts optimized for their high-protein, high-lipid diets. Sharks are also generally known for eating large meals on an infrequent basis, potentially going days, or even weeks, without a meal (Wetherbee et al. 1987; Cortés et al. 2008; Armstrong and Schindler 2011). Hence, in order to acquire ample nutrients from their infrequent meals, sharks must have mechanisms of slowing the rate of digesta transit to allow sufficient time for digestion and nutrient absorption, yet there has been minimal investigation into shark nutritional physiology. Although the field of comparative nutritional physiology is relatively young (e.g., Karasov and Diamond 1983; Diamond and Karasov 1987; Karasov and Martinez del Rio 2007), much has been learned about gut function in ecological and evolutionary contexts, albeit mostly about terrestrial organisms because of research in biomedical and livestock fields (Choat and Clements 1998; Clements et al. 2009). Within marine biology, far more advances have been made concerning the nutritional physiology of teleost fishes (e.g., German 2011) than for sharks. New methods of investigation have been developed, as well as new theories and models that could be applied to sharks (e.g., German et al. 2015; Clements et al. 2017). The most recent reviews of elasmobranch digestive physiology (Cortés et al. 2008; Bucking 2016; Ballantyne 2016) lament the dearth of data available on shark digestion, and thus, make logical connections to the recent advances in the understanding of teleost nutritional physiology, where there have been efforts to integrate diet with digestive tract function and metabolism. Here, we review what is currently known regarding shark feeding mechanisms, digestive morphology, digestive strategies, digestive biochemistry, and gastrointestinal microbiomes. In each of these categories, we make comparisons to what is known about teleost nutritional physiology and describe how similar techniques can be used in shark research. We also identify knowledge gaps and provide suggestions to continue the progression of the field, ending with a summary of new directions that should be addressed in future studies regarding the nutritional physiology of sharks.
Feeding mechanisms
Much of our knowledge of shark feeding mechanics is founded in observation, dissections, and muscle histology. In recent years, the feeding mechanisms of teleost fishes have been studied using modern techniques such as high-speed video kinematics, Video Reconstruction of Moving Morphology (VROMM), X-ray Reconstruction of Moving Morphology (XROMM), and biorobotic models (Shamur et al. 2016; Longo et al. 2016; Laurence-Chasen et al. 2016; Gidmark et al. 2015; Camp et al. 2015; Camp and Brainerd 2014; Kenaley and Lauder 2016; Corn et al. 2016; Wilga and Ferry 2016). These types of methodologies allow researchers to determine the exact skeletal elements and muscles involved in the different feeding mechanisms. They also give us the data necessary to quantify and model exactly how these cranio-facial elements move (in terms of volume of the buccal cavity, angles of skeletal elements, length of muscle, etc.) before, during, and after a feeding event. For example, Camp et al. (2015) used XROMM to show that the power required for buccal cavity expansion during suction feeding in the large mouth bass is generated primarily by axial swimming muscles rather than smaller cranial muscles. This research changed the perspective of musculoskeletal function in ray-finned fishes (over 30,000 species) and opened doors for further investigation using XROMM. Examples of videos created from XROMM can be found at: http://www.xromm.org/movies. Despite the success of these methods when studying the feeding mechanics of teleosts, they have yet to become common in investigations of shark feeding. Adapting some of these methods, particularly high-speed video kinematics and VROMM, to be used in large tank systems, or even in the field, would provide important details on shark feeding mechanics that heretofore have gone un-studied.
As a result of their broad range of diet types, sharks have highly diverse methods for feeding (Navia et al. 2007). The great diversity of feeding mechanisms that are exhibited by sharks can generally be sorted into three main categories: bite and retain, suction, and ram feeding (Motta and Wilga 2001). The biting mechanism is likely the most studied. Sharks are historically known for their sharp teeth and strong jaws that aid in the prey capture process (Cortés et al. 2008; Wilga and Ferry 2016). Shark jaws generally comprise ten main cartilaginous elements: the chondrocranium (consisting of the rostral cartilage, nasal capsule, pre-orbital process, post-orbital process, and otic capsule) the palatoquadrate, Meckel’s cartilage, the hyomandibula, and ceratohyals (Fig. 2, Motta and Wilga 1995; Wilga and Ferry 2016). The teeth are part of the jaw apparatus and most sharks have “replacement” dentition, meaning they continually make new teeth to replace older ones (Smith et al. 2013). Using dynamic testing models, Corn et al. (2016) showed that shark tooth performance (in terms of durability and cutting ability) varies between species due to structural differences of different tooth types. Overall, we do not understand whether there is a match between dentition, feeding mechanism, and diet. Unlike many bony fishes, sharks lack pharyngeal teeth to act as a secondary mechanical digestive method (trituration) and therefore, very few sharks actually chew their prey and generally swallow their food whole (or in very large sections, Fig. 2, Gajić 2013). However, there are a few select durophagous species, such as bonnethead (Sphyrna tiburo), nurse (Ginglymostoma cirratum), and horn sharks (Heterodontus francisci) that crush and grind their prey with molariform teeth set behind their triangular jaw teeth (or maxillary and mandibular) used for securing the prey (Fig. 1, Wilga and Motta 2000). Usually these species consume molluscs and crustaceans, which have hardened shells or exoskeletons, and therefore, require mechanical disruption of the food before swallowing (Kolmann et al. 2016).

(Revised from Motta and Wilga 1995 by J.S. German)
A left lateral view of the ten main cartilaginous elements that make up the neurocranium and jaws of a lemon shark, Negaprion brevirostris.
Suction is the most common mode of prey capture among both bony fishes and sharks (Frazetta and Prange 1987). In fact, even sharks that primarily use either the bite and retain feeding mechanism or ram feeding will also employ some suction during prey capture (Ferry-Graham and Lauder 2001). Since ocean water is dense and viscous, when a predator moves forward to capture a prey item, it will produce a bow-wave, which will push the prey away from the predator. To adjust for this action, suction, caused by rapidly expanding the oral cavity to create negative pressure relative to the water around the shark, is used to draw the prey closer to the predator, termed compensatory suction (Ferry-Graham and Lauder 2001). During inertial suction, prey is captured exclusively by being pulled into the oral cavity by the localization and direction of the suction forces created by slow moving predators such as nurse sharks (Motta and Wilga 2001; Motta et al. 2008; Wilga and Ferry 2016). However, the effectiveness of suction is limited by distance, with models predicting that the force of suction dissipates within just a few centimeters or millimeters, depending on shark size (Muller and Osse 1984). Nurse sharks show effective suction distances of approximately 3 cm (Motta et al. 2008). As a result, suction is commonly used in conjunction with ram prey capture. The basking shark (Cetorhinus maximus), whale shark (Rhincodon typus), and megamouth shark (Megachasma pelagios) are the only known shark species to have independently evolved pure ram feeding as their main feeding mechanism (Fig. 1, Cortés et al. 2008). The basking shark opens its jaws wide, spreads its gill arches apart, and swims forward through the water column which allows water and plankton, their prey of choice, to pass directly over their gills (Fig. 3), trapping the small zooplankton in the process (Paig-Tran et al. 2011). In the mouth is a larger filter apparatus composed of parallel plates that connect the internal gill openings to holobranchs. The filter apparatus is extremely efficient for intake of prey ranging in size from copepods (1–2 mm) to anchovies (2–40 cm, Fig. 3). It has also been established that filter mesh size and shark swimming speed will impact the efficiency of filter feeding (Paig-Tran and Summers 2013). Whale sharks employ the same mode of filter feeding as basking sharks; however, they possess epibranchial filter pads made up of a reticulated mesh instead of distinct parallel plates (Fig. 3). They have also been observed using a suction-gulping behavior (Nelson 2004; Motta et al. 2010). The megamouth shark has never been observed feeding, but it is hypothesized that their light lip coloration is used to lure deep-water prey (Nakaya 2001). It is also known that they have a large gape, proportionally small gill openings (compared to the basking shark), and long bucco-pharyngeal cavities suggesting that they use engulfment feeding similar to mechanisms seen in some whales (Nakaya et al. 2008). Their raker structures have a denticular texture that catches and traps their prey upon entry (Fig. 3, Paig-Tran and Summers 2013).
The filtering apparatuses of filter-feeding sharks. Illustration of structures are on the left, and SEM micrographs of surfaces are on the right (representing boxed areas on illustrations). a Rhincodon typus epibranchial (EB) filter pad, which includes a reticulated mesh (RM). b Gill rakers of Cetorhinus maximus, showing a stiff raker and smooth raker surface. c Gill raker of Megachasma pelagios showing denticle surface of the raker.
Digestive anatomy and physiology
In recent years, CT scan and MRI technology has been used to create 3D images of teleost fish skeletal elements, muscles, tendons, and connective tissues (Summers and Hayes 2016; Wainwright and Lauder 2016; Wu et al. 2015). Use of CT scanning in feeding investigations of Potamotrygon motoro (freshwater stingray) has shown that asymmetrical jaw motion is sufficient for mastication, which contradicts what is known about mammalian chewing (Kolmann et al. 2016). CT scans have also been used to investigate functional trade-offs between feeding apparatuses and other cranial structures in Eusphyra blochii (winghead shark, Mara et al. 2015). Wu et al. (2015) used MRI images of grass carp, tilapia, turbot, pompano, and large yellow croaker to create 3D models to study the mechanism of fat accumulation in these economically important fishes and found distinct patterns of adipose tissue distribution. The accumulation of excess fat in cultured fishes impairs the ability to metabolize lipids and impacts overall fish health. Using MRI technology, researchers can gain insight into the distribution, shape, and volume of adipose tissues in live fish and use this information to construct optimal nutrition plans for cultured fish (Wu et al. 2015). While sharks do not accumulate adipose tissue, they do store fat in the liver and using MRI technology could still be used to create 3D models to investigate the effects that varying diets have on fat accumulation. These technologies have also been used for human medical research (including intestinal studies) and diagnostics for decades (Pei-You et al. 2015; Murphy et al. 2015). CT scans have been 80% effective at accurately diagnosing small intestine diseases (Pei-You et al. 2015) and have provided digital 3D models of the human digestive tract. If we begin to incorporate these modern methods into studies of shark digestive physiology, we can begin to understand the mechanisms involved in digestive tract form and function. Phylogenetic approaches have also greatly advanced investigations of digestive anatomy form and function in teleost fishes. For example, German et al. (2016) used phylogenetic and comparative genomic analyses to show that a digestive phenotype (elevated amylase activity) in prickleback fishes can be achieved through multiple mechanisms such as increased gene copy number, or elevated expression of fewer genes. These analyses, if used in studies of shark digestion, could provide information about the genetic underpinnings responsible for digestive phenotypes. Taking a metabolomics approach to studies of shark physiology could also open doors to evaluating digestive health using metabolites such as amino acids, fatty acids, sugars, etc. (Karsten and Rice 2004; Sole et al. 2008, 2010; Viana et al. 2008; Dove et al. 2012). Dove et al. (2012) used metabolomics to identify biomarkers in the blood of captive whale sharks that vary with changing health conditions. Understanding critical metabolites and how they vary in differing conditions can aid in improving conceptual models of shark metabolism. This approach has also been used by Sole et al. (2008) in wild environments to monitor marine pollution by identifying changes in lipid peroxidation levels in Merluccius merluccius (European hake) and Galeus melastomus (Blackmouth catshark). Metabolomics has the potential to be a useful tool in identifying how biological and ecological variables impact the health of both teleost and shark populations.
Sharks make use of both mechanical and biochemical processes to break down their food and absorb nutrients. The initial mechanical site of digestion is the orobranchial cavity which consists of the mouth and the pharynx (Holmgren and Nilsson 1999). Food then moves into the esophagus: a short, broad, tube consisting of striated muscle that secretes mucus in order to transport food to the stomach. The beginning of the stomach is obvious histologically due to the sudden change from the stratified epithelium of the esophagus to the columnar mucus cells of the stomach walls (Fig. 4, Wilson and Castro 2011). Most sharks have a large, siphonal (J-shaped) stomach (Holmgren and Nilsson 1999); however, there are some exceptions. For example, the bonnethead shark has a straight (I-shaped) stomach (Fig. 5, Jhaveri et al. 2015). Some teleost fishes have exhibited cecal (Y) shaped stomachs; however, siphonal and straight are the most common forms among sharks (Wilson and Castro 2011). Despite the varying shapes, the overarching purpose of the stomach in sharks, like most vertebrates, is initial storage and biochemical digestion, particularly of protein (Motta and Wilga 1995). The stomach mucosa contains gastric pits (foveola) that lead to the gastric glands (Wilson and Castro 2011). The two main types of gastric cells are columnar cells and oxynicopeptic cells (Fig. 4). Columnar cells line the stomach surface and secrete the mucous used to protect the epithelium from acidic gastric juices. Most sharks possess oxynicopeptic gland cells as the single cell type responsible for the secretion of gastric acid and pepsinogen. Under acidic conditions, pepsinogen (the zymogen) is converted into pepsin, a proteolytic enzyme that plays a critical role in the breakdown of proteins (Fig. 4, Holmgren and Nilsson 1999; Wilson and Castro 2011). At least one species, the sixgill shark (Hexanchus griseus), exhibits mammalian-like parietal and zymogen secreting cells (Michelangeli et al. 1988); however, this has not been observed in any other shark species to date.

From Wilson and Castro (2011), used with permission
Longitudinal sections of the tubular gastric gland of Scyliorhinus canicula. a Immunofluorescent localization of H+/K+-ATPase (green) apically and Na+: K+:Cl- cotransporter (red) basolaterally with representative cross-sections (A–F). Nuclei are counterstained with DAPI (blue) and differential interference contrast images are overlaid for tissue orientation. b Eosin staining revealing granular (pepsinogen) staining in the lower tubule.

Adapted from Jhaveri et al. 2015
Adult bonnethead shark, Sphyrna tiburo, with its “I” shaped digestive tract.
In vertebrates, gastric acid secretion is controlled by both nervous and hormonal inputs (Lloyd and Debas 1994; Furuse and Dockray 1995; Olsson and Holmgren 2001; Schubert 2015). The initial stimulus for increased acid production is expansion of the stomach wall, followed by secretagogues that are secreted by the stomach itself (Holmgren and Nilsson 1999). Gastrin, histamine, and acetylcholine have all been shown to induce increased acid production in teleosts (Holmgren and Nilsson 1999). Gastrin and histamine have been identified in shark tissues, and both induce increased gastric acid secretion, although the response is somewhat lower than expected and the role of these hormones is not fully understood (Hogben 1967; Vigna 1983). Cholecystokinin (CCK) has also been identified in select shark species. CCK is a peptide hormone released in response to proteins and fats entering the intestine, which slows gastric emptying, allowing for longer periods of digestion and absorption of nutrients in the intestine. CCK is also known to suppress appetite due to the presence of a sufficient amount of food already in the intestine. Distinct CCK peptides have been identified in spiny dogfish (Squalus acanthias), the porbeagle (Lamna cornubica), and the shortfin mako shark (Isurus oxyrinchus) but no studies have definitively determined if CCK aids in the control of gastric evacuation rates of sharks in a similar fashion to teleosts (Fig. 1, Goran et al. 1988; Aldman et al. 1989; Konturek et al. 1994; Johnsen et al. 1997; Oliver and Vigna 1996; Olsson et al. 1999).
Sharks have evolved the ability to regulate acid secretion during varying levels of fasting, similar to other fishes and tetrapods, including humans (Day et al. 2014; Sachs et al. 1994). However, there appears to be two patterns of regulation with some species secreting acid continuously and others intermittently (Papastamatiou and Lowe 2005). Nurse sharks (G. cirratum) maintain pH values of 2–3 when digesting, while pH intermittently increases above 8 during periods of fasting (Papastamatiou and Lowe 2005). Leopard sharks (Triakis semifasciata), blacktip reef sharks (Carcharhinus melanopterus), and spiny dogfish (S. acanthias) each maintain stomach pH levels of 1–2, even when fasting (Papastamatiou and Lowe 2004; Papastamatiou et al. 2007; Wood et al. 2007); this lack of stomach pH regulation likely correlates more with feeding frequency than with phylogeny, as these species are not closely related to one another (Fig. 1). Continuous secretion of gastric acid is most likely energetically expensive because of the ATP investment in hydrogen-potassium-ATPase activity; however, the constant presence of acid in the stomach may increase gastric evacuation rates of subsequent meals, since any digesta present in the stomach can be digested to smaller particle sizes and processed more rapidly, which could be important for frequently-feeding sharks (Papastamatiou 2007). More rapid reduction in particle size would increase gastric evacuation rates, allowing a faster return of appetite (Sims et al. 1996). Variation of gastric acid secretion is a topic that is starting to be explored in other vertebrates such as snakes which, like sharks, are generally infrequent feeders (Secor et al. 2012). It has been shown that infrequently feeding snake species rapidly upregulate and downregulate intestinal function with the beginning and ending of every meal. This is hypothesized to reduce the expenditure of energy when the animal is in the post-absorptive state (Secor et al. 2012).
Mixing of gastric fluids with prey, mechanical digestion and evacuation of digested products to the intestine requires the coordinated efforts of gastric motility. Patterns of motility and the factors regulating them have been poorly studied in sharks. As with acid secretion, motility is likely regulated via both nervous and hormonal control (Holmgren et al. 1985; Andrews and Young 1988; Holmgren and Nilsson 1999). Gastric evacuation studies and measurements of motility or electrical impedance in free swimming individuals suggest that there is a lag in both evacuation of prey and gastric motility following feeding; likely to allow the accumulation of gastric fluids and for the stomach to expand and accommodate additional prey items (Bush and Holland 2002; Papastamatiou et al. 2007; Meyer and Holland 2012).
Distal to the stomach is the intestine. While mammalian digestive physiology uses the terms duodenum, jejunum, and ileum to describe the sections of the small intestine (Silverthorn 2013), we argue that these terms are not fitting for describing the shark digestive tract. This is due to the fact that average length, diameter, absorptive surface area, the presence of folds and microvilli, and cell types in the different sections of the small intestine in mammals are not comparable to what is known about the intestinal sections of sharks (Kararli 1995). The duodenum is defined by Brunner’s glands and the division between the duodenum and the jejunum is the ligament of Treitz which forms a distinct kink. The jejunum and ileum can be separated by the presence of m-cells in the ileum. These distinctions of cell types in different regions of the shark gut have not been made (Kararli 1995); instead, distinctions between various regions of shark intestines have been made by differentiating between stratified columnar and cubodial muscle tissue (Chatchavalvanich et al. 2006). Therefore, we suggest the terms proximal, spiral, and distal intestines when discussing shark digestive anatomy.
The proximal intestine is the location of the opening of the ductus choledochus (bile and pancreatic ducts), and thus, is likely the site of polymer digestion. The assumed role of the spiral intestine in sharks is to increase surface area for nutrient absorption and to slow food passage to allow more time for digestion (Wetherbee et al. 1987; Chatchavalvanich et al. 2006; Jhaveri et al. 2015; Bucking 2016). However, little is known about the absorptive properties of the spiral intestine in comparison to other regions of the gastrointestinal tract. The anatomy of the spiral intestine varies amongst species; however, four main variations have been identified based on the direction of the spirals, where the spirals connect with tissue, and whether the intestine is morphologically “scroll” or “spiral” in shape (Fig. 6, Parker 1885). There can be as few as two and as many as fifty spirals, depending on the species (Holmgren and Nilsson 1999). Histological studies on the spiral intestine of the freshwater stingray (Himantura signifier) show that the spiral intestine does in fact increase the surface area of the intestine; however, there was no quantification of its absorptive properties (Chatchavalvanich et al. 2006). Peptide transporter 1 (PepT1), a protein that has been identified as being responsible for the translocation of peptides released during digestion in vertebrates, was identified within the stomach and scroll intestine epithelial lining of bonnethead sharks and as such, is likely correlated with important roles in dietary peptide absorption within these organs (Hart et al. 2016). Additionally, re-absorption of sodium, chloride, water and urea has been measured across the intestine of Squalus acanthias after a feeding event (Anderson et al. 2015). Continuing this type of work on regulation of protein metabolism and absorption in the guts of sharks may explain why they exhibit different feeding and digestive strategies (Ballantyne 2016). There is mounting evidence that the spiral intestine slows transit rate. Retrograde spontaneous muscle peristalsis was observed in the intestines of catsharks (Scyliorhinidae, Fig. 1) and it was suggested as a possible mechanism for retaining food in the intestine for a longer period of time (Andrews and Young 1993). Additionally, 86% of intestinal content mass was found in the scroll intestines of bonnethead sharks, suggesting that the flow of digesta does in fact slow at this location along the gut (Jhaveri et al. 2015). The spiral intestine is connected to the distal intestine (including the rectum), which has a thicker muscular wall since feces accumulates in this section, until pressure on the rectal walls causes nerve impulses to pass to the brain. The brain then sends messages to the voluntary muscles in the anus to relax, permitting expulsion into the cloaca (the genital and urinary ducts also open here; Holmgren and Nilsson 1999).

Images from Wilson and Castro (2011), used with permission
Spiral intestines of elasmobranchs. a Illustrations of various types of spiral intestines from a ventral cut-away view. (i) columnar spiral (Raia spp.), (ii) funnels oriented posteriorly (Raia spp.), (iii) funnels oriented anteriorly (Raia spp.), (iv) cylindrical spiral (Sphyrna spp.). b Light micrograph of a longitudinal section through the spiral intestine of Scyliorhinus caniculae. IW intestinal wall, IF intestinal fold, SF spiral fold. The epithelium consists of columnar cells interspersed with goblet-type mucous cells identified with periodic acid Schiff (PAS) staining. c Longitudinal section of spiral intestine of S. caniculae stained for Na+/K+-ATPase (green), an important ion pump for secondary active transport, and a basolateral membrane marker.
The spiral intestine illustration originally published by Parker in 1885 has been revised and re-cited by shark feeding and nutritional physiology studies for over 130 years (Fig. 6, Bertin 1958; Holmgren and Nilsson 1999; Wilson and Castro 2011). It is clearly time to adopt a new method for visually depicting these unique structures, and to discern how the spiral intestine functions. Additionally, the majority of our morphological quantifications of the spiral intestine have come from dissections and histological images. While these methods no doubt have provided useful information, they are damaging to the structure of the spiral intestine and do not provide an adequate way to visualize whole-scale flow of digesta or quantify volume of intestinal space and surface area of tissue folds. With CT scanning technology becoming more accessible to researchers, we should begin to investigate the morphometrics of the spiral intestine using this technique in order to update our understanding of these unique structures.
Little is also known about intestinal motility. While many teleosts rely on peristalsis of the intestine to move digesta through, to our knowledge, this has not been investigated in sharks (Grove and Campbell 1979). Quantifying the contraction capabilities of the shark proximal, spiral, and distal intestines would aid in establishing digesta transit rates at various points throughout the gut, rather than simply measuring evacuation rate of digesta as a whole (Wetherbee et al. 1987). This information, paired with further quantification of the volumetric flow rate and absorptive properties of the entire shark gut, and the spiral intestine in particular, would reveal how this unique gut type contributes to the economical design of the gastrointestinal tract as a whole.
Digestive strategies and efficiency
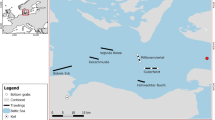
In the past few decades biologists have implemented chemical reactor theory (CRT), a concept historically used in chemical engineering, to study animal digestion. The concept of CRT involves the introduction of chemical reactants (in this case, substrates and enzymes) to a system in which biochemical reactions can occur (digestive tract) to generate products; in the case of a digestive tract in an animal, these products (e.g., glucose, amino acids, fatty acids) can be absorbed and used by the animal for growth, reproduction, tissue maintenance, further acquisition of chemical reactants, etc. (Fig. 7, Penry and Jumars 1987; Horn and Messer 1992; Wolesensky and Logan 2006). The various regions of the gut (e.g., stomach, proximal intestine, spiral intestine, distal intestine) may each function differently in terms of CRT, and this is a function of the compartmentalization of the gut (Fig. 7), which will affect transit time of material through the system (German 2011). In the case of sharks, which all possess a stomach, the chemical reactants must first move into the stomach, through a pyloric sphincter to the intestine, which leads into the spiral intestine (Cortés 1996). This allows for control of transit rate and the passage of only smaller sized particles from the stomach. Larger, proteinaceous food items will likely remain in the stomach, which functions as a continuous-stirred tank reactor (CSTR) in the framework of CRT (Fig. 7, Karasov and Douglas 2013), for a longer period of time until particle sizes are reduced enough to pass through the pyloric sphincter into the intestine. The intestine in most teleost fishes functions as a plug-flow reactor (PFR), meaning that there is little axial mixing along the intestine (Karasov and Douglas 2013; German 2011), and the same would be assumed for the proximal intestine of sharks. Because the flow of material through the spiral intestine has not been visualized, the function of this gut region within CRT remains a mystery, although there is some evidence of slowed flow in this region (Andrews and Young 1993; Jhaveri et al. 2015).
An illustration of the gastrointestinal tract of a the bonnethead shark and b the Greenland shark. The stomach, proximal intestine, spiral intestine, and distal intestine are described as either a continuous-stirred-tank reactor (CSTR), a plug-flow reactor (PFR), or a combination of the two for each shark. The pyloric cecum of the Greenland shark is also depicted. The spiral intestine illustration for each shark is enlarged and the black arrows depict presumed flow paths of digesta. For the spiral intestine of (a) it is presumed that flow can occur through a central lumen as a PFR, or through various “side” lumens, also as PFRs. For (b), flow may occur through a central lumen as a PFR, or can remain within the lumen areas created by the spiral tissue folds as a CSTR. CT scans of spiral intestines suggest that a central lumen may exist, indicating that flow does not always have to follow the entire spiral configuration. The arrow below the enlarged spiral intestine image depicts a nutrient gradient and transitions from high (black) to low (light grey) nutrient concentration. CT scan images of spiral intestines for (a) the bonnethead shark and (b) the pacific spiny dogfish are included below the illustrations. The pacific spiny dogfish spiral intestine is used as an approximation to the Greenland shark spiral intestine.
Along with the efficiency of compartmentalization and the morphology of the digestive tract, visceral temperature, meal size, food type (both physical digestibility and organic makeup), and body size are most likely to determine transit rate of digesta through the digestive system (Cortés et al. 2008). It has been well established that increased stomach temperatures accelerate gastric evacuation in teleosts and sharks (Brett and Groves 1979; Cortés and Gruber 1996; Bush and Holland 2002). There are also endothermic sharks, such as Carcharodon carcharias (white shark), Isurus oxyrinchus (shortfin mako), Isurus paucus (longfin mako), Lamna nasus (porbeagle), and Lamna ditropis (salmon shark) which are all in the Lamnidae family (Fig. 1), that maintain elevated body core temperatures (and specifically stomach temperatures) compared to the surrounding seawater (McCosker 1987; Sepulveda et al. 2004; Goldman et al. 2004). Gastric evacuation times have not been measured in any of these species although they are predicted to be faster than similar sized ectothermic species. Gastric evacuation rates of shortfin mako sharks have been observed to be less than 6 h (Sepulveda and Bernal, pers. obs., as cited in Bernal and Lowe 2016). Some sharks are also thought to optimize the digestive process behaviorally by selecting either warmer or cooler habitats within which to digest (e.g., Sims et al. 2006, Papastamatiou et al. 2015). These could include animals moving vertically in the water column, selecting specific habitats (e.g. sand or mudflats), or both. The behavioral selection may vary based on whether the shark needs to optimize digestive efficiency, or feeding rates.
Meal size influence on gastric evacuation rate appears to vary depending on the species. Prey size was found to have no significant effect on the gastric evacuation time of spiny dogfish (S. acanthias, Jones and Green 1977). Conversely, scalloped hammerheads (Sphyrna lewini) showed an increased gastric evacuation rate with increased meal size (Bush and Holland 2002). Of course, the prey items of sharks differ widely in their physical structure and biochemical composition and therefore food type and quality is presumed to have an impact on gastric evacuation rate. Studies have shown that prey items consisting of hard, chitin-containing exoskeletons (crustaceans) or calcified shells (molluscs) generally take longer to traverse the gut than softer prey (Jackson et al. 1987; Cortés et al. 2008). It has also been found that increased lipid content of prey items prolong gastric evacuation times (Fänge and Grove 1979). This prolonged retention of high-quality prey items (those rich in lipid and protein) is presumably to increase the time for nutrient absorption, and therefore, increase the overall digestive efficiency (Cortés et al. 2008), consistent with slowed gut motility caused by increased CCK secretion in response to amino acids and lipids in the intestine. Additionally, few studies have investigated the effects that consumption of low-quality prey items (those low in protein and lipid, such as plant material) may have on gut transit times in sharks. These types of investigations could provide insight into how and why sharks may specialize on specific prey items, particularly lower-quality items such as plankton, chitin-rich prey, and plant material. There has been little advancement in our understanding of the effect of shark size on gastric evacuation rates, likely because of the difficulties of housing large sharks in captivity; however, the dynamics of nutrient ecology of adults may be very different than that of juveniles.
Sharks generally consume a high-quality diet (high in protein) and may feed infrequently depending on the species (Cortés et al. 2008). As with other fishes, which exist on a ‘feast or famine’ regime, sharks may be digestion limited in that they spend more time digesting than they do searching for food (Jeschke 2007; Armstrong and Schindler 2011; Papastamatiou et al. 2015). Similar to other fishes that consume infrequent meals, shark overall gut length tends to be shorter than their body length (German 2011). One possible reason for needing a smaller gut is to allow room for the large liver that sharks possess, which can be up to 25% of the animal’s body weight (Baldridge 1970; Navarro-Garcia et al. 2000). These large livers play a crucial role in buoyancy and energy storage, due to their ability to store low-density oils (Baldridge 1970; Navarro-Garcia et al. 2000).
In order to understand what sharks are capable of absorbing from their prey, it is important to consider the nutrients available in the food type and what is assimilated during digestion (German 2011). The greatest loss that occurs during digestion is the emission of feces (Fig. 8). While most studies have focused on the digestive efficiency of bony fishes, the same methods used to measure efficiency could be applied to many shark species as well (Fig. 8). Digestive efficiency has historically been measured by feeding fish a set amount of food in a controlled environment and then collecting the fish feces to measure energy and/or nutrient content (Wetherbee and Gruber 1993; Hume 2005; Karasov and Martinez del Rio 2007). Both apparent and true assimilation efficiency can be quantified using this approach. Apparent assimilation efficiency does not account for fecal material containing endogenous materials (such as intestinal mucus and cells) while true assimilation efficiency does account for these types of materials (German 2011; Wetherbee and Gruber 1993; Hume 2003). Apparent assimilation efficiency of sharks has been shown to be similar to that of teleosts although it has only been measured in a few species (Wetherbee and Gruber 1993). The round stingray (Urobatis halleri) was found to be equally as capable as carnivorous teleosts at effectively absorbing nutrients from food, having efficiencies of 75–90% for organic matter (Paig-Tran and Lowe 2010). Juvenile lemon sharks (N. brevirostris) were found to have organic matter digestive efficiencies ranging from 76 to 88% depending on the form and quantity of food (Wetherbee and Gruber 1993). Atlantic stingrays (Dasyatis sabina) and the whitespotted bamboo shark (Chiloscyllium plagiosum), had organic matter absorption efficiencies of 70%, although the total gastric evacuation times were 20 and 40% shorter, respectively, than for lemon sharks (Di Santo and Bennett 2011). This was likely due to the effects of a larger meal size on faster gut transit rate, and thus, lower overall digestive efficiency in the Di Santo and Bennett (2011) study.

Modified from German (2011)
The movement of energy throughout the shark during digestion. Fe, Ne, and Ge represent endogenous energy (lost from the shark directly). Ff, Nf, and Gf correspond to food energy lost in feces, nitrogenous wastes, or gases, respectively. Some portion of metabolizable energy is lost in the form of heat.
Digestive efficiency of teleost fishes has recently been discussed in what is known as a “rate vs. yield” theoretical framework (Fig. 9, Sibly 1981; German et al. 2015). At one end of the spectrum, there is a rate-maximizing strategy, characterized by high intake of food, fast movement of digesta through the gut, and little dependence on microbial digestion (Sibly 1981; German et al. 2015). Rate-maximizers generally only assimilate components of their food that can easily be digested by endogenous enzymes. These fishes use high-intake of food to compensate for the loss of unprocessed nutrients in their fecal material. At the other end of the spectrum is the yield-maximizing strategy which is characterized by less frequent food intake, slower transit of digesta through the gut, and more of a dependence on digestive assistance from microbial communities in the gut (German et al. 2015). Although the “rate vs. yield” framework has largely been used to differentiate amongst herbivorous fishes and their digestive strategies, this framework could also potentially be used to categorize shark digestive strategies (Wetherbee et al. 1987; German et al. 2015). Evidence for a yield-maximizing strategy has recently been found in bonnethead sharks, (S. tiburo, Jhaveri et al. 2015), but given the broad range of diet types, feeding mechanisms, and feeding frequencies exhibited by sharks, it is likely that not all sharks will fall so clearly at one end of the spectrum, and may vary their strategies temporally in response to food availability and quality.
Cumulative nutrient gained (solid black line) by a shark as a function of time spent processing a meal (modified from German et al. 2015; Jhaveri et al. 2015). The slope of the black line labeled “Max Rate” is the maximum rate at which the nutrient can be absorbed from the meal. A rate-maximizing strategy is characterized by a line tangential to the curve (red line “R”), with defecation of gut contents occurring at time 1 (t1). A portion of the nutrient consumed is lost in the feces (“Wastage”), but at t1 the animal can take a new meal. This is the Rate-maximizing strategy with high-intake. Maximum yield (blue line “Y”) is attained by extending processing time to time 2 (t2), however, this is done at the cost of reduced digestive rate. In animals with lower intake, this strategy tends to involve longer retention times of food in the gut and can include microbial fermentation in the hindgut (especially in herbivorous vertebrates)
One technique that can be used to assess dietary assimilation is stable isotope analysis (Bucking 2016). Stable isotope analysis has proven to be useful in providing insight into the dietary patterns of birds, mammals, and teleost fish (Hobson and Clark 1992; Dalerum and Angerbjorn 2005; Trueman et al. 2005; Suzuki et al. 2005; Vollaire et al. 2007; Zuanon et al. 2007; Gamboa-Delgado et al. 2008; Martinez del Rio et al. 2009; Weidel et al. 2011; Nielsen et al. 2015); however, few studies have used this technique when evaluating dietary assimilation in elasmobranchs (e.g., Logan and Lutcavage 2010; Bethea et al. 2011; Kim et al. 2012; Churchill et al. 2015; Li et al. 2015; Carlisle et al. 2017). The use of stable isotope analysis in studies of shark trophic ecology and foraging location has become common; however, it is still a relatively novel technique in studies of shark digestive physiology and the assimilation of specific nutrients. Stable isotope analysis quantifies the incorporation of carbon (C), nitrogen (N), sulfur (S), and sometimes hydrogen (H) from a food source into the tissues of a consumer by measuring naturally occurring variations in isotopic signatures (Hesslein et al. 1993; Wolf et al. 2012; Martínez del Rio and Carleton 2012; Newsome et al. 2015). An isotopic signature is the ratio of the heavy-to-light forms of non-radiogenic isotopes of a particular element, determined by mass spectrometry relative to a standard with a known signature (Martinez del Rio and Wolf 2005; Post 2002; Post et al. 2007). In nature, C and N are approximately 99% 12C and 14N respectively, but the ratio of heavy-to-light isotopes (e.g., 13C/12C) varies in different organisms. When a consumer assimilates nutrients from a particular food source, they generally incorporate the C or N isotopic signature of that food source, and thus, the isotopic signature of a consumer’s tissues can be used to estimate assimilated dietary items (Caut et al. 2009; German and Miles 2010; Kim et al. 2012; Caut et al. 2013; Newsome et al. 2015). However, there is usually some quantitative difference between the host tissue and its diet, known as a tissue-diet discrimination factor, or trophic shift, which varies by element, tissue type, taxon, and diet, amongst other factors (Reich et al. 2008; Caut et al. 2009; Martinez del Rio et al. 2009; German and Miles 2010; Caut et al. 2013). For N, tissue-diet discrimination is usually in the positive direction, allowing N isotopic signatures to be used to understand an animal’s trophic level (Caut et al. 2009; Bethea et al. 2011). In marine systems, C isotopic signatures can be used to infer foraging location, as coastal seagrass (δ13C = ~−10‰) and oceanic phytoplankton (δ13C = ~−20‰) signatures are different, and therefore, the tissues of animals, including sharks, that consume resources from these different foodwebs will have different carbon signatures (Fry 2006; Reich et al. 2007; Bethea et al. 2011). However, if the goal is to use stable isotopic analysis for explicitly understanding shark diet, then it becomes more complicated.
Indeed, understanding the isotopic signatures of consumer tissues is not a simple process, as the 13C and 15N signatures of consumer tissues can change during metabolic processes, and can vary by organism and even by individuals based on diet quality, intake, nutrient assimilation efficiency, body size, growth, age, activity level, protein turnover, tissue type, etc., making it complex to analyze accurately (Peterson and Fry 1987; Martinez del Rio et al. 2009). More controlled laboratory studies of isotopic incorporation and tissue-diet discrimination in sharks are needed to better understand how the isotopic signatures of large predators relate to prey items. To our knowledge, there are two such studies in the literature: one on leopard sharks (Triakis Semifasciata; Kim et al. 2012), and one on the nursehound shark (Scyliorhinus stellaris; Caut et al. 2013). Kim et al. (2012) noted isotopic incorporation rates that varied amongst tissues types (plasma solutes < red blood cells < muscle tissue) in T. semifasciata, as well as tissue-diet discrimination factors that varied by more than 30% with tissue and diet. These results are consistent with papers on fish (e.g., Caut et al. 2009; German and Miles 2010). However, Caut et al. (2013) found variation in isotopic incorporation of S. stellaris on different diets, with rates being faster in plasma solutes than in red blood cells on a fish diet, but, rather surprisingly, the opposite for 15N incorporation rates in sharks fed a mussel diet (i.e., red blood cells < plasma solutes). Like Kim et al. (2012), Caut et al. (2013) observed that tissue-diet discrimination varied with tissue type and diet. A third study examined tissue-diet discrimination of large sand tiger sharks (Carcharias taurus) from a public aquarium that had consumed a known diet (Hussey et al. 2010), also finding variation among tissues. What all of these studies suggest is that knowledge of isotopic incorporation rates, tissue-diet discrimination factors, and the isotopic signatures of prey is required if one is to discern specific dietary items of wild sharks using stable isotopic analyses (Caut et al. 2013). In the laboratory, the use of isotopically-labeled dietary items (e.g., Dennis et al. 2010; German and Miles 2010) and compound specific stable isotope analyses (e.g., Newsome et al. 2011, 2014) provide the potential to discern which parts of specific prey a shark can assimilate, and how the shark uses those resources metabolically, therefore providing new avenues of research in shark nutritional physiology, especially for smaller taxa, or juveniles of large ones.
Another technique that can be used to assess dietary assimilation is fatty acid profiling (Kelly and Scheibling 2012; Cnudde et al. 2015; Bucking 2016; Clements et al. 2017). Fatty acids are released from lipids during digestion and absorbed into the blood stream. Similar to stable isotope analysis, fatty acid profiling requires knowledge of the fatty acid signature of prey items and a known long-term feeding history of the predator, making it a difficult technique to use in field studies or in studies focusing on organisms of which there is little dietary information (Pethybridge et al. 2010; Beckmann et al. 2013; Bucking 2016). However, laboratory studies have shown that liver and muscle fatty acid profiles indicated dietary shifts (Beckmann et al. 2014). Moreover, fatty acid profiling can lead to new hypotheses regarding dietary breadth in different species, even for those with knowledge gaps regarding their nutritional ecology (Clements et al. 2017). For instance, some dietary items, like diatoms, have very specific fatty acid profiles (Kelly and Scheibling 2012), and thus, fatty acid signatures indicative of diatom consumption in the tissues of a consumer likely indicate that they assimilated diatomaceous fatty acids, whether that was an assumed dietary item for that species or not. Along these lines, it is suspected that one benefit to fatty acid profiling is that unique signature patterns in marine fish and invertebrates potentially allows for the identification of specific individual prey species (Iverson et al. 2002; Kelly and Scheibling 2012) and that different tissues may assimilate fatty acids at different rates (Beckmann et al. 2013). While stable isotope analysis and fatty acid profiling are still generally new to the field of shark nutritional physiology, their careful incorporation into future studies in concert with traditional digestibility and stomach content analysis could provide a well-rounded investigation of nutrient assimilation in elasmobranchs.
Enzyme activity in the gastrointestinal tract
To date, no studies have confirmed a conclusive list of endogenously produced digestive enzymes in any shark species, although the recently sequenced elephant shark (Callorhinchus milii; a Holocephalan) genome should increase our development of future enzymatic libraries (Venkatesh et al. 2014). It has been assumed that sharks generally have the same variety of digestive enzymes as other fishes, with their most active enzymes being pepsin, a protein-degrading enzyme in the stomach (Papastamatiou 2007), and aminopeptidase and trypsin in the intestine (Jhaveri et al. 2015). The enzymatic capacities of the spiral intestine have only been quantified for adult bonnethead sharks (Fig. 10, Jhaveri et al. 2015). Aminopeptidase activity levels (indicating protein digestion) peak in the mid-spiral intestine, and are approximately 10x higher than activities in the intestinal tissues of carnivorous teleost fishes (Jhaveri et al. 2015), consistent with dense staining for peptide transporters in this gut region (Hart et al. 2016). Additionally, bonnethead trypsin activities are qualitatively similar to the high activity levels in mako sharks (Jhaveri et al. 2015; Newton et al. 2015). This high level of activity suggests that bonnethead sharks are highly efficient at digesting proteins. Lipase activity (indicating lipid digestion) in bonnetheads is not uncommonly elevated in comparison to teleosts; however, the broad distribution of lipolytic activity along the gut suggests that these sharks most likely digest lipids with great efficiency and regularity (Fig. 10, Jhaveri et al. 2015).

Adapted from Jhaveri et al. (2015)
Aminopeptidase (a) and lipase (b) activities in the tissues of different regions of the gastrointestinal tract of Sphyrna tiburo (bonnethead shark). Activities are mean ± SEM. Aminopeptidase or lipase activities were compared among gut regions independently with ANOVA followed by Tukey’s HSD multiple comparisons test. Regional enzymatic activity values for an enzyme that share a letter are not significantly different from one another (P > 0.05). Because aminopeptidase is a brushborder enzyme, it was not measured in the pancreas. SI spiral intestine, and colon is also called “distal intestine” in the text.
Sharks may be well suited for digesting proteins and lipids, but their ability to digest carbohydrates (starch, maltose, laminarin, laminaribiose, chitin, chitobiose) is not well known (Karasov and Martinez del Rio 2007; Jhaveri et al. 2015; Crane et al. 1979; Kuz’mina 1990; Newton et al. 2015). Bonnethead sharks have relatively low levels of carbohydrate degrading enzymes (maltase) in their intestines; however, there is a spike in β-glucosidase in the distal intestine (Jhaveri et al. 2015). β-glucosidase activity was observed to be 2x higher in the bonnethead shark than in herbivorous teleost fish such as the monkeyface prickleback (Cebidichthys violaceus), suggesting their ability to digest the products of cellulose and laminarin degradation (German et al. 2015). Additionally, elevated levels of chitin-degrading enzymes have been found in some shark guts (Fänge et al. 1979). Bonnethead sharks also possess elevated N-acetyl-β-d-glucosaminidase (NAG) activity in the distal intestine (Jhaveri et al. 2015). Since bonnetheads consume chitin containing crustaceans in their diets (Bethea et al. 2007), this is not entirely surprising. What is surprising is that NAG activity was 5× higher in bonnetheads than in carnivorous, omnivorous, and detritivorous teleost fishes (German et al. 2015). Since the NAG spike occurred in the distal intestine, this is suggestive of a microbial origin. There have been limited numbers of studies investigating chitin-degrading enzymes in other shark species, although there was successful cDNA cloning of a stomach chitinase in blue sharks (Prionace glauca, Suzuki et al. 2014). Additionally, a digestive chitinase gene is observed on contig_19775 in the elephant shark genome (see Venkatesh et al. 2014). The activity levels of each of these enzyme types will likely depend on the diet of the shark, which will vary among species (Karasov and Martinez del Rio 2007).
Enzymatic activity in the digestive tract of teleost fishes has been used to investigate dietary specialization, has been linked to molecular characterization of gene expression, aided in quantifying nutrient utilization and in establishing absorption efficiencies, has been used in constructing efficient aquaculture techniques, and has aided in explanations of ontogenetic shifts in diet (Song et al. 2016; German et al. 2010a, b; Ruan et al. 2010; Guerreiro et al. 2010; Moro et al. 2016; German et al. 2016). Despite the vast role that enzymatic activity experiments have played in nutrition studies of teleosts and the ample resources on the methodology (German et al. 2004; Clements et al. 2009), there have been limited investigations in sharks (Bucking 2016). There should therefore be a push to replicate these types of biochemical enzyme assays and digestibility work on other shark species, beyond the bonnethead (e.g., Newton et al. 2015), in order to start to make these dietary links to digestive functionality and truly grasp what sharks are digesting and excreting into their environments.
Gastrointestinal microbiome
Even less is known about enzymes that are produced in the shark gut by endosymbiotic microorganisms. The intestinal microbial community can vary depending on multiple factors that affect the host such as the temperature and salinity of the environment, developmental stage, digestive physiology, and diet choices (Givens et al. 2015). Since so many factors are capable of changing the microbiome of an individual, it is difficult to study the endosymbiotic relationship in a controlled environment that will likely vary from the microbial community of the wild organism. Using culture-dependent methods, it has been shown that the sharpnose (Rhizoprionodon terraenovae), spinner (Carcharhinus brevipinna), and sandbar sharks (Carcharhinus plumbeus) have microbiomes that consist of Cetobacterium sp., Proteobacterium sp. and Vibrio sp., with Proteobacterium ribotypes dominating the core group of all three of these shark species (Givens et al. 2015). Additional studies have used culture-independent methods to find that Actinobacteria, Firmicutes (Clostridium sp.), Fusobacteria (Cetobacterium sp.), and other Proteobacteria (Campylobacter sp. and Vibrio sp.) are also important members of the shark gut microbiome (Givens et al. 2015). A comparison of the shark microbiome (limited to the above species) to that of bony fishes indicates that there is less species diversity and abundance in the tested shark gut microbiomes than in the tested bony fishes (Table 1, Givens et al. 2015). The exact role of each of the microbe taxa is still largely unknown in sharks (Givens et al. 2015). It has been suggested that microbes are responsible for vitamin production (LeBlanc et al. 2013); however, nothing is known specifically about the vitamin requirements of sharks. The role of microbial fermentation also has received very little exploration in sharks, but there has been some investigation in teleosts. Clements et al. (2017) provide short-chain fatty acid (SCFA) profiles for various reef fishes that indicate fermentation of resources that differ considerably in macronutrient composition, including amino-acids, which is a good indication of protein fermentation in the hind-gut. As such, measurements of SCFA concentrations in the guts of sharks could also indicate microbial fermentation of amino acids. Additionally, further advancement of stable isotope analysis should focus on the contribution of symbiotic microbes to the amino acid homeostatsis of sharks. Newsome et al. (2011) used stable isotope analysis of Nile tilapia to show that 13C values of amino acids resembled 13C values of carbohydrates at low protein intakes, which is consistent with assimilation of essential amino acids of microbial origin by tilapia fed low-protein diets. These types of studies need to be explored in sharks in order to determine how they incorporate the macromolecules consumed through their diverse diets into their tissues and how their microbiome may play a role in this process.
The role of gastrointestinal microbiota in teleosts is a relatively new field (Clements et al. 2014), but already studies have investigated what species of microbes are present in the guts of teleosts, what roles they have at different stages of the gut, how microbial communities change onotogenetically, spatially between habitat types, and due to dietary changes, and how manipulating the gut microbiota can benefit fish health management (Nayak 2010; Lauzon et al. 2014; Sullam et al. 2014; Zarkasi et al. 2016; Ringo et al. 2016). Further investigation into this topic is crucial in developing the relationship between microbes and digestion in the shark gut.
Summary of new directions
It is notable that the latest reviews on elasmobranch nutritional physiology (Cortés et al. 2008; Ballantyne 2016; Bucking 2016) also point out the limited advancements in this field, as much of the focus has been on feeding mechanics (e.g., Wilga and Ferry 2016). Meanwhile, nutritional physiology of teleost fishes has continued to expand (even outside of aquacultural studies). Many of the topics and techniques used in investigations of teleost nutritional physiology can now be applied to cartilaginous fishes.
We suggest the following as directions of study in shark nutritional physiology:
-
(1)
Studies of shark feeding mechanics should involve the use of high-speed video kinematics, VROMM, XROMM, and biorobotic models to better quantify the movements of muscle and skeletal elements (e.g., Corn et al. 2016) for each of the commonly identified feeding mechanisms (bite and retain, suction, and ram). These advances in methodology have been highly successful in quantifying the feeding mechanics of teleosts and could be used in large tank or enclosure settings, as well as potentially in the field.
-
(2)
Despite years of scientists citing reviews that state that the spiral intestine is the most important organ in the nutrient absorption process and that it slows the rate of transit through the gut, there is little quantitative evidence to support these contentions (Wilson and Castro 2011; Jhaveri et al. 2015; Chatchavalvanich et al. 2006; Hart et al. 2016). This needs to be addressed if we are to further the field of shark digestive physiology. We need to move beyond dissection photographs and illustrations and use CT scan technology to create 3D renderings of the various spiral intestine structures. These renderings give us the ability to visualize flow, and make quantitative assessments of the intestinal volume and tissue surface area. This information, paired with quantification of the contractive capabilities, volumetric flow rate, and absorptive properties of the entire shark gut, and the spiral intestine in particular, would reveal how this unique gut type contributes to the economical design of the gastrointestinal tract as a whole. The spiral intestine may also present a new model for chemical reactor theory. Does the spiral intestine function as a PFR, CSTR, or some mixture of the two (Fig. 7)?
-
(3)
We propose that the “rate vs. yield” theoretical framework currently used to describe teleost digestive strategies should be applied to sharks as well (e.g., Jhaveri et al. 2015). Like teleosts, sharks consume a broad range of diet types, inhabit a broad range of habitats, and feed with different frequencies. As such, they likely encompass a broad range of digestive strategies and efficiencies that could be described using this framework. Dietary specialization within this framework could be coupled to stable isotope analysis (e.g., Lujan et al. 2011) and fatty acid profiling (e.g., Clements et al. 2017) to further our understanding of shark dietary diversity (Bucking 2016). Additionally, more studies of isotopic turnover rates and tissue-diet discrimination in sharks are necessary in order to make sense of field isotopic data.
-
(4)
Few studies have explored the role of digestive enzymes in the guts of sharks. There is much to be learned from identifying and classifying the enzymes in each region of the digestive tract, or what enzymes are even present in shark genomes (e.g., Castro et al. 2014; Venkatesh et al. 2014). Such information can be used to pinpoint exactly which nutrients are being used by sharks and where their breakdown is occurring within the gut. This may also provide insight about shark vitamin and mineral requirements. It has been assumed that their needs are similar to other vertebrates (iron, calcium, B vitamins, lipid soluble vitamins, etc.) but this has yet to be explored in a shark species (Halver 2002; Teles 2012). Many of the techniques to measure enzymatic activity are already being used to explore the digestive physiology of other organisms, and therefore, the methodology could be readily applied to sharks as well (German 2011), especially using incidental mortalities from survey work (e.g., Bethea et al. 2007; Jhaveri et al. 2015; Newton et al. 2015).
-
(5)
Understanding which enzymes are of microbial origin versus endogenously derived would also aid in developing the field of digestive physiology from a biochemical standpoint. Since microbiomes are prone to change based on the surrounding environment and individual physiology, it is likely that sharks, particularly migratory sharks, have access to variable sources of exogenous enzymatic activity and nutrient input at varying points throughout their lifetime. This could greatly impact their digestive success and food choices as they develop. As for endogenous enzyme production, exploration in gene expression (transcriptomics) would provide insight to which genes activate the secretion of different enzymes for different shark species. There are many studies of teleost genomics and transcriptomics (Whitehead et al. 2011; Qian et al. 2014; German et al. 2016; Calduch-Giner et al. 2016), but few in sharks (Dowd et al. 2008; Pinhal et al. 2012; Wyffels et al. 2014; Venkatesh et al. 2014; Mulley et al. 2014), and none on the gut in sharks. Understanding which genes are expressed in various shark species would reveal the molecular underpinnings leading to dietary specialization. Ontogenetic shifts in gene expression and enzyme activity have also not been explored (except in teleost fishes) and could be extremely informative given that many species have dramatic shifts in diet as they grow. More genomic studies of Chondrichthyians, and especially in sharks, should be a top priority given that there is currently very little genomic information available for elasmobranchs (Leucoraja erinacea; Wyffels et al. 2014) and holocephalans (Callorhinchus milii; Venkatesh et al. 2014).
Sharks are undeniably still a mystery in many respects, especially with regards to their nutritional physiology. We know that they share some similarities with other, well-studied, carnivorous vertebrates, such as teleosts. However, there are still knowledge gaps in the topics of feeding mechanics, functional morphology of the digestive tract (the spiral intestine in particular), digestive biochemistry, and gastrointestinal/microbiota relationships. As important as sharks are presumed to be ecologically, there is a definite need for future research to investigate their nutritional physiology.
References
Aldman G, Jonsson AC, Jensen J, Holmgren S (1989) Gastrin/CCK-like peptides in the spiny dogfish, squalus acanthias-concentrations and actions in the gut. Comp Biochem Physiol C-Pharmacol Toxicol Endocrinol 91(1):103–108
Anderson WG, McCabe C, Brandt C, Wood CM (2015) Examining urea flux across the intestine of the spiny dogfish, Squalus acanthias. Comp Biochem Physiol 181:71–78
Andrews PLR, Young JZ (1988) The effect of peptides on the motility of the stomach, intestine and rectum in the skate. Comp Biochem Physiol CB9:343–348
Andrews PLR, Young JZ (1993) Gastric motility patterns for digestion and vomiting evoked by sympathetic nerve stimulation and 5-hydroxytryptamine in the dogfish Scyliorhinus canicula. Philos Trans R Soc Lond 342:363–380
Armstrong JB, Schindler DE (2011) Excess digestive capacity in predators reflects a life of feast and famine. Nature 476:84–87
Baldridge H (1970) Sinking factors and average densities of Florida sharks as a function of liver buoyancy. Copeia 4:744–754
Ballantyne JS (2016) Metabolism of elasmobranchs (Jaws II). In: Shadwick RE, Farrell AP, Brauner CJ (eds) Physiology of elasmobranch fishes: internal processes. Elsevier, London
Beckmann CL, Mitchell JG, Seuront L, Stone DA, Huveneers C (2013) Experimental evaluation of fatty acid profiles as a technique to determine dietary composition in benthic elasmobranchs. Physiol Biochem Zool 86:266–278
Beckmann CL, Mitchell JG, Stone DA, Huveneers C (2014) Inter-tissue differences in fatty acid incorporation as a result of dietary oil manipulation in Port Jackson Sharks (Heterodontus portusjacksoni). Lipids 49(6):577–590
Bernal D, Lowe CG (2016) Field studies of elasmobranch physiology. In: Shadwick RE, Farrell AP, Brauner CJ (eds) Physiology of elasmobranch fishes: structure and interaction with environment. Elsevier, London
Bertin L (1958) Appareil digestif. In: Grasse PP (ed) Traite de Zoologic, vol 13. Mason, Paris, pp 1248–1302
Bethea DM, Hale L, Carlson JK, Cortés E, Manire CA, Gelsleichter J (2007) Geographic and ontogenetic variation in the diet and daily ration of the bonnethead shark, Sphyrna tiburo, from the eastern Gulf of Mexico. Mar Biol 152:1009–1020
Bethea DM, Carlson JK, Hollensead LD, Papastamatiou YP, Graham BS (2011) A comparison of the foraging ecology and bioenergetics of the early life-stages of two sympatric hammerhead sharks. Bull Mar Sci 87(4):873–889
Brett JR, Groves TDD (1979) Physiological energetics. In: Hoar WS, Randall DJ, Brett JR (eds) Fish physiology, vol 8. Academic Press, New York, pp 280–352
Bucking C (2016) Feeding and digestion in elasmobranchs: tying diet and physiology together. In: Shadwick RE, Farrell AP, Brauner CJ (eds) Physiology of elasmobranch fishes: structure and interaction with environment. Elsevier, London
Bush AC, Holland KH (2002) Food limitation in a nursery area: estimates of daily ration in juvenile scalloped hammerheads, Sphyrna lewini, in Kanéohe Bay, O’ahu. J Exp Mar Biol Ecol 278:157–178
Calduch-Giner JA, Sitja-Bodadilla A, Perez-Sanches J (2016) Gene expression profiling reveals functional specialization along the intestinal tract of a carnivorous teleostean fish (Dicentrarchus labrax). Front Physiol. doi:10.3389/fphys.2016.00359
Camp AL, Brainerd EL (2014) Role of axial muscles in powering mouth expansion during suction feeding in largemouth bass (Micropterus salmoides). J Exp Biol 217(8):1333–1345
Camp AL, Roberts TJ, Brainerd EL (2015) Swimming muscles power suction feeding in largemouth bass. Proc Natl Acad Sci USA 112(28):8690–8695
Cant J, McBride B, Croom W Jr (1996) The regulation of intestinal metabolism and its impact on whole animal energetics. J Anim Sci 74:2541–2553
Carlisle AB, Litvin SY, Madigan DJ, Lyons K, Bigman JS, Ibarra M, Bizzarro JJ (2017) Interactive effects of urea and lipid content confound stable isotope analysis in elasmobranch fishes. Can J Fish Aquat Sci 74(3):419–428
Castro L, Goncalves O, Mazan S, Tay B, Venkatesh B, Wilson J (2014) Recurrent gene loss correlates with the evolution of stomach phenotypes in gnathostome history. Proc R Soc B 281:2013–2669
Caut S, Angulo E, Courchamp F (2009) Variation in discrimination factors (Delta N-15 and Delta C-13): the effect of diet isotopic values and applications for diet reconstruction. J Appl Ecol 46(2):443–453
Caut S, Jowers MJ, Michel L, Lepoint G, Fisk AT (2013) Diet- and tissue-specific incorporation of isotopes in the shark Scyliorhinus stellaris, a North Sea mesopredator. Mar Ecol Prog Ser 492:185–198
Chatchavalvanich K, Marcos R, Poonpirom J, Thongpan A, Rocha E (2006) Histology of the digestive tract of the freshwater stingray Himantura signifer Compagno and Roberts, 1982 (Sharkii, Dasyatidae). Anat Embryol 211:507–518
Choat JH, Clements KD (1998) Vertebrate herbivores in marine and terrestrial environments: a nutritional ecology perspective. Annu Rev Ecol Syst 29:375–403
Churchill DA, Heithaus MR, Vaudo JJ, Grubbs RD, Gastrich K, Castro JI (2015) Trophic interactions of common elasmobranchs in deep-sea communities of the Gulf of Mexico revealed through stable isotope and stomach content analysis. Deep-Sea Res II 115:92–102
Clements KD, Raubenheimer D, Choat JH (2009) Nutritional ecology of marine herbivorous fishes: ten years on. Funct Ecol 23(1):79–92
Clements KD, Angert ER, Montgomery WL, Choat JH (2014) Intestinal microbiota in fishes: what’s known and what’s not. Mol Ecol 23:1891–1898
Clements KD, German DP, Piche J, Tribollet A, Choat JH (2017) Integrating ecological roles and trophic diversification on coral reefs: multiple lines of evidence identify parrotfishes as microphages. Biol J Linn Soc
Cnudde C, Moens T, Werbrouck E, Lepoint G, Van Gansbeke D, De Troch M (2015) Trophodynamics of estuarine intertidal harpacticoid copepods based on stable isotope composition and fatty acid profiles. Mar Ecol Prog Ser 524:225–239
Compagno L (2008) Pelagic shark diversity. In: Camhi MD, Pikitch EK (eds) Sharks of the open ocean: biology, fisheries and conservation. Blackwell, Oxford, pp 14–23
Corn KA, Farina SC, Brash J, Summers AP (2016) Modeling tooth-prey interactions in sharks: the importance of dynamic testing. R Soc Open Sci 3:160141. doi:10.1098/rsos.160141
Cortés E (1996) A critical review of methods of studying fish feeding based on analysis of stomach contents: application to shark fishes. Can J Fish Aquat Sci 54:726–738
Cortés E, Gruber SH (1996) Gastric evacuation in the young lemon shark, Negaprion brevirostris, under field conditions. Environ Biol Fishes 35:205–212
Cortés E, Papastamatiou Y, Carlson J, Ferry-Graham L, Wetherbee B (2008) An overview of the feeding ecology and physiology of shark fishes. In: Cyrino J, Bureau D, Kapoor B (eds) Feeding and digestive functions in fishes. Science Publishers, New Hampshire
Crane R, Boge G, Rigal A (1979) Isolation of brushborder membranes in vesivular form from the intestinal spiral valve of the small dogfish (Scyliorhinus canicula). Biochim Biophys Acta 554:264–267
Dalerum F, Angerbjorn A (2005) Resolving temporal variation in vertebrate diets using naturally occurring stable isotopes. Oecologia 144:647–658
Day RD, Tibbetts IR, Secor SM (2014) Physiological responses to short-term fasting among herbivorous, omnivorous, and carnivorous fishes. J Comp Physiol B 184:297–512
Dennis CA, MacNeil MA, Rosati JY, Pitcher TE, Fisk AT (2010) Diet discrimination factors are inversely related to δ15N and δ13C values of food for fish under controlled conditions. Rapid Commun Mass Spectrom 24:3515–3520
Di Santo V, Bennett WA (2011) Is post-feeding thermotaxis advantageous in shark fishes? J Fish Biol 78:195–207
Diamond JM, Karasov WH (1987) Adaptive regulation of intestinal nutrient transporters. Proc Natl Acad Sci USA 84(8):2242–2245
Dove ADM, Leisen J, Zhou M, Byrne JJ, Lim-Hing K, Webb HD, Gelbaum L, Viant MR, Kubanek J, Fernandez FM (2012) Biomarkers of whale shark health: a metabolomic approach. PLoS ONE. doi:10.1371/journal.pone.0049379
Dowd WW, Wood CM, Kajimura M, Walsh PJ, Kültz D (2008) Natural feeding influences protein expression in the dogfish shark rectal gland: a proteomic analysis. Comp Biochem Physiol D Genomics Proteomics 3:118–127
Fänge R, Grove D (1979) Digestion. In: Hoar WS, Randall DJ (eds) Fish physiology, vol 8. Academic Press, New York, pp 161–260
Fänge R, Lundblad G, Lind J, Slettengren K (1979) Chitinolytic enzymes in the digestive system of marine fishes. Mar Biol 53:317–321
Ferry-Graham LA, Lauder GV (2001) Aquatic prey capture in ray-finned fishes: a century of progress and new directions. J Morphol 248:99–119
Frazetta TH, Prange CD (1987) Movements of cephalic components during feeding in some requiem sharks (Carcharhiniformes: Carcharhinidae). Copeia 1987:979–993
Fry B (2006) Stable isotope ecology. Springer, New York
Furuse M, Dockray GJ (1995) The regulation of gastrin-secretion in the chicken. Regul Pept 55(3):253–259
Gajić A (2013) Comparative odontology of selachians (Chondricthyes: Sharkii): development and morphological characteristics of teeth. Presented at the 17th annual Symposium of Biology Students in Europe. Abstract
Gamboa-Delgado J, Canavate JP, Zerolo R, Le Vay L (2008) Natural carbon stable isotope ratios as indicators of the relative contribution of live and inert diets to growth in larval Senegalese sole (Solea senegalensis). Aquaculture 280(1–4):190–197
German DP (2011) Digestive efficiency. In: Farrell AP (ed) Encyclopedia of fish physiology: from genome to environment, vol 3. Academic Press, San Diego, pp 1596–1607
German DP, Miles RD (2010) Stable carbon and nitrogen incorporation in blood and fin tissue of the catfish Pterygoplichthys disjunctivus (Siluriformes, Loricariidae). Environ Biol Fishes 89:117–133
German DP, Horn MH, Gawlicka A (2004) Digestive enzyme activities in herbivorous and carnivorous prickleback fishes (Teleostei: Stichaeidae): ontogenetic, dietary, and phylogenetic effects. Physiol Biochem Zool 77(5):789–804
German DP, Nagle BC, Villeda JM, Ruiz AM, Thomson AW, Contreras S, Evans DH (2010a) Evolution of herbivory in a carnivorous clade of minnows (Teleostei: Cyprinidae): effects on gut size and digestive physiology. Physiol Biochem Zool 83(1):1–18
German DP, Neuberger DT, Callahan MN, Lizardo NR, Evans DH (2010b) Feast to famine: the effects of food quality and quantity on the gut structure and function of a detritivorous catfish (Teleostei: Loricariidae). Comp Biochem Physiol A 155:281–293
German DP, Sung A, Jhaveri P, Agnihotri R (2015) More than one way to be an herbivore: convergent evolution of herbivory using different digestive strategies in prickleback fishes (family Stichaeidae). Zoology 118:161–170
German DP*, Foti DM*, Heras J, Amerkhanian H, Lockwood BL (2016) Elevated gene copy number does not always explain elevated amylase activities in fishes. Physiol Biochem Zool 89:277–293
Gidmark NJ, Taylor C, LoPresti E, Brainerd E (2015) Functional morphology of Durophagy in Black Carp, Mylopharyngodon piceus. J Morphol 276(12):1422–1432
Givens C, Ransom B, Bano N, Hollibaugh J (2015) Comparison of the gut microbiomes of 12 bony fish and 3 shark species. Mar Ecol Prog Ser 518:209–223
Goldman KJ, Anderson SD, Latour RJ, Musick JA (2004) Homeothermy in adult salmon sharks, Lamna ditropis. Environ Biol Fishes 71:403–411
Goran A, Jonsson A, Jensen J, Holmgren S (1988) Gastrin/CCK-like peptides in the spiny dogfish, Squalus acanthias; concentrations and actions in the gut. Comp Biochem Phys 92(1):103–108
Grove DJ, Campbell G (1979) The role of extrinsic and intrinsic nerves in the co-ordination of gut motility in the stomachless flatfish Rhombosolea tapirina and Ammotretis rostrata Guenther. Comp Biochem Physiol C 63(1):143–159
Guerreiro I, de Vareilles M, Pousao-Ferreira P, Rodrigues V, Dinis MT, Ribeiro L (2010) Effect of age-at-weaning on digestive capacity of white seabream (Diplodus sargus). Aquaculture 300(1–4):194–205
Halver JE (2002) The vitamins. In: Halver JE, Hardy RW (eds) Fish nutrition, 3rd edn. Academic Press, San Diego, pp 61–141
Hart HR, Evans AN, Gelsleichter J, Ahearn GA (2016) Molecular identification and functional characteristics of peptide transporters in the bonnethead shark (Sphyrna tiburo). J Comp Physiol B 186(7):855–866
Hesslein RH, Hallard KA, Ramlal P (1993) Replacement of sulfur, carbon, and nitrogen in tissue of growing broad whitefish (Coregonus nasus) in response to a change in diet traced by δ34S, δ13C, and δ15N. Can J Fish Aquat Sci 50:2071–2076
Hobson KA, Clark RG (1992) Assessing avian diets using stable isotopes I: turnover of 13C in tissues. Condor 94:181–188
Hogben CAM (1967) Response of the isolated dogfish gastric mucosa to histamine. Proc Soc of Exp Biol Med 124:890–893
Holmgren S, Nilsson S (1999) Digestive system. In: Hamlett WC (ed) Sharks, skates, and rays: the biology of shark fishes. The Johns Hopkins University Press, Baltimore, pp 144–173
Holmgren S, Grimes D, Brayton P, Colwell R, Gruber S (1985) Vibrios as autochthonous flora of neritic sharks. Syst Appl Microbiol 6:221–226
Horn MH, Messer KS (1992) Fish guts as chemical reactors: a model of the alimentary canals of marine herbivorous fishes. Mar Biol 113:527–535
Hume I (2003) Nutrition of carnivorous marsupials. In: Jones M, Dickman C, Archer M (eds) Predators with pouches: the biology of carnivorous marsupial. CSIRO Publishing, Collingwood, Australia, pp 221–227
Hume I (2005) Concepts of digestive efficiency. In: Starck J, Wang T (eds) Physiological ecological adaptations to feeding vertebrates. Science Publishers, Enfield, pp 43–58
Hussey NE, Brush J, McCarthy ID, Fisk AT (2010) δ15N and δ13C diet—tissue discrimination factors for large sharks under semi-controlled conditions. Comp Biochem Physiol A 155:445–453
Iverson SJ, Frost KJ, Lang SLC (2002) Fat content and fatty acid composition of forage fish and invertebrates in Prince William Sound, Alaska: factors contributing to among and within species variability. Mar Ecol Prog Ser 241:161–181
Jackson S, Dubby DC, Jenkins JFG (1987) Gastric digestion in marine vertebrate predators: in vitro standards. Funct Ecol 1:287–291
Jeschke JM (2007) When carnivores are “full and lazy”. Oecologia 152(2):357–364
Jhaveri P, Papastamatiou Y, German DP (2015) Digestive enzyme activities in the guts of bonnethead sharks (Sphyrna tiburo) provide insight into their digestive strategy and evidence for microbial digestion in their hindguts. Comp Biochem Physiol A 189:76–83
Johnsen AH, Jonson L, Rourke IJ, Rehfeld JF (1997) Sharks express separate cholecystokinin and gastrin genes. Proc Natl Acad Sci USA 94(19):10221–10226
Jones BC, Green GH (1977) Food and feeding of spiny dogfish (Squalus acanchias) in British Columbia waters. J Fish Res Board Can 43:2067–2078
Kararli T (1995) Comparison of the gastrointestinal anatomy, physiology, and biochemistry of human and commonly used laboratory animals. Biopharm Drug Dispos 16:351–380
Karasov WH (1992) Tests of the adaptive modulation hypothesis for dietary control of intestinal nutrient transport. Am J Physiol 263:R496–R502
Karasov WH, Diamond J (1983) Adaptive regulation of sugar and amino acid transport by vertebrate intestine. Am J Physiol 245:G443–G462
Karasov WH, Douglas AE (2013) Comparative and digestive physiology. Compr Physiol 3:741–783
Karasov WH, Martinez del Rio C (2007) Physiological ecology: how animals process energy, nutrients, and toxins. Princeton University Press, Princeton
Karsten AH, Rice CD (2004) c-Reactive protein levels as a biomarker of inflammation and stress in the Atlantic sharpnose shark (Rhizoprionodon terraenovae) from three southeastern USA estuaries. Mar Environ Res 58(2–5):747–751
Kelly JR, Scheibling RE (2012) Fatty acids as dietary tracers in benthic food webs. Mar Ecol Prog Ser 446:1–22
Kenaley CP, Lauder GV (2016) A biorobotic model of the suction-feeding system in largemouth bass: the roles of motor program speed and hyoid kinematics. J Exp Biol 219:2048–2059
Kim SL, Martinez del Rio C, Casper D, Koch PL (2012) Isotopic incorporation rates for shark tissues from a long-term captive feeding study. J Exp Biol 215(14):2495–2500
Kolmann MA, Welch K, Summers AP, Lovejoy NR (2016) Always chew your food: freshwater stingrays use mastication to process tough insect prey. Proc R Soc B Biol Sci 283:20161392. doi:10.1098/rspb.2016.1392
Konturek JW, Thor P, Maczka M, Stoll R, Domschke W, Konturek SJ (1994) Role of cholecystokinin in the control of gastric emptying and secretory response to a fatty meal in normal subjects and duodenal ulcer patients. Scand J Gastroenterol 29(7):583–590
Kuz’mina V (1990) Characteristics of enzymes involved in membrane digestion in shark fishes. Zhur Evolyut Biokhim Fiziolog 26:161–166
Laurence-Chasen JD, Jimenez YE, Knorlein BJ, and Brainerd EL (2016) Video Reconstruction of Moving Morphology (VROMM) for studies of suction feeding in ray-finned fishes. Conference: Annual Meeting of the Society-for-Integrative-and-Comparative-Biology (SICB). Integrative and Comparative Biology, 56(1): E321, Meeting Abstract: P2.175, Portland, OR
Lauzon HL, Perez-Sanchez T, Merrifield DL, Ringo E, Balcazar JL (2014) Probiotic applications in cold water fish species. Aquac Nutr Gut Health Probiotics Prebiotics 9:223–252
LeBlanc J, Milani C, Savoy de Giori G, Sesma F, van Sinderen D, Ventura M (2013) Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol 24:160–168
Li Y, Zhang Y, Hussey NE, Dai X (2015) Urea and lipid extraction treatment effects on δ15N and δ13C values in pelagic sharks. Rapid Commun Mass Spectrom 30:1–8
Lloyd KCK, Debas HT (1994) Peripheral regulation of gastric acid secretion. In: Johnson LR (ed) Physiology of the gastrointestinal tract. Plenum Publishing Corporation, New York, pp 229–334
Logan JM, Lutcavage ME (2010) Stable isotope dynamics in elasmobranch fishes. Hydrobiologia 644:231–244
Longo SJ, McGee MD, Oufiero CE, Waltzek TB, Wainwright PC (2016) Body ram, not suction, is the primary axis of suction-feeding diversity in spiny-rayed fishes. J Exp Biol 219(1):119–128
Lujan NK, German DP, Winemiller KL (2011) Do wood grazing fishes partition their niche? Morphological and isotopic evidence for trophic segregation in Neotropical Loricariidae. Funct Ecol 25:1327–1338
Mara KR, Motta PJ, Martin AP, Hueter RE (2015) Constructional morphology within the head of hammerhead sharks (Sphyrnidae). J Morphol 276(5):526–539
Martin RA (2003) Biology of sharks and rays. Illustrations. World Wide Web Publication, www.elasmo-research.org/copyright.htm
Martine A, Fuhrman F (1995) The relationship between summated tissue respiration and metabolic rate in the mouse and dog. Physiol Biochem Zool 28:18–34
Martínez del Rio C, Carleton SA (2012) How fast and how faithful—the dynamics of isotopic incorporation into animal tissues. J Mamm 93:353–359
Martinez del Rio C, Wolf N (2005) Mass-balance models for animal isotopic ecology. In: Starck MA, Wang T (eds) Physiological and ecological adaptations to feeding in vertebrates. Science Publishers, Enfield, New Hampshire, pp 141–174
Martinez del Rio C, Wolf N, Carleton SA, Gannes LZ (2009) Isotopic ecology ten years after a call for more laboratory experiments. Biol Rev 84:91–111
McCosker JE (1987) The white shark, Carcharodon carcharias, has a warm stomach. Copeia 1987:195–197
Meyer CG, Holland KN (2012) Autonomous measurement of ingestion and digestion processes in free-swimming sharks. J Exp Biol 215:3681–3684
Michelangeli F, Ruiz MC, Dominquez MG, Parthe V (1988) Mammalian like differentiation of gastric cells in the shark Hecanchus griseus. Cell Tissue Res 251:225–227
Moro GV, Silva TSC, Zanon RB, Cyrino JEP (2016) Starch and lipid in diets for dourado Salminus brasiliensis (Cuvier 1816): growth, nutrient utilization and digestive enzymes. Aquac Nutr 22(4):890–898
Motta PJ, Wilga CAD (1995) Anatomy of the feeding apparatus of the lemon shark, Negaprion brevirostris. J Morphol 226:309–329
Motta PJ, Wilga CAD (2001) Advances in the study of feeding behaviors, mechanisms, and mechanics of sharks. Environ Biol Fish 60:131–156
Motta PJ, Hueter RE, Tricas TC, Summers AP, Huber DR, Lowry D, Mara KR, Matott MP, Whitenack LB, Wintzer AP (2008) Functional morphology of the feeding apparatus, feeding constraints, and suction performance in the nurse shark. J Morphol 269:1041–1055
Motta PJ, Maslanka M, Hueter RE, Davis RL, de la Parra R, Mulvany SL, Habegger ML, Strother JA, Mara KR, Gardiner JM, Tyminski JP, Zeigler LD (2010) Feeding anatomy, filter-feeding rate, and diet of whale sharks, Rhincodon typus during surface ram filter feeding off the Yucatan Peninsula, Mexico. Zoology 113:199–212
Muller M, Osse JWM (1984) Hydrodynamics of suction feeding in fish. Trans Zool Soc Lond 37:51–135
Mulley JF, Hargreaves AD, Hegarty MJ, Heller RS, Swain MT (2014) Transcriptomic analysis of the lesser spotted catshark (Scyliorhinus canicula) pancreas, liver and brain reveals molecular level conservation of vertebrate pancreas function. BioMed Cent. doi:10.1186/1471-2164-15-1074
Murphy KP, Crush L, Twomey M, McLaughlin PD, Mildenberger IC, Moore N, Bye J, O’Connor OJ, McSweeney SE, Shanahan F, Maher MM (2015) Model-based iterative reconstruction in CT enterography. AJR Am J Roentgenol 205(6):1173–1181
Nakaya K (2001) White band on upper jaw of megamouth shark, Megachasma pelagios, and its presumed function (Lamniformes: Megachasmidae). Bull Fac Sci Hokkaido Univ Sapporo 52:125–129
Nakaya K, Matsumoto R, Suda K (2008) Feeding strategy of the megamouth shark Megachasma pelagios (Lamniformes: Megachasmidae). J Fish Biol 73(1):17–34
Navarro-Garcia G, Aguilar-Pacheco R, Cordova-Vallejo B, Suarez-Ramirez J, Bolanos A (2000) Lipid composition of the liver oil of shark species from the Caribbean and gulf of California waters. J Food Compos Anal 13(5):791–798
Navia A, Mejia-Falla P, Giraldo A (2007) Feeding ecology of shark fishes in coastal waters of the Columbian Eastern Tropical Pacific. BMC Ecol 7:8
Nayak SK (2010) Role of gastrointestinal microbiota in fish. Aquac Res 41(11):1553–1573
Nelson JD (2004) Distribution and foraging ecology by Whale Sharks (Rhincodon typus) within Bahia de los Angeles, Baja California Norte, Mexico. M.Sc. Thesis. San Diego State University, CA, USA
Newsome SD, Fogel ML, Kelly L, Martinez del Rio C (2011) Contributions of direct incorporation from diet and microbial amino acids to protein synthesis in Nile tilapia. Funct Ecol 25(5):1051–1062
Newsome SD, Wolf N, Peters J, Fogel ML (2014) Amino acid δ13C analysis shows flexibility in the routing of dietary protein and lipids to the tissue of an omnivore. Integr Comp Biol 54(5):890–902
Newsome SD, Sabat P, Wolf N, Rader JA, Martinez del Rio C (2015) Multi-tissue δ2H analysis reveals altitudinal migration and tissue-specific discrimination patterns in Cinclodes. Ecosphere 6(11):1–18
Newton K, Wraith J, Dickson K (2015) Digestive enzyme activities are higher in the shortfin mako shark, Isurus oxyrinchus, than in ectothermic sharks as a result of visceral endothermy. Fish Physiol Biochem 41:887–898
Nielsen JM, Popp BN, Winder M (2015) Meta-analysis of amino acid stable nitrogen isotope ratios for estimating trophic position in marine organisms. Oecologia 178:631–642
Oliver AS, Vigna SR (1996) CCK-X receptors in the endothermic mako shark (Isurus oxyrinchus). Gen Comp Endocrinol 102(1):61–73
Olsson C, Holmgren S (2001) The control of gut motility. Comp Biochem Physiol A-Mol Integr Physiol 128(3):481–503
Olsson C, Aldman G, Larsson A, Holmgren S (1999) Cholecystokinin affects gastric emptying and stomach motility in the rainbow trout Oncorhynchus mykiss. J Exp Biol 202(2):161–170
Paig-Tran EWM, Lowe C (2010) Elemental and energy assimilation in the round stingray, Urobatis halleri. Annu Meet Soc Integr Comp Biol 50(1):E227
Paig-Tran EWM, Summers AP (2013) Comparison of the structure and composition of the branchial filters of suspension feeding sharks. Anat Rec 297(4):701–715
Paig-Tran EWM, Bizzarro JJ, Strother JA, Summers AP (2011) Bottles as models: predicting the effects of varying swimming speed and morphology on size selectivity and filtering efficiency in fishes. J Exp Biol 214:1643–1654
Papastamatiou Y (2007) The potential influence of gastric acid secretion during fasting on digestion time in leopard sharks (Triakis semifasciata). Comp Biochem Physiol A 147:37–42
Papastamatiou YP, Lowe CG (2004) Postprandial response of gastric pH in leopard sharks (Triakis semifasciata) and its use to study foraging ecology. J Exp Biol 207(Pt2):225–232
Papastamatiou Y, Lowe C (2005) Variations in gastric acid secretion during periods of fasting between two species of shark. Comp Biochem Physiol A 141:210–214
Papastamatiou Y, Purkis S, Holland K (2007) The response of gastric pH and motility to fasting and feeding in free-swimming blacktip reef sharks, Carcharhinus melanopterus. J Exp Mar Biol Ecol 345:129–140
Papastamatiou YP, Watanabe YY, Bradley D, Dee LE, Weng K, Lowe CG, Caselle JE (2015) Drivers of daily routines in an ectothermic marine predator: Hunt warm, rest warmer? PLoS ONE 10:e0127807
Parker TJ (1885) On the intestinal spiral valve in the genus Raja. Zool Soc Lond Trans 11:49–61
Pei-You G, Jun-Xia L, Feng-Li L, Liang-Ming Z, Hai-Zhu X, Yan-Bin S (2015) Retrospective comparison of computed tomography enterography and magnetic resonance enterography in diagnosing small intestine disease. J Pak Med Assoc 65(7):710–714
Penry DL, Jumars PA (1987) Modeling animal guts as chemical reactors. Am Nat 129(1):69–96
Peterson BJ, Fry B (1987) Stable isotopes in ecosystem studies. Annu Rev Ecol Syst 18:293–320
Pethybridge H, Daley R, Virtue P, Nichols P (2010) Lipid composition and partitioning of deepwater chondrichthyans: inferences of feeding ecology and distribution. Mar Biol 157(6):1367–1384
Pinhal D, Shivji MS, Nachtigall PG, Chapman DD, Martins C (2012) A streamlined DNA tool for global identification of heavily exploited coastal shark species (genus Rhizoprionodon). PLoS ONE 7(4):e34797
Post DM (2002) Using stable isotopes to estimate trophic position: models, methods, and assumptions. Ecol. 83:703–718
Post DM, Layman CA, Arrinton DA, Takimoto G, Quattrochi J, Montana CG (2007) Getting to the fat of the matter: models, methods and assumptions for dealing with lipids in stable isotope analysis. Oecologia 152:179–189
Qian X, Ba Y, Zhuang Q, Zhong G (2014) RNA-seq technology and its application in fish transcriptomics. OMICS 18(2):98–110
Reich KJ, Bjorndal KA, Bolten AB (2007) The ‘lost years’ of green turtles: using stable isotopes to study cryptic lifestages. Biol Let 3:712–714
Reich KJ, Bjorndal KA, Martinez del Rio C (2008) Effects of growth and tissue type on the kinetics of (13)C and (15)N incorporation in a rapidly growing ectotherm. Oecologia 155(4):651–663
Ringo E, Zhou Z, Vecino JLG, Wadsworth S, Romero J, Krojdahl A, Olsen RE, Dimitroglou A, Foey A, Davies S, Owen M, Lauzon HL, Martinsen LL, De Schryver P, Bossier P, Perstad S, Merrifield DL (2016) Effect of dietary components on the gut microbiota of aquatic animals. A never-ending story? Aquac Nutr 22(2):219–282
Ruan GL, Li Y, Gao ZX, Wang HL, Wang WM (2010) Molecular characterization of trypsinogens and development of trypsinogen gene expression and tryptic activities in grass carp (Ctenopharyngodon idellus) and topmouth culter (Culter alburnus). Comp Biochem Physiol B Biochem Mol Biol 155(1):77–85
Sachs G, Prinz C, Loo D, Bamberg K, Besancon M, Shin JM (1994) Gastric acid secretion: activation and inhibition. Yale J Biol Med 67:81–95
Schubert ML (2015) Functional anatomy and physiology of gastric secretion. Curr Opin Gastroenterol 31(6):479–485
Secor S, Taylor J, Grosell M (2012) Selected regulation of gastrointestinal acid-base secretion and tissue metabolism for the diamondback water snake and Burmese python. J Exp Biol 215:185–196
Sepulveda CA, Kohin S, Chan C, Vetter R, Graham JB (2004) Movement patterns, depthpreferences, and stomach temperatures of free-swimming juvenile mako sharks, Isurus oxyrinchus, in the Southern California Bight. Mar Biol 145:191–199
Shamur E, Zilka M, Hassner T, China V, Liberzon A, Holzmen R (2016) Automated detection of feeding strikes by larval fish using continuous high-speed digital video: a novel method to extract quantitative data from fast, sparse kinematic events. J Exp Biol 219(11):1608–1617
Sibly RM (1981) Strategies of digestion and defecation. In: Townsend CR, Callow P (eds) Physiological ecology: an evolutionary approach to resource use. Sinauer, Sunderland, pp 109–139
Silverthorn D (ed) (2013) Human physiology: an integrated approach, vol 6. Pearson, Boston, p 699
Sims D, Davies S, Bone Q (1996) Gastric emptying rate and return of appetite in lesser spotted dogfish, Scyliorhinus canicula. J Mar Biol. Assn UK 76:479–491
Sims D, Wearmouth VJ, Southall EJ, Hill JM, Moore P, Rawlinson K, Hutchinson N, Budd GC, Righton D, Metcalfe JD, Nash JP, Morritt D (2006) Hunt warm, rest cool: bioenergetic strategy underlying diel vertical migration in a benthic shark. J Anim Ecol 75:176–190
Smith MM, Johanson Z, Underwood C, Diekwisch TGH (2013) Pattern formation in development of chondrichthyan dentitions: a review of an evolutionary model. Hist Biol Int J Paleobiol 25(2):127–142
Sole M, Lobera G, Aljinovic B, Rios J, de la Parra LMG, Maynou F, Cartes JE (2008) Cholinesterases activities and lipid peroxidation levels in muscle from shelf and slope dwelling fish from the NW Mediterranean: its potential use in pollution monitoring. Sci Total Environ 402:306–317
Sole M, Anto M, Baena M, Carrasson M, Cartes JE, Maynou F (2010) Hepatic biomarkers of xenobiotic metabolism in eighteen marine fish from NW Mediterranean shelf and slope waters in relation to some of their biological and ecological variables. Mar Environ Res 70(2):181–188
Song Z, Wang J, Qiao H, Li P, Zhang L, Xia B (2016) Ontogenetic changes in digestive enzyme activities and the amino acid profile of starry flounder Platichthys stellatus. Chin J Oceanol Limnol 34(5):1013–1024
Sullam KE, Dalton CM, Russell JA, Kilham SS, El-Sabaawi R, German DP, Flecker AS (2014) Changes in digestive traits and body nutritional composition accommodate a trophic niche shift in Trinidadian guppies. Oecologia 177(1):245–257
Summers AP, Hayes M (2016) CT scans. Retrieved from www.osf.io/ecmz4
Suzuki KW, Kasai A, Nakayama K, Tanaka M (2005) Differential isotopic enrichment and half-life among tissues in Japanese temperate bass (Lateolabrax japonicus) juveniles: implications for analyzing migration. Can J Fish Aquat Sci 62(3):671–678
Suzuki T, Kakizaki H, Ikeda M, Matsumiya M (2014) Molecular cloning of the novel chitinase gene from blue shark (Prionace glauca; Chondrichthyes) stomach. J Chitin Chitosan Sci 2(2):143–148
Teles OA (2012) Nutrition and health of aquaculture fish. J Fish Dis 35:83–108
Trueman CN, McGill RAR, Guyard PH (2005) The effect of growth rate on tissue-diet isotopic spacing in rapidly growing animals. An experimental study with Atlantic salmon (Salmo salar). Rapid Commun Mass Spectrom 19(22):3239–3247
Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, Swann JB, Ohta Y, Flajnik MF, Sutoh Y, Kasahara M, Hoon S, Gangu V, Roy SW, Irimia M, Korzh V, Kondrychyn I, Lim ZW, Tay BH, Tohari S, Kong KW, Ho S, Lorente-Galdos B, Quilez J, Marques-Bonet T, Raney BJ, Ingham PW, Tay A, Hillier LW, Minx P, Boehm T, Wilson RK, Brenner S, Warren WC (2014) Elephant shark genome provides unique insights into gnathostome evolution. Nature 505:174–179
Viana TP, Inacio AF, de Albuquerque C, Linde-Arias AR (2008) Biomarkers in a shark species to monitor marine pollution: Effects of biological parameters on the reliability of the assessment. Mar Environ Res 66:171
Vigna S (1983) Evolution of endocrine regulation of gastrointestinal function in lower vertebrates. Am Zool 23:729–738
Vollaire Y, Banas D, Marielle T, Roche H (2007) Stable isotope variability in tissues of the Eurasian perch Perca fluviatilis. Comp Biochem Physiol A-Mol Integr Physiol 148(3):504–509
Wainwright D, Lauder GV (2016) Three-dimensional analysis of scale morphology in bluegill sunfish, Lepomis macrochirus. Zoology 119(3):182–195
Weidel BC, Carpenter SR, Kitchell JF, Vander Zanden MJ (2011) Rates and components of carbon turnover in fish muscle: insights from bioenergetics models and a whole-lake 13C addition. Can J Fish Aquat Sci 68:387–399
Wetherbee B, Gruber S (1993) Absorption efficiency of the lemon shark negaprion brevirostris at varying rates of energy intake. Copeia 2:416–425
Wetherbee B, Gruber S, Ramsey A (1987) X-radiographic observations of food passage through digestive tracts of lemon sharks. Trans Am Fish Soc 116:763–767
Wetherbee BM, Gruber SH, Cortés E (1990) Diet, feeding habits, digestion, and consumption in sharks, with special reference to the lemon shark, Negaprion brevirostris. NOAA Tech Rep NMFS 90:29–47
Whitehead A, Galvez F, Zhang S, Williams LM, Oleksiak MF (2011) Functional genomics of physiological plasticity and local adaptation in killfish. J Hered 102(5):499–511
Wilga CD, Ferry LA (2016) Functional anatomy and biomechanics of feeding in elasmobranchs. In: Shadwick RE, Farrell AP, Brauner CJ (eds) Physiology of elasmobranch fishes: internal processes. Elsevier, London
Wilga CD, Motta PJ (2000) Durophagy in sharks: Feeding mechanics of the hammerhead Sphyrna tiburo. J Exp Biol 201:1345–1358
Wilson J, Castro L (2011) Morphological diversity of the gastrointestinal tract in fishes. Fish Physiol 30:1–55
Wolesensky W, Logan JD (2006) Chemical reactor models of digestion modulation. In: Burk AR (ed) Focus on ecology research, pp 197–247
Wolf N, Newsome SD, Fogel ML, Martinez del Rio C (2012) An experimental exploration of the incorporation of hydrogen isotopes from dietary tissues into avian tissues. J Exp Biol 215:1915–1922
Wood CM, Kajimura M, Bucking C, Walsh PJ (2007) Osmoregulation, ionoregulation and acid-base regulation by the gastrointestinal tract after feeding in the shark (Squalus acanthias). J Exp Biol 210:1335–1349
Wu JL, Zhang JL, Du XX, Shen YJ, Lao X, Zhang ML, Chen LQ, Du ZY (2015) Evaluation of the distribution of adipose tissues in fish using magnetic resonance imaging (MRI). Aquaculture 448:112–122
Wyffels J, King BL, Vincent J, Chen C, Wu CH, Polson SW (2014) SkateBase, an shark genome project and collection of molecular resources for chondrichthyan fishes. F1000 Research, v1; ref status: indexed, http://f1000r.es/445
Zarkasi KZ, Taylor RS, Abell GC, Tamplin ML, Glencross BD, Bowman JP (2016) Atlantic salmon (Salmo salar L.) gastrointestinal microbial community dynamics in relation to digesta properties and diet. Microb Ecol 71(3):589–603
Zuanon JAS, Pezzato AC, Ducatti C, Barros MM, Pezzato LE, Passos JRS (2007) Muscle delta C-13 change in nile tilapia (Oreochromis niloticus) fingerlings fed lants grain-based diets. Comp Biochem Physiol A-Mol Integr Physiol 147(3):761–765
Acknowledgements
We would like to thank the comparative physiology group at UCI for providing guidance and advice, particularly J. Heras, B. Wehrle, and A. Frederick. J. German and A. Dingeldein helped with image creation and analysis. CT scanning facility and assistance provided by A. Summers and the University of Washington at Friday Harbor Laboratories.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Leigh, S.C., Papastamatiou, Y. & German, D.P. The nutritional physiology of sharks. Rev Fish Biol Fisheries 27, 561–585 (2017). https://doi.org/10.1007/s11160-017-9481-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11160-017-9481-2