Abstract
In the present paper, the mathematical modeling of highly sensitive enzyme biosensor kinetics is discussed. The standard method of inverting a Laplace transform according to the Heaviside expansion theorem is applied to solve the coupled nonlinear time-dependent reaction–diffusion equations for the Michaelis–Menten expression that describes the concentrations of the substrate and product within the enzymatic layer. The analytical expressions for the concentration of the substrate and product have been derived for all values of the rate constant. A numerical simulation is also reported using the MATLAB software program. Our analytical results are compared with our simulation results. The analytical results show good agreement with those obtained using numerical method.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Analytical chemistry, specifically electroanalytical chemistry, plays an important role in the fields of biochemistry, pharmaceuticals, chemistry, environment science and food production. In recent decades, biosensors have emerged from the laboratories into the everyday lives of many millions of people around the world. An electroanalytical biosensor based on the enzyme-catalysis is an analytical device that uses biological enzymes to detect the presence of chemical molecules.
In the biosensor field, electrochemical techniques, such as cyclic voltammetry (CV), and electrochemical impedance spectroscopy (EIS), have been proven to be advantageous for developing new methods for the determination of pharmaceutical [1, 2], environmental [3] and food [4] samples. Therefore, several researchers have pursued the investigation of the equivalent circuit model for the interface that consists of the electrode geometry and electrolyte parameter. Recently, an ultrasensitive enzyme biosensor was developed [5,6,7,8,9,10,11].
Preparing a novel ultrasensitive biosensor may be very expensive, as it may require many laboratory experiments. Therefore, it is wise to conduct computational experiments prior to physical experiments, which necessitates mathematical modeling and simulations of the biosensor. In chemical engineering, modeling and simulation are important tools for engineers and scientists to better understand the behavior of chemical processes [12,13,14,15,16,17]. Modeling methods are very useful for the design and optimization of chemical plants, for process control, and for training of operators and operational planning [18,19,20,21,22].
Mathematical models of enzyme biosensor responses can be created by solving partial differential equations (PDE) of the substrate diffusion and the biocatalytical conversion with the initial and the boundary conditions. Theoretical modeling of time dependent nonlinear differential equations for the electrochemical enzyme biosensors implies the use of a nonlinear term related to the Michaelis–Menten kinetic scheme, which can be solved analytically and numerically. For this purpose, this paper derives an analytical expression for the concentrations of the substrate and product for application in highly sensitive enzyme biosensors using the standard method of inverting a Laplace transform according to the Heaviside expansion theorem.
Kinetics and mathematical model
Kinetics model
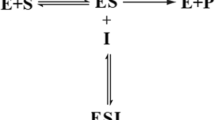
Electrochemical biosensors are constructed, for many cases, using a three-electrode system with a modified enzyme as the working electrode, Ag/AgCl as the reference electrode and platinum as the counter electrode. The use of electrochemical impedance spectroscopy (EIS) in this field may be applied to discriminate and quantify the different processes that determine a biosensor performance, such as the Ohmic resistance Rs, charge transfer resistance Rct, diffusion resistance Zw, and capacitance Cdl (Fig. 1). However, the kinetic model for an enzyme action, first elucidated by Michaelis and Menten (Eq. 1) suggests the binding of the free enzyme (E) to the reactant or substrate (S) forming an enzyme–substrate complex (ES). This complex undergoes a transformation, which releases the product (P) and enzyme (E) [23,24,25]. Note that substrate binding is reversible, but product release is not (Fig. 1).
Here k1, k2 and k3 are the rate constants.
Mathematical model of highly sensitive biosensor
In biosensors, coupling of the substrate transport with the diffusion is described by Fick’s law with the enzyme-catalyzed reaction in the enzyme layer, which leads to the following equations:
Here x represents space between \(0\) and \(d\), with \(d\) is the thickness of the enzyme layer; \(c_{s} (x,t)\) and \(c_{p} (x,t)\) are the molar concentrations of the substrate \(S\) and the product \(P\) in the enzyme layer, respectively; \(V_{m}\) is the maximal enzymatic rate; \(k_{M}\) is the Michaelis constant; \(d\) is the thickness of the enzyme layer; and \(D_{s}\) and \(D_{p}\) are the diffusion coefficients.
For highly sensitive enzyme biosensors, the amount of substrate is small. Thus, the amount of the substrate \(c_{s}\) is negligible compared to the magnitude of The Michaelis constant, \(k_{M}\). Then, Eqs. 2 and 3 will be reduced to the following form:
With
Dimensionless form of the problem
The following parameters are used to convert the above Eqs. 4 and 5 into their normalized forms. We make the above nonlinear partial differential equations in a dimensionless form by defining the following parameters:
Then, Eqs. 4 and 5 reduce to the following dimensionless form:
The dimensionless initial and boundary conditions are:
Analytical solution of the reaction–diffusion problem for highly sensitive biosensor
By applying Laplace transformation to the partial differential Eqs. 7 and 8 and using the conditions Eq. 9, the following transformed differential equations are obtained. The expansion method and inversion of the Laplace transform are used to solve this system.
Analytical solution of kinetic of substrate concentration
The Laplace transform reduces the partial differential equation in Eq. 7 to the an ordinary differential equation (Eq. 10):
With boundary conditions:
The general solution of this equation is:
With boundary conditions at \(X = 0\) and \(X = 1\), the solution becomes:
We observe that the solution becomes a new function \(F\left( {X,s} \right)\):
The standard method of inverting a Laplace transform makes use of the residue theorem [26]. According the Heaviside expansion theorem [27], the inverse transform of F(s) are given by the residue theorem. That is, let:
Here, we have:
On the other hand, the solution of inverting a Laplace transform is:
If \(S_{n}\): is a simple pole of F(s), then \(\rho_{n} (t)\) is given by:
Here \(Q^{'} \left( {s_{n} } \right)\), denoted \(\frac{dQ}{dt}\), is evaluated at the singular point of interest.
Recall that:
Therefore, the Q simple zero is located at:
-
The first simple pole when S = 0
-
The second pole: \(e^{{\sqrt {s + a} }} + e^{{ - \sqrt {s + a} }} = 0\)
When \(s_{n} = \frac{{ - \pi^{2} \left( {2n + 1} \right)^{2} - 4a}}{4}\)
Applying Eq. 19 gives:
We can use these results to invert a Laplace transform for the analytical solution of the substrate concentration. The analytical solution is given by:
With: \(\lambda_{n} = \pi \left( {n + 1/2} \right)\)
Analytical solution of the kinetic product concentration
We need to introduce a new function \(C_{M} \left( {X,T} \right)\) as:
We obtain a system of ordinary differential equations:
By applying the Laplace transformation in Eq. 26:
With
The analytical solution of Eq. 27 with the initial and boundary conditions of Eq. 28 in the form:
The analytical solution of Eq. 29 using the same method for solving Eq. 14 (case a = 0) is given by:
Using Eqs. 23, 25 and 30, we have:
After rearrangement, the solution of the dimensionless product concentration is given by:
Results and discussion
Analytical solution validation
The analytical solutions are validated for a specific set of values. In this paper, the analytical data are validated through the numerical data obtained from the numerical modeling with the MATLAB program. The function pdex4 in MATLAB, which is a function of solving the initial boundary value problems for parabolic partial differential equations [28], is used to solve Eqs. 7 and 8 for the corresponding boundary conditions in Eq. 9.
Fig. 2 shows the response of the enzyme biosensor for various substrate and product concentrations, accepting two cases a = 0.1 and a = 1 with different times of t = 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1. In all the cases, the percentage of deviation for the analytical solution from the numerical result are calculated to be less than 1%. This conforms that the modeled data with analytical solution are very much similar to numerical data.
Reaction–diffusion effect
The mathematical model presented here in Eq. 23 for the substrate and Eq. 32 for the product utilizes well-developed enzyme-catalyzed reaction diffusion equations, which were applied to a highly sensitive enzyme biosensor. The mathematical solution results in Eqs. 23 and 32 indicate that the factor \(a = \frac{{ V_{m} d^{2} }}{{D k_{M} }}\) is the principal factor that controls the biosensor response.
In the case of biosensors, the diffusion modulus or Damkohler number (a) compares the rate of the enzyme reaction \((\frac{{ V_{m} }}{{ k_{M} }})\) with the diffusion \((\frac{ D }{{d^{2} }})\) through the enzyme layer [29].
Figs. 3 and 4 show the concentration profiles for different values of the Damkohler number. Small values of the Damkohler number indicate that the surface reaction dominates and that a significant amount of the reactant diffuses well into the membrane without reacting.
It can be seen from Fig. 3 that with the decrease in the Damkohler number, there is an increase in the dimensionless substrate concentration degradation.
As shown in Fig. 4, concentration of the product decreases when there is a decrease in the Damkohler number.
Conclusions
The mathematical model of a highly sensitive enzyme biosensor can be successfully used to investigate the response of biosensors when the enzyme reacts with its substrate to produce new product. An approximate analytical expression of substrate and final product has been derived using the standard method of inverting a Laplace transform according to the Heaviside expansion theorem to solve the coupled nonlinear time-dependent reaction–diffusion equations for the Michaelis–Menten expression that describes the concentrations of the substrate and the product within the enzymatic layer. Our results are compared with the numerical simulations in MATLAB, showing a good agreement is found between the two sets of results. The analytical method is an extremely simple approach and is promising to better understand the ultrabiosensor model.
References
Gil ED, de Melo GR (2010) Electrochemical biosensors in pharmaceutical analysis. Braz J Pharm Sci 46:375–391
Djaalab E, Samar MEH, Zougar S, Kherrat R (2018) Electrochemical biosensor for the determination of amlodipine besylate based on gelatin-polyaniline iron oxide biocomposite film. Catalysts 8:233
Wanekaya AK, Chen W, Mulchandani A (2008) Recent biosensing developments in environmental security. J Environ Monit 10:703–710
Viswanathan S, Radecka H, Radecki J (2009) Electrochemical biosensors for food analysis. Monatsh Chem 140:891–899
Sethuraman V, Muthuraja P, Raj JA, Manisankar P (2016) A highly sensitive electrochemical biosensor for catechol using conducting polymer reduced graphene oxide–metal oxide enzyme modified electrode. Biosens Bioelectron 84:112–119
Zehani N, Fortgang P, Saddek LM, Baraket A, Arab M, Dzyadevych SV, Kherrat R, Jaffrezic-Renault N (2015) Highly sensitive electrochemical biosensor for bisphenol A detection based on a diazonium-functionalized boron-doped diamond electrode modified with a multi-walled carbon nanotubetyrosinase hybrid film. Biosens Bioelectron 74:830–835
Wang X, Hu J, Zhang G, Liu S (2014) Highly selective fluorogenic multianalyte biosensors constructed via enzyme-catalyzed coupling and aggregation-induced emission. J Am Chem Soc 136(28):9890–9893
Tang H, Yan F, Lin P, Xu J, Chan HLW (2011) Highly sensitive glucose biosensors based on organic electrochemical transistors using platinum gate electrodes modified with enzyme and nanomaterials. Adv Func Mater 21(12):2264–2272
Zheng Z, Zhou Y, Li X, Liu S, Tang Z (2011) Highly-sensitive organophosphorous pesticide biosensors based on nanostructured films of acetylcholinesterase and CdTe quantum dots. Biosens Bioelectron 26(6):3081–3085
Lu J, Drzal LT, Worden RM, Lee I (2007) Simple fabrication of a highly sensitive glucose biosensor using enzymes immobilized in exfoliated graphite nanoplatelets nafion membrane. Chem Mater 19(25):6240–6246
Lee D, Lee J, Kim J, Kim J, Na HB, Kim B, Kim HS (2005) Simple fabrication of a highly sensitive and fast glucose biosensor using enzymes immobilized in mesocellular carbon foam. Adv Mater 17(23):2828–2833
Lente G (2017) Advanced data analysis and modelling in chemical engineering. Reac Kinet Mech Cat 120(2):417–420
Yang KZ, Twaiq F (2017) Modelling of the dry reforming of methane in different reactors: a comparative study. Reac Kinet Mech Cat 122(2):853–868
Elizalde I, Trejo F, Muñoz JA, Torres P, Ancheyta J (2016) Dynamic modeling and simulation of a bench-scale reactor for the hydrocracking of heavy oil by using the continuous kinetic lumping approach. Reac Kinet Mech Cat 118(1):299–311
Izadbakhsh A, Khatami A (2014) Mathematical modeling of methanol to olefin conversion over SAPO-34 catalyst using the percolation concept. Reac Kinet Mech Cat 112(1):77–100
Marečić M, Jović F, Kosar V, Tomašić V (2011) Modelling of an annular photocatalytic reactor. Reac Kinet Mech Cat 103(1):19–29
Le TD, Lasseux D, Nguyen XP, Vignoles G, Mano N, Kuhn A (2017) Multi-scale modeling of diffusion and electrochemical reactions in porous micro-electrodes. Chem Eng Sci 173:153–167
Ingham J (2000) Chemical engineering dynamics: an introduction to modelling and computer simulation. Wiley-VCH, Weinheim
Yeo YK (2017) Chemical engineering computation with MATLAB, 1st edn. CRC Press, Boca Raton, FL
AL Malah K (2013) MATLAB numerical methods with chemical engineering applications. McGraw Hill, New York
Buzzi-Ferraris G, Manenti F (2013) Nonlinear systems and optimization for the chemical engineer: Solving numerical problems. Wiley-VCH, Weinheim
Ross G (1987) Computer programming examples for chemical engineers. Elsevier, Amsterdam
English BP, Min W, Van Oijen AM, Lee KT, Luo G, Sun H, Cherayil BJ, Kou SC, Xie XS (2006) Ever-fluctuating single enzyme molecules: Michaelis–Menten equation revisited. Nat Chem Biol 2(2):87–94
Dóka É, Lente G (2012) Stochastic mapping of the Michaelis–Menten mechanism. J Chem Phys 136(5):054111
Kou SC, Cherayil BJ, Min W, English BP, Xie XS (2005) Single-molecule Michaelis–Menten equations. J Phys Chem B 109:19068–19081
Loney NW (2006) Applied mathematical methods for chemical engineers, 2nd edn. CRS Press, Boca Raton
Mathews JH, Howell RW (2012) Complex analysis for mathematics and engineering, 6th edn. Jones and Bartlett Learning, LLC, Bullington
MATLAB & Simulink, www.mathworks.com/help/matlab/ref/pdepe.html. Accessed 09 July, 2018
Villadsen J, Nielsen J, Liden G (2011) Bioreaction engineering principles. Springer, Dordrecht
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Djaalab, E., Samar, M.E.H. & Zougar, S. Mathematical modeling of the kinetics of a highly sensitive enzyme biosensor. Reac Kinet Mech Cat 126, 49–59 (2019). https://doi.org/10.1007/s11144-018-1516-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11144-018-1516-8