Abstract
The paper extended the TMDD model to drugs with more than two (N > 2) identical binding sites (N-to-one TMDD). The quasi-steady-state (N-to-one QSS), quasi-equilibrium (N-to-one QE), irreversible binding (N-to-one IB), and Michaelis–Menten (N-to-one MM) approximations of the model were derived. To illustrate properties of new equations and approximations, N = 4 case was investigated numerically. Using simulations, the N-to-one QSS approximation was compared with the full N-to-one TMDD model. As expected, and similarly to the standard TMDD for monoclonal antibodies (mAb), N-to-one QSS predictions were nearly identical to N-to-one TMDD predictions, except for times of fast changes following initiation of dosing, when equilibrium has not yet been reached. Predictions for mAbs with soluble targets (slow elimination of the complex) were simulated from the full 4-to-one TMDD model and were fitted to the 4-to-one TMDD model and to its QSS approximation. It was demonstrated that the 4-to-one QSS model provided nearly identical description of not only the observed (simulated) total drug and total target concentrations, but also unobserved concentrations of the free drug, free target, and drug-target complexes. For mAb with a membrane-bound target, the 4-to-one MM approximation adequately described the data. The 4-to-one QSS approximation converged 8 times faster than the full 4-to-one TMDD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
It has been shown that monospecific biologics that bind to a single target receptor with high affinity is not optimal for many therapeutic applications. For example, multivalent binding is needed for optimal IgG antibody-dependent cellular cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC) to improve target cell killing [1, 2]. Furthermore, biologics that can bind to multiple receptors can neutralize pathogens, diseased cells, and soluble proteins much better than monospecific biologics [3, 4]. As a result, many avidity-based biologics that can bind multiple receptors have been developed to overcome the therapeutic limitation of monospecific biologics [4, 5].
A commonly used target-mediated drug disposition (TMDD) model [6] and its approximations [7, 8] assume that both, the drug and the target have only one binding site (one-to-one binding). In [9], the TMDD model and its approximations were derived for drugs that have two identical binding sites (two-to-one binding). This was an important extension as most therapeutic monoclonal IgG antibodies (mAbs) belong to this class [10, 11]. Here we extend the TMDD model and its approximations to drugs that have more than two (N > 2) identical binding sites (N-to-one binding). This extension can be helpful for modeling IgA antibodies that have 2 or 4 binding sites [12], IgM antibodies that have 10 or 12 binding sites [13], or engineered antibodies or other biologics with more than 2 binding sites [2, 5, 14].
While the TMDD model with one-to-one binding assumption describes biologics sufficiently accurately in most cases, development of mathematical models that describe N-to-one binding more mechanistically may facilitate understanding of drug-target interactions and their influence on pharmacokinetic and pharmacodynamic properties of the system. To simplify notations, the models for N-to-one binding will be referred to as N-to-one TMDD, N-to-one quasi-steady state (QSS), N-to-one quasi-equilibrium (QE), N-to-one irreversible binding (IB), and N-to-one Michaelis–Menten (MM) models.
Theoretical
TMDD model for a drug with N binding sites
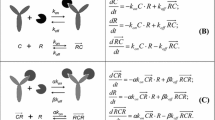
A schematic representation of binding interactions for the drug that has N = 4 identical binding sites with the target that has one binding site is presented in Fig. 1. We assume that all sites are identical, i.e. binding constants kon and koff are the same for all sites and are independent of the number of occupied sites. The free drug (C) and the free target (R) are defined as the drug and the target that are not bound to each other. The binding processes form drug-target complexes, RkC, k = 1, …, N, where k is the number of drug binding sites occupied by the target. When k < N, these complexes correspond to the partially bound drug. The complex RNC corresponds to the fully bound drug, with all binding sites occupied by the target. For future derivations it is convenient to extend the notation to k = 0 and k = N + 1 and define R0C = C and RN+1C = 0.
Concentrations of the total drug (Ctot) and the total target (Rtot) can be then expressed as
We assume that the drug is described by a two-compartment model with combined linear elimination and target-mediated drug disposition/elimination following intravenous (IV) and subcutaneous (SC) dosing and that elimination rate of all drug-target complexes (kint) is the same and is independent of the type of the complex (number of binding sites occupied). We also assume that binding constants are the same for all binding sites and they do not depend on the occupancy of the other binding sites. Then, equations of the system can be written as 4 equations that describe total drug and total target concentrations:
and N equations that describe drug-target complexes:
Here kel is the linear elimination rate constant, kpt and ktp are inter-compartment rate constants, kon, koff, and kint are the binding, dissociation, and internalization (elimination of the complex) rate constants; kdeg and ksyn are the degradation (elimination of the target) and target production rate constants; Vc is the central compartment volume; In(t) is the infusion rate; FSC is the absolute bioavailability of the subcutaneous dose. Initial conditions correspond to the case where the free drug that is not present endogenously is administered as a subcutaneous dose D1 and bolus dose D2.
Quasi-steady-state approximation
Similarly to TMDD equations with one-to-one binding, the quasi-steady-state approximation of the TMDD N-to-one system can be derived by assuming that the free (unbound) drug C, the free (unbound) target R, and the drug-target complexes RkC are in quasi-steady-state [8], where binding rates are balanced by the sum of dissociation and internalization rates on the scale of the other processes:
The QSS approximation is then described by Eqs. (3)-(6) supplemented by the algebraic Eqs. (1)-(2) and (8).
A recent work by Ng and Bauer [15] demonstrated that the QSS system of differential and algebraic equations can be successfully solved numerically. However, it is rather challenging and requires significant expertise in numerical methods. Here we demonstrate an example of analytical solution of algebraic part of the system, allowing for simple and fast numerical implementation.
We aim to find the algebraic solution of Eqs. (1)-(2) and (8), that is, to express all quantities C, R, RkC, k = 1,…,N as functions of Ctot and Rtot defined in (1)-(2).
First, we will derive equation for R. To do this, we compute the sum of Eqs. (8) multiplying each by the respective k (number of binding sites occupied by the target). The resulting equation is:
Then, tedious but straightforward calculations presented in Appendix 1 result in the equation:
Here KD = koff/kon is the dissociation rate constant and KIB = kint/kon is the irreversible binding rate constant introduced in [9].
This equation is identical to the corresponding expression for one-to-one binding if we interpret \(N{\cdot C}_{tot}\) as the total number of binding sites in the system. One can solve this equation for R to find:
This is the key equation of the QSS approximation, but it is only half of the solution. We also need expressions for C and RkC as functions of R, Ctot, and Rtot.
As shown in Appendix 2, RNC can be expressed as
while the remaining elements can be found recursively as follows:
Note that free drug concentration C can be found from (14) when k = 0, or, equivalently, from the equation:
Thus, the N-to-one QSS approximation is described by differential Eqs. (3)-(6) supplemented by algebraic Eqs. (11)-(15). Equations (12)-(14) are explicitly written for N = 4 in Appendix 3.
When internalization rate of the complexes kint is equal to the degradation rate of the free target kdeg, the total target concentration Rtot is a constant parameter of the system, and the Eq. (6) is not needed.
Quasi-equilibrium approximation
QE equations can be obtained from the corresponding QSS equations by setting KIB = 0. They can be used to resolve binding equation for the in-vitro experiment with known drug and target concentrations, Ctot and Rtot, respectively. In this case, distribution of species is described by the system:
In fact, in this case the system can be simplified (Appendix 4) and presented in the form:
This solution is intuitively obvious. When probability of the binding site being occupied in equal to R/(KD + R), and probability of the site being free is KD/(KD + R), then Eqs. (20) show the probability of k sites being occupied and (N-k) sites being free.
Irreversible binding approximation
The irreversible binding approximation can be obtained from the QSS approximation by assuming koff = 0 (resulting in KD = 0). Binding equations in this case have the form
In this case the system can be simplified and presented in the form (see Appendix 5):
Michaelis–Menten approximation
To further simplify the QSS equations, we will assume that the free drug concentration is approximately equal to the total drug concentration and that the free target concentration R is small relative to \({({K}_{IB}+K}_{D})\). In this case, we can derive (see Appendix 6)
Since R is small and RkC is of the order Rk, we can conclude that
and that there is negligible concentration of complexes with k > 1.
Disregarding the quadratic term when solving Eq. (10) for R we get:
and then
Substituting Ctot – C and Rtot – R by R1C in (4) and (6), keeping only the terms up to the order of R, and assuming that Ctot is approximately equal to C, we can arrive at equations of the Michaelis–Menten approximation:
These equations are equivalent to the MM approximation of the standard TMDD system where KSS = (KD + KIB)/N.
As before, if kint is equal to kdeg, the total target concentration is a constant parameter of the system, and the last equation can be removed. Then, the system is described by the two-compartment model with parallel linear and Michaelis–Menten elimination.
Investigation of the N-to-one model
To investigate the N-to-one model and its approximations, several simulation studies were performed for N = 4 binding sites. NONMEM 7.5.1® software [16] was used for simulations, estimation of parameters, and computation of predictions. FOCEI estimation method was used for model fitting.
First, the case of “slow elimination of complexes”, with parameters typical for mAbs and soluble targets was evaluated with two examples.
In example 1, typical predictions of the free drug, the free target, the total drug, the total target, and each of the 4 drug target complexes were computed for the full 4-to-one TMDD model and the 4-to-one QSS approximation to compare the curves at different doses and times. Four consecutive intravenous (IV) doses of 10, 50, 100, and 500 nmol were administered 21 days apart. The parameters from Table 1 (“True” values of fixed-effect parameters) were used for simulation; all variance parameters were set to zero.
In example 2, population PK simulation/estimation was performed. The population PK data set that imitated data from combined typical phase 1 and phase 2 studies (Table 2) was used to simulate concentrations of various quantities from the full 4-to-one TMDD model. The parameters from Table 1 (“True” values) were used for simulations. The simulated concentrations of the free drug and of the total target were then used to fit the following models:
-
a.
Full 4-to-one TMDD model with kon parameter estimated.
-
b.
Full 4-to-one TMDD model with kon parameter fixed at the true value.
-
c.
QSS 4-to-one approximation.
For these models, the estimated parameters were compared with the true values. Predictions of all quantities (including unobserved ones) were also computed and compared with the true (i.e. simulated from the full 4-to-one TMDD model) values. Conversion times were recorded. For all estimations, the true values were used as the initial values, thus the ability of models to converge was not tested and conversion times may have been optimistic (especially for the full model).
Then, the case of “fast elimination of complexes”, with parameters typical for mAbs and membrane-bound targets where Michaelis–Menten equations should be valid was evaluated. The same simulations as in example 1 of the “slow elimination of complexes” case were performed for the 4-to-one full TMDD, QSS, and MM models. The parameters from Table 3 were used in the simulations.
Results
For the “slow elimination of complexes” case, typical predictions of the full 4-to-one TMDD model and the 4-to-one QSS approximation (Fig. 2) were almost identical for all quantities.
In the “fast elimination of complexes” case, kint is equal to kdeg, so Rtot is constant and only C, R, and R1C are changing with time. As shown in Fig. 3, both 4-to-one QSS and the 4-to-one Michaelis–Menten models were able to reproduce the predictions of the full 4-to-one TMDD model for all these quantities.
In the population study for the “slow elimination of complexes” case, the full 4-to-one TMDD model (Model a) converged and provided accurate estimates for all model parameters except kon (Table 1). Model b. where kon was fixed to the true value converged and provided accurate estimates for all the other model parameters. The 4-to-one QSS model provided accurate estimates for all the parameters of the QSS model.
Model fit (objective function value [OFV]) and run times of all the models are shown in Table 1. The run time of the full 4-to-one TMDD model with the fixed kon value was approximately 8 times longer than that of the 4-to-one QSS model. The run time of the full 4-to-one TMDD model with the estimated kon value was approximately 21 times longer than that of the 4-to-one QSS model. Each function evaluation of the full model was taking approximately 9–10 times longer of the QSS model due to longer model integration time.
Discussion
Equations of the TMDD model and its QSS, QE, IB, and MM approximations were derived for drugs with N > 2 binding sites. The N-to-one QSS model was the most general approximation. The N-to-one QE and N-to-one IB approximations can be obtained from N-to-one QSS by setting KIB = 0 or KD = 0, respectively. The QSS 4-to-one model correctly estimated model parameters and predicted drug and target concentrations over time when it was fitted to the data simulated from the full 4-to-one TMDD model.
While advances in computer power and the software made numerical solution of the full N-to-one TMDD model possible, they did not (and could not) resolve the identifiability issue: binding parameters of the system cannot be determined from the routinely available data. Therefore, when the parameter kon was fixed at the true value, the full model converged much faster and was able to estimate all parameters correctly, while the model with the estimated value of kon took 2.5 times longer time to converge and was not able to estimate this parameter precisely even though dense sampling in the range of interest (immediately after the end of infusion) was available.
The system of Eqs. (1)-(4) was presented using differential equations for the total drug and total target concentrations rather than in the (equivalent and more traditional) form that uses differential equations for free drug and free target concentrations. The advantage of this form is that the large terms (terms that contain the parameter kon) are localized in equations for the RkC complexes rather than distributed throughout the entire system. We demonstrated that this is convenient for theoretical analysis of the system. Authors’ experience with numerical investigations using the full TMDD model indicated that this version is also more stable numerically. We speculate that this is because the processes with high derivatives (that involve fast binding) are localized in Eq. (7) while derivatives in Eqs. (1)-(4) are relatively small. This may allow more stable integration of the part of the system that describes slow-changing variables.
The run time of the 4-to-one QSS model was about 8 times faster than that of the full 4-to-one TMDD model with fixed kon, and it was about 21 times faster than that of the full 4-to-one TMDD model with the estimated kon value. All estimations used the true values as initial values. Further study is needed to determine and compare the ability of these models to obtain precise and robust parameter estimates from the real study data. The NONMEM code of the full 4-to-1 TMDD model, the QSS approximation of this model, and the corresponding Michelis-Menten approximation are provided in Appendix 7, 8, and 9, respectively.
We should repeat the limitation of the suggested analytical approach. We assumed that binding constants kon and koff are the same for all sites and they are independent of the number of occupied sites. Indeed, the parameters kon and koff are expected to be similar due to structural symmetry (as in most cases they are replicas of the same structure). However, it is unknown how much these parameters (and other parameters of the drug-target complexes, including kint) change when some or many of the binding sites are occupied, thus changing the structure (protein folding) of the drug-target complexes and their molecule weight. If this assumption does not hold, the analytical approach is intractable. For example, this approach does not describe systems with cooperative or allosteric binding.
Data availability
No datasets were generated or analysed during the current study.
References
Diebolder CA, Beurskens FJ, de Jong RN, Koning RI, Strumane K, Lindorfer MA, Voorhorst M, Ugurlar D, Rosati S, Heck AJ, van de Winkel JG, Wilson IA, Koster AJ, Taylor RP, Saphire EO, Burton DR, Schuurman J, Gros P, Parren PW (2014) Complement is activated by IgG hexamers assembled at the cell surface. Science 343(6176):1260–1263
Patel KR, Roberts JT, Barb AW (2019) Multiple variables at the leukocyte cell surface impact Fc gamma receptor-dependent mechanisms. Front Immunol 10:223. https://doi.org/10.3389/fimmu.2019.00223
Rujas E, Kucharska I, Tan YZ, Benlekbir S, Cui H, Zhao T, Wasney GA, Budylowski P, Guvenc F, Newton JC, Sicard T, Semesi A, Muthuraman K, Nouanesengsy A, Aschner CB, Prieto K, Bueler SA, Youssef S, Liao-Chan S, Glanville J, Christie-Holmes N, Mubareka S, Gray-Owen SD, Rubinstein JL, Treanor B, Julien JP (2021) Multivalency transforms SARS-CoV-2 antibodies into ultrapotent neutralizers. Nat Commun 12(1):3661. https://doi.org/10.1038/s41467-021-23825-2
Oostindie SC, Lazar GA, Schuurman J, Parren P (2022) Avidity in antibody effector functions and biotherapeutic drug design. Nat Rev Drug Discov 21(10):715–735
Overdijk MB et al (2020) Dual Epitope Targeting and Enhanced Hexamerization by DR5 Antibodies as a Novel Approach to Induce Potent Antitumor Activity Through DR5 Agonism. Mol Cancer Ther 19(10):2126–2138
Mager DE, Jusko WJ (2001) General pharmacokinetic model for drugs exhibiting target-mediated drug disposition. J Pharmacokinetic and Pharmacodynamic 28:507–532
Mager DE, Krzyzanski W (2005) Quasi-equilibrium pharmacokinetic model for drugs exhibiting target-mediated drug disposition. Pharm Res 22(10):1589–1596
Gibiansky L, Gibiansky E, Kakkar T, Ma P (2008) Approximations of the target-mediated drug disposition model and identifiability of model parameters. J Pharmacokinet Pharmacodyn 35(5):573–591
Gibiansky L, Gibiansky E (2017) Target-mediated drug disposition model for drugs with two binding sites that bind to a target with one binding site. J Pharmacokinet Pharmacodyn. https://doi.org/10.1007/s10928-017-9533-1
Kuester K, Kloft C: Pharmacokinetics of Monoclonal Antibodies, (2006) pp. In: MEIBOHM B, (ed) Pharmacokinetics and Pharmacodynamics of Biotech Drugs: Principles and Case Studies in Drug Development. WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim, pp 45–91
Wand W, Wang EQ, Balthasar JP (2008) Monoclonal antibody pharmacokinetics and pharmacodynamics. Clin Pharmacol Ther 84(5):548–558
Leusen JH (2015) IgA as therapeutic antibody. Mol Immunol 68(1):35–39. https://doi.org/10.1016/j.molimm.2015.09.005
Keyt BA, Baliga R, Sinclair AM, Carroll SF, Peterson MS (2020) Structure, Function, and Therapeutic Use of IgM Antibodies. Antibodies (Basel) 9(4):53. https://doi.org/10.3390/antib9040053.PMID:33066119;PMCID:PMC7709107
Ljungars A, Schiött T, Mattson U et al (2020) A bispecific IgG format containing four independent antigen binding sites. Sci Rep 10:1546. https://doi.org/10.1038/s41598-020-58150-z
Ng CM, Bauer RJ (2023) General quasi-equilibrium multivalent binding model to study diverse and complex drug-receptor interactions of biologics. submitted to J Pharmacokinet Pharmacodyn. Preprint. https://doi.org/10.21203/rs.3.rs-3877678/v1
Beal SL, Sheiner LB, Boeckmann AJ, Bauer R (eds) (1989–2022) NONMEM Users Guides. Icon Development Solutions, Ellicott City, MD, USA
Strang G (2023) Introduction to Linear Algebra, Sixth Edition. ISBN : 978–17331466–7–8
Author information
Authors and Affiliations
Contributions
L.G., C.N., and E.G. contributed to the design and implementation of the research, to the analysis of the results and to the writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix 1: Derivation of Eq. (10)
From (8) we have:
Introducing the dissociation rate constant KD = koff/kon and the irreversible binding rate constant KIB = kint/kon one can arrive at
For any function F(k), where we use the notation
we can derive:
and
Therefore,
and
Using Eqs. (36) and (37) in Eq. (33) we get:
Collecting the terms for \({R}_{k}C\) and \({R}_{k}C\cdot R\) we have
Using the definitions of \({C}_{tot}\) and \({R}_{tot}\) from Eqs. (1) and (2), we then have
Equation (10) immediately follows.
Appendix 2: Derivation of Eq. (12)
Derivation of this section is based on the matrix linear algebra that can be found in many textbooks. We can recommend [17].
The system of Eqs. (8) for k = 2, …, N together with Eq. (2) can be presented in the matrix form \(A*x=b\).
We will present the case of N = 4, but it will be clear from the derivation that the same procedure is valid for any N.
For N = 4, A is the (N + 1)-by-(N + 1) matrix of the form:
and x and b are the vectors:
This equation has a solution \(x={A}^{-1}*b\), where \({A}^{-1}\) is the inverse matrix of \(A\).
We are interested in the expression for \({R}_{4}C\), the last element of x. Since all elements of vector b except for the first are zero,\({R}_{4}C={({A}^{-1})}_{N+\mathrm{1,1}}\cdot {C}_{tot}\), where \({({A}^{-1})}_{N+\mathrm{1,1}}\) is the (N + 1,1) element of the matrix \({A}^{-1}.\)
This element of the inverse matrix is equal to.
\({\left({A}^{-1}\right)}_{N+\mathrm{1,1}}={C}_{1,N+1}\)/det(A), where \({C}_{1,N+1}\) is the co-factor of the (1,N + 1) element of the matrix A.
\({C}_{1,N+1}\) is equal to \({(-1)}^{N+2}{\text{det}}(X)\), where \(X\) is the N-by-N matrix of the form:
The determinant of a triangular matrix is equal to the product of its diagonal elements. Therefore, \({\text{det}}\left(X\right)=N!\cdot {R}^{N}\). Obviously, \({(-1)}^{N+2}={(-1)}^{N}.\)
Thus,
\({R}_{4}C\) =\({(-1)}^{N}\bullet\) \(N!\cdot {R}^{N}\cdot {C}_{tot}\) /det(A).
To complete the derivation we need to show that
To compute \(det\left(A\right)\) we will reduce the matrix A to the triangular form using a sequence of matrix transformations that do not change the determinant, i.e., adding one column (or row) of the matrix with some coefficients to the other column (row) of the matrix. The sequence of these transformations and all intermediate matrices are presented below.
We start with (N + 1)-by-(N + 1) matrix of the form:
Subtract column (4) from column (5):
Subtract column (3) from column (4):
Subtract column (2) from column (3):
Subtract column (1) from column (2):
Now we can remove the first row and the first column of the matrix without changing the determinant:
Add row (4) to row (3):
Add row (3) to row (2):
Add row (2) to row (1):
The resulting matrix has a very simple form that can be easily generalized to any N. We then continue as follows:
Add rows (1), (2), (3) to row (4); add rows (1), (2) to row (3); add row (1) to row (2):
Subtract column 4 from columns (1), (2), (3):
Add rows (1) and (2) to row (3):
Subtract column (3) multiplied by 3 from column (1) and then subtract column (3) multiplied by 2 from column (2):
Add row (1) multiplied by 2 to row (2):
Subtract column (2) multiplied by 3 from column (1):
The determinant of the triangular matrix is equal to the product of its diagonal elements. Thus, the proof for N = 4 is complete. The derivation is general and can be repeated for any N.
Appendix 3: Specific examples
The expressions for 4-to-one binding:
Appendix 4: Derivation of the QE equations
We will use recursion to prove Eqs. (26). First, the base of the recursion for k = N and k = N-1 can be computed as:
Let us now assume that we proved Eq. (26) for all values of k greater than K. Then
Thus, we proved it for k = K and, by recursion, for any k ≥ 0.
Appendix 5: Derivation of the IB equations
We will use recursion to prove Eq. (31). First, the base of the recursion for k = N and k = N-1 can be computed as:
Let us now assume that we proved (31) for all values of k > K. Then
Thus, we proved it for k = K and, by recursion, for any k ≥ 0.
Appendix 6: Derivation of the Michaelis–Menten equations
We will use recursion to prove Eq. (26). First, the base of the recursion for k = N and k = N-1 can be computed starting from Eqs. (12) and (13) and assuming that R is small relative to \({({K}_{IB}+K}_{D})\) as:
Assuming that Eq. (26) is valid for all k > K, from Eq. (14) we have:
Removing the terms of the order of \({R}^{K+1}\), we have
Thus, we proved it for k = K and, by recursion, for any k ≥ 0.
Appendix 7: Nonmem code of the full TMDD model with 4-to-1 binding
$PROBLEM 101est, full TMDD model with 4 binding sites.
$INPUT C = DROP,ID,TIME,AMT,DV,LDV = DROP,EVID,MDV,CMT,DOSE,TYPE.
$DATA../../Data/DerivedData/SimulatedNonmemData101.csv IGNORE = C.
$ABBREV DERIV2 = NO.
$SUBROUTINES ADVAN13 TOL = 9.
$MODEL.
NCOMP = 8.
$PK.
N = 4.
CL = THETA(1)*EXP(ETA(1)).
V1= THETA(2)*EXP(ETA(2))
Q = THETA(3)*EXP(ETA(3)).
V2 = THETA(4)*EXP(ETA(4))
K10= CL/V1
K12= Q/V1
K21= Q/V2
F1= THETA(5)
KA = THETA(6)*EXP(ETA(5)).
KON = THETA(7).
KOFF = THETA(8).
KINT = THETA(9)*EXP(ETA(6)).
KSYN = THETA(10)*EXP(ETA(7)).
KDEG = THETA(11)*EXP(ETA(8)).
BASE = KSYN/KDEG.
KSS = (KOFF + KINT)/KON.
KD = KOFF/KON.
A_0(4) = BASE.
$DES.
RC = A(5).
R2C = A(6)
R3C = A(7)
R4C = A(8)
CTOT = A(2)/V1.
RCTOT = R4C + R3C + R2C + RC.
C = CTOT-RCTOT.
RTOT = A(4).
RPTOT = 4*R4C + 3*R3C + 2*R2C + RC.
R = RTOT-RPTOT.
DADT(1) = -KA*A(1); Total Drug depot amount.
; Total Drug central amount.
DADT(2) = KA*A(1)-K10*C*V1-KINT*RCTOT*V1-K12*C*V1 + K21*A(3).
DADT(3) = K12*C*V1-K21*A(3); Free Drug second compartment amount.
DADT(4) = KSYN—KDEG*R—KINT*RPTOT; Total target central concentration.
; Drug-Target Complex RC, R2C, R3C, R4C concentrations:
DADT(5) = (N-0)*KON*C*R—(1*KOFF + KINT + (N-1)*KON*R)*RC + 2*KOFF*R2C.
DADT(6) = (N-1)*KON*RC*R- (2*KOFF + KINT + (N-2)*KON*R)*R2C + 3*KOFF*R3C.
DADT(7) = (N-2)*KON*R2C*R—(3*KOFF + KINT + (N-3)*KON*R)*R3C + 4*KOFF*R4C.
DADT(8) = (N-3)*KON*R3C*R—(4*KOFF + KINT + (N-4)*KON*R)*R4C.
$ERROR.
RRC = A(5).
RR2C = A(6).
RR3C = A(7).
RR4C = A(8).
CCTOT = A(2)/V1.
RRCTOT = RR4C + RR3C + RR2C + RRC.
CC = CCTOT-RRCTOT.
RRTOT = A(4).
RRPTOT = 4*RR4C + 3*RR3C + 2*RR2C + RRC.
RR = RRTOT-RRPTOT.
Y = CCTOT*EXP(EPS(1)).
IF(TYPE.EQ.2) Y = RRTOT*EXP(EPS(2)).
IPRED = Y.
$THETA.
(0,0.3); 1 CL
(0,3.00); 2 V1
(0,0.2); 3 Q
(0,3.0); 4 V2
(0,0.7); 5 F1
(0,0.5); 6 KA
(0,20,30); 7 KON
(0,2); 8 KOFF
(0,0.2); 9 KINT
(0,1);10 KSYN
(0,10);11 KDEG
$OMEGA.
0.04 ;1 CL
0.04 ;2 V1
0.04 ;3 Q
0.04 ;4 V2
0.04 ;5 KA
0.04 ;6 KINT
0.04 ;7 KSYN
0.04 ;8 KDEG
$SIGMA.
0.0225
0.04
$EST MAXEVAL = 99999 METHOD = 1 INTER PRINT = 10 NOABORT.
NOTHETABOUNDTEST NOOMEGABOUNDTEST NOSIGMABOUNDTEST
$COV PRINT = E UNCONDITIONAL MATRIX = S.
$TABLE ID TIME IPRED DOSE CCTOT RRCTOT RR4C RR3C RR2C RRC.
CC RRTOT RRPTOT RR ONEHEADER NOPRINT FILE = ../101est.tab.
Appendix 8: Nonmem code of the QSS approximation of the TMDD model with 4-to-1 binding.
$PROBLEM 102est, QSS TMDD model with 4 binding sites.
$INPUT C = DROP,ID,TIME,AMT,DV,LDV = DROP,EVID,MDV,CMT,DOSE,TYPE.
$DATA../../Data/DerivedData/SimulatedNonmemData101.csv IGNORE = C.
$ABBREV DERIV2 = NO.
$SUBROUTINES ADVAN13 TOL = 9.
$MODEL.
NCOMP = 4.
$PK.
N = 4.
CL = THETA(1)*EXP(ETA(1)).
V1 = THETA(2)*EXP(ETA(2))
Q = THETA(3)*EXP(ETA(3))
V2 = THETA(4)*EXP(ETA(4))
K10 = CL/V1
K12 = Q/V1
K21 = Q/V2
F1 = THETA(5)
KA = THETA(6)*EXP(ETA(5)).
KON = THETA(7).
KOFF = THETA(8).
KINT = THETA(9)*EXP(ETA(6)).
KSYN = THETA(10)*EXP(ETA(7)).
KDEG = THETA(11)*EXP(ETA(8)).
BASE = KSYN/KDEG.
KSS = (KOFF + KINT)/KON.
KD = KOFF/KON.
KIB = KINT/KON.
A_0(4) = BASE.
$DES.
CTOT = A(2)/V1.
RTOT = A(4).
R = 0.5*(-(N*CTOT + KIB + KD-RTOT) + sqrt((N*CTOT + KIB + KD-RTOT)**2 + 4*(KIB + KD)*RTOT)).
R4C = CTOT*(R/(KIB + KD + R))*(2*R/(KIB + 2*KD + 2*R))*(3*R/(KIB + 3*KD + 3*R))*(4*R/(KIB + 4*KD + 4*R))
R3C = R4C*(KIB+4*KD)/R
R2C = R3C*(KIB+3*KD+1*R)/(2*R)-R4C*4*KD/(2*R)
RC = R2C*(KIB + 2*KD + 2*R)/(3*R)-R3C*3*KD/(3*R).
RCTOT = R4C + R3C + R2C + RC.
C = CTOT-RCTOT.
RPTOT = 4*R4C + 3*R3C + 2*R2C + RC.
DADT(1) = -KA*A(1); Total Drug depot amount.
; Total Drug central amount.
DADT(2) = KA*A(1)-K10*C*V1-KINT*RCTOT*V1-K12*C*V1 + K21*A(3).
DADT(3) = K12*C*V1-K21*A(3); Free Drug second compartment amount.
DADT(4) = KSYN—KDEG*R—KINT*RPTOT; Total target central concentration.
$ERROR.
CCTOT = A(2)/V1.
RRTOT = A(4).
RR = 0.5*(-(N*CCTOT + KIB + KD-RRTOT) +
sqrt((N*CCTOT + KIB + KD-RRTOT)**2 + 4*(KIB + KD)*RRTOT)).
RR4C = CCTOT*(RR/(KIB + KD + RR))*(2*RR/(KIB + 2*KD + 2*RR))*
(3*RR/(KIB + 3*KD + 3*RR))*(4*RR/(KIB + 4*KD + 4*RR)).
RR3C = RR4C*(KIB + 4*KD)/RR.
RR2C = RR3C*(KIB + 3*KD + 1*RR)/(2*RR)-RR4C*4*KD/(2*RR).
RRC = RR2C*(KIB + 2*KD + 2*RR)/(3*RR)-RR3C*3*KD/(3*RR).
RRCTOT = RR4C + RR3C + RR2C + RRC.
CC = CCTOT-RRCTOT.
RRPTOT = 4*RR4C + 3*RR3C + 2*RR2C + RRC.
Y = CCTOT*EXP(EPS(1)).
IF(TYPE.EQ.2) Y = RRTOT*EXP(EPS(2)).
IPRED = Y.
$THETA.
(0,0.3); 1 CL
(0,3.00); 2 V1
(0,0.2); 3 Q
(0,3.0); 4 V2
(0,0.7); 5 F1
(0,0.5); 6 KA
20 FIX ; 7 KON
(0,2); 8 KOFF.
(0,0.2); 9 KINT
(0,1);10 KSYN
(0,10);11 KDEG
$OMEGA.
0.04 ;1 CL
0.04 ;2 V1
0.04 ;3 Q
0.04 ;4 V2
0.04 ;5 KA
0.04 ;6 KINT
0.04 ;7 KSYN
0.04 ;8 KDEG
$SIGMA.
0.0225
0.04
$EST MAXEVAL = 99,999 METHOD = 1 INTER PRINT = 10 NOABORT.
NOTHETABOUNDTEST NOOMEGABOUNDTEST NOSIGMABOUNDTEST
$COV PRINT = E UNCONDITIONAL MATRIX = S.
$TABLE ID TIME IPRED DOSE CCTOT RRCTOT RR4C RR3C RR2C RRC.
CC RRTOT RRPTOT RR ONEHEADER NOPRINT FILE = ../102est.tab.
Appendix 9: Nonmem code of the Michelis-Menten approximation of the TMDD model with 4-to-1 binding.
$PROBLEM 103est, MM TMDD model with 4 binding sites.
$INPUT C = DROP,ID,TIME,AMT,DV,LDV = DROP,EVID,MDV,CMT,DOSE,TYPE.
$DATA../../Data/DerivedData/SimulatedNonmemData101.csv IGNORE = C.
$ABBREV DERIV2 = NO.
$SUBROUTINES ADVAN13 TOL = 9.
$MODEL.
NCOMP = 4.
$PK
N=4
CL=THETA(1)*EXP(ETA(1))
V1=THETA(2)*EXP(ETA(2))
Q=THETA(3)*EXP(ETA(3))
V2=THETA(4)*EXP(ETA(4))
K10=CL/V1
K12=Q/V1
K21=Q/V2
F1=THETA(5)
KA=THETA(6)*EXP(ETA(5))
KON=THETA(7)
KOFF=THETA(8)
KINT=THETA(9)*EXP(ETA(6))
KSYN=THETA(10)*EXP(ETA(7))
KDEG=THETA(11)*EXP(ETA(8))
BASE=KSYN/KDEG
KSS=(KOFF+KINT)/KON
KD=KOFF/KON
KIB=KINT/KON
A_0(4)=BASE
$DES
C = A(2)/V1
RTOT = A(4)
R=RTOT*(KIB+KD)/(KIB+KD+4*C)
DADT(1) =-KA*A(1); Free Drug depot amount
; Free Drug central amount
DADT(2) = KA*A(1)-K10*C*V1-KINT*A(4)*4*C*V1/(KIB+KD+4*C)-K12*C*V1+K21*A(3)
DADT(3) = K12*C*V1-K21*A(3) ; Free Drug second compartment amount
DADT(4) = KSYN-KDEG*R-KINT*A(4)*4*C/(KIB+KD+4*C) ; Total target central concentration
$ERROR
CC = A(2)/V1
RRTOT = A(4)
RRC = RRTOT*4*CC/(KIB+KD+4*CC)
RR = RRTOT*(KIB+KD)/(KIB+KD+4*CC)
Y = CC*EXP(EPS(1))
IF(TYPE.EQ.2) Y = RRTOT*EXP(EPS(2))
IPRED = Y
$THETA
(0,0.3); 1 CL
(0,3.00); 2 V1
(0,0.2) ; 3 Q
(0,3.0) ; 4 V2
(0,0.7) ; 5 F1
(0,0.5) ; 6 KA
20 FIX ; 7 KON
(0,2 ) ; 8 KOFF
(0,0.2) ; 9 KINT
(0,1) ;10 KSYN
(0,10) ;11 KDEG
$OMEGA
0.04 ;1 CL
0.04 ;2 V1
0.04 ;3 Q
0.04 ;4 V2
0.04 ;5 KA
0.04 ;6 KINT
0.04 ;7 KSYN
0.04 ;8 KDEG
$SIGMA
0.0225
0.04
$EST MAXEVAL=99999 METHOD=1 INTER PRINT=10 NOABORT
NOTHETABOUNDTEST NOOMEGABOUNDTEST NOSIGMABOUNDTEST
$COV PRINT=E UNCONDITIONAL MATRIX=S
$TABLE ID TIME IPRED DOSE RRC CC RRTOT RR ONEHEADER NOPRINT FILE=../103est.tab
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gibiansky, L., Ng, C.M. & Gibiansky, E. Target-mediated drug disposition model for drugs with N > 2 binding sites that bind to a target with one binding site. J Pharmacokinet Pharmacodyn (2024). https://doi.org/10.1007/s10928-024-09917-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10928-024-09917-8