Abstract
Genome-wide association studies (GWAS) are useful to facilitate crop improvement via enhanced knowledge of marker-trait associations (MTA). A GWAS for grain yield (GY), yield components, and agronomic traits was conducted using a diverse panel of 239 soft red winter wheat (Triticum aestivum) genotypes evaluated across two growing seasons and eight site-years. Analysis of variance showed significant environment, genotype, and genotype-by-environment effects for GY and yield components. Narrow sense heritability of GY (h 2 = 0.48) was moderate compared to other traits including plant height (h 2 = 0.81) and kernel weight (h 2 = 0.77). There were 112 significant MTA (p < 0.0005) detected for eight measured traits using compressed mixed linear models and 5715 single nucleotide polymorphism markers. MTA for GY and agronomic traits coincided with previously reported QTL for winter and spring wheat. Highly significant MTA for GY showed an overall negative allelic effect for the minor allele, indicating selection against these alleles by breeders. Markers associated with multiple traits observed on chromosomes 1A, 2D, 3B, and 4B with positive minor effects serve as potential targets for marker assisted breeding to select for improvement of GY and related traits. Following marker validation, these multi-trait loci have the potential to be utilized for MAS to improve GY and adaptation of soft red winter wheat.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Identification of marker-trait associations (MTA) is a first step toward marker-assisted selection (MAS), which has become an important tool for accelerating varietal improvement and rate of genetic gain (Moose and Mumm 2008; Wang et al. 2014b). Whole-genome mapping approaches such as genome-wide association studies (GWAS) have recently become a popular alternative to bi-parental quantitative trait loci (QTL) mapping for identifying MTA in plant populations, due in large part to recent advances in high-throughput sequencing and genotyping platforms that have decreased cost and increased discovery of marker polymorphisms (Patel et al. 2015; Ruggieri et al. 2014; Thomson 2014).
GWAS use the concept of linkage disequilibrium (LD), the non-random co-segregation of alleles at multiple loci, to survey genomic regions that render significant variation to phenotypes (Breseghello and Sorrells 2006a; Flint-Garcia et al. 2003). A primary advantage of GWAS is exploitation of recombination events that have occurred over an individual’s evolutionary history using a diverse population (Myles et al. 2009), consequently resulting in a higher mapping resolution compared to a bi-parental approach (Zhu et al. 2008). Additionally, GWAS allows for a much larger gene pool to be surveyed and screened for genetic variation in traits of interest (Neumann et al. 2011; Zhao et al. 2011).
Previous studies have established the usefulness of GWAS in identifying regions affecting variation for GY and adaptation traits in bread wheat (Triticum aestivum). Wang et al. (2014a) reported MTA for kernel hardness, kernel weight, grain protein concentration, grain volume, and plant height in a diverse set of 94 wheat lines. Prior to this, Neumann et al. (2011) conducted GWAS for 20 agronomic traits in a winter wheat core collection using diversity array technology (DArT) markers where significant MTA were detected for plant height, GY, and disease resistance. Sukumaran et al. (2014) and Lopes et al. (2015) identified genomic regions associated with GY and yield-related traits in a wheat association mapping initiative (WAMI) panel consisting of 287 elite lines of spring wheat from CIMMYT, Mexico. Sehgal et al. (2017) recently identified regions affecting GY and yield stability and their epistatic interactions using a large elite panel of CIMMYT spring wheat genotypes under multiple environments.
Hoffstetter et al. (2016) identified important loci governing GY and other economic traits in an elite collection of soft red winter wheat (SRWW) lines adapted to the northeastern US while Addison et al. (2016) determined genomic regions affecting GY potential utilizing a bi-parental approach in a population derived from two elite SRWW cultivars. Except for these studies, reports on MTA for GY and related traits for US soft winter wheat remain limited and hence there is a need to identify yield-related QTL in current soft red winter wheat germplasm. The objectives of this study were to perform GWAS for GY and agronomic traits and to examine population structure and linkage disequilibrium of a diverse panel of SRWW lines adapted to the southern region of the US using genome-wide SNP markers. Information from this research will serve as a valuable resource for genetic improvement of GY and related traits via marker-assisted selection approaches.
Materials and methods
Plant material and experimental design
The association mapping panel (AMP) used for this study consisted of 239 inbred lines of SRWW, including cultivars from the SunGrains® (Southeastern University Grains) small grain breeding and genetics group, publicly and privately developed cultivars, and genotypes adapted to the southeastern region of the US. Trials were drill seeded in seven row plots (1.5 m width × 4.5 m length) at a rate of 118 kg of seed hectare−1. The AMP was evaluated in a total of eight high yield potential site-years that included two environments in the 2013–2014 season and six environments in the 2014–2015 season. Locations included Fayetteville (FAY14, FAY15), Marianna (MAR15), Stuttgart (STU14, STU15), Keiser (KEI15) and Rohwer (ROH15), in the state of Arkansas; and Okmulgee, in the state of Oklahoma (OKL15), US. All locations belong to the west south central US region of SRWW commercial production.
The AMP was sown in an augmented incomplete block design (Federer and Raghavarao 1975; Federer and Crossa 2012), with two repeated check lines (Jamestown and Pioneer Brand 26R20) with unreplicated lines on each location. The random nature of the new treatments and blocking variables are considered in augmented designs resulting in a more efficient analysis (Federer et al. 2001). In all locations except for OKL15, the experimental field was divided into 24 incomplete blocks, each containing 10 different AMP genotypes and both checks. For OKL15, unequal incomplete block sizes, k, were used, where k = 10 for IB 1–19; k = 20 for IB 20–23 and k = 18 for IB 24. Planting and harvest dates and trial management varied based on recommendations at each location for maximizing yield potential, but included routine fungicide applications to control foliar diseases.
Trait measurements
Grain yield (GY) in kg ha−1 was recorded by harvesting whole plots, weighing the grain, and adjusting values to 13% moisture content. Heading date (HD) was recorded as the date when 50% of plants from the whole plot had fully visible spikes and reported in Julian Days. Plant height (PH) was recorded from the soil surface to tip of the spike, excluding awns when present. Kernel weight (KW) was determined by counting 1000 seeds using a Seedburo® 801 seed counter (Chicago, IL, USA). Peduncle length (PL) was measured as the length of the uppermost internode, in cm, averaged across ten culms plot−1. Spike length (SL) was taken as the measurement from the base to tip of the spike (excluding awns), in cm, averaged across ten spikes plot−1. Kernel number spike−1 (KNS) and kernel weight spike−1 (KWS) were estimated by hand-harvesting 50 spike-bearing culms from each plot at maturity prior to harvesting of whole plots.
Statistical analysis
Phenotypic data were analyzed following procedures described by Wolfinger et al. (1997) for analysis of augmented designs using PROC MIXED in SAS v.9.4 (SAS Institute 2011). Genotypes, incomplete blocks, environments, incomplete blocks nested within environments and genotype-by-environment interactions were regarded as random effects. Adjusted means represented as least square means (LSM) for each genotype were estimated using a restricted maximum likelihood (REML) approach for each site-year. Narrow sense heritability (h 2) was calculated for each trait using TYPE3 sum of squares from the adjusted means, with the formula:\(h^{2} = \frac{{\sigma_{G}^{2} }}{{\sigma_{G }^{2} + \sigma_{{\frac{GEI}{e}}}^{2} + \sigma_{{\frac{E}{er}}}^{2} }}\),where \(\sigma_{G }^{2}\), \(\sigma_{GEI }^{2}\) and \(\sigma_{E }^{2}\) variances due to genotype, genotype-by-environment, and error, respectively; and e and r are the number of environments and replications. Associations between traits and environments were explored using principal component analysis (PCA) with the contribution of each variable to the first two principal components (PC) illustrated using bi-plots. The PROC CORR procedure in SAS v.9.4 was used to calculate correlation of normalized means of phenotypes across environments.
SNP marker genotyping
DNA was isolated from each sample following a CTAB extraction procedure modified from Pallotta et al. (2003). Samples were genotyped using the Illumina 9K iSelect assays for wheat previously described by Cavanagh et al. (2013) through the USDA-ARS Eastern Regional Small Grain Genotyping Laboratory, Raleigh, NC. Marker data polymorphisms of 8632 SNPs were scored using the GenomeStudio® software (Illumina, San Diego, USA). After filtering, 5715 polymorphic markers with minor allele frequency (MAF) ≥ 0.04% and less than 10% missing data remained and were used to perform GWAS. SNPs with low MAF were included to capture rare allele variants (MAF < 0.01) which could potentially explain additional variability within the measured traits (Lee et al. 2014).
In addition to the 9K iSelect assay, the AMP was genotyped using KASP® allele-specific SNP markers (LGC Genomics, UK) diagnostic for height (Rht-B1, Rht-D1), vernalization (Vrn-A1 and Vrn-B1) and photoperiod (Ppd-B1, Ppd-D1) loci (Guedira et al. 2014, 2016). Reactions were performed in a total volume of 5 µL [2.5 µL KASP® mix and 2.5 µL DNA sample (50 ng)], following manufacturer’s instructions with minor modifications. Conditions for thermal cycling were as follows: 94 °C for 15 min; 94 °C for 20 s and 65–58 °C (decrement of 0.8 °C per cycle) for 9 cycles; 94 °C 20 s and 57 °C for one minute for 25 cycles; 35 °C for 3 min and a plate read step. An additional thermal cycling step (94 °C for 20 s followed by 57.0 °C for one minute for 2 cycles; and 35 °C for one minute and a plate read step) was used as needed to improve accuracy and precision of clustering.
Linkage disequilibrium, population structure, and genetic diversity
Coefficients of linkage disequilibrium (LD), represented by the square of allele frequency correlations, r 2 (Weir and Cockerham 1996), were calculated using the program TASSEL 5.2.33 (Bradbury et al. 2007). Imputation for missing genotype data was done using a numeric, Euclidean-based distance method in TASSEL, with minimum and maximum allele frequencies set to 0.05 and 1.0, respectively. Pairwise r 2 values were plotted against genetic distance (in cM; based on genetic linkage map by Cavanagh et al. (2013)) and a locally weighted polynomial regression (LOESS) curve (Cleveland 1979) was fitted on the LD plot using RStudio® (R Development Core Team 2010) using the ‘loess’ function. Critical values were estimated by performing a square root transformation of corresponding r 2 estimates for unlinked marker pairs (distance > 50 cM) and then taking the 95th percentile of this distribution (Breseghello and Sorrells 2006b). The intersection of LOESS line and r 2 critical value was regarded as the distance where LD starts to decay (Laido et al. 2014; Nielsen et al. 2014). A p < 0.005 was considered the significance threshold for marker pairs to be in LD with each other.
Population stratification was assessed using the program STRUCTURE (Pritchard et al. 2000) applying an admixture model, a burn-in of 10,000 iterations followed by 10,000 Monte Carlo Markov Chain (MCMC) replicates and number of clusters (K) set in the range 2–10, with number of replications per K equal to 10. The true number of clusters which best fit the data was inferred using the Evanno criterion, which uses an ad hoc statistic ∆K based on rate of change in the log probability of data between successive values of K (Evanno et al. 2005). Likelihood scores and results from STRUCTURE were collated and visualized using the program STRUCTURE Harvester (Earl 2012). Bar plots for membership coefficients, Q for the AMP were plotted using the ‘pophelper’ package (Francis 2016) in RStudio®.
Analysis of molecular variance (AMOVA; Excoffier et al. 1992) was conducted using a ploidy independent infinite allele model (ρ) tested under 999 permutations implemented in the software Genodive (Meirmans and Van Tienderen 2004). Rho (ρ) is an analogue of the population differentiation coefficient (Fixation index, Fst) and is independent of the organism’s ploidy level (Meirmans and Van Tienderen 2004). Fixation indices and pairwise Gst values of subpopulations were calculated using STRUCTURE and Genodive programs, respectively. Fst estimates the correlation of alleles within the same subgroup relative to the entire population (Chao et al. 2010) while G st compares heterozygosity within and between populations, considering a correction for a bias resulting from sampling a limited number of populations (Nei 1987).
GWAS for GY and agronomic traits
Association analyses was performed employing several model selections for a compressed mixed linear model (CMLM) implemented in the Genome Association Prediction Integrated Tool (GAPIT) (Lipka et al. 2012) package in RStudio®. Models included: (1) a naïve model, where only the kinship, K information, and no correction for population structure were applied (K only model); (2) a K-PC model (Zhao et al. 2007) where kinship information together with the first three principal components (PC) were included for GWAS; and (3) a K-Q approach, where a centered IBS (Identical by State) kinship method (Endelman and Jannink 2012) in TASSEL 5.2.33 and a population structure matrix derived from STRUCTURE were included in the model as fixed effects to address population structure. In addition to these models, marker scores for Rht and Vrn loci were included under the K and K-PC as covariates to correct their effects in identifying GY related MTA (Lopes et al. 2015).
The mixed model used to account for genetic relatedness in the AMP was as follows:
where y is the vector of observed phenotype; µ is the mean; x is the genotype of the SNP; β is the effect of the SNP; u is the random effects due to genetic relatedness with Var (u) = \(\sigma_{\text{g}}^{2}\) K and Var (e) = \(\sigma_{\text{e}}^{2}\); K is the kinship matrix across all genotypes (Kang et al. 2008; Lopes et al. 2015). CMLM tests one marker at a time and considers the u and K matrices as the mean additive genetic relatedness between individuals to model polygenetic effects (Lipka et al. 2012).
A total of five combined datasets were used for GWAS, namely BLUP trait values calculated from adjusted means across all environments (ABLUP); BLUP values derived from 2014 site-years (BLUP14); BLUP from the 2015 site-years (BLUP15); BLUP from northern environments across two years (Fayetteville, Keiser, AR; Okmulgee, OK; NBLUP), and from southern environments across the two years (Stuttgart, Marianna, Rohwer, AR; SBLUP).
The most reliable model for GWAS was identified by performing a tenfold cross validation (CV) under a ridge regression best linear unbiased prediction (rrBLUP) model (Endelman 2011) for the most heritable trait on an ABLUP dataset, where kinship, K represented as a marker relationship matrix and scores for Q and PC as covariates were fitted on the model. A value of p < 0.0005 was considered the threshold for defining significant SNP due to deviations of observed quantile–quantile (QQ) plots and to further reduce Type I errors (Hoffstetter et al. 2016; Lopes et al. 2015). Manhattan plots were visualized using the ‘qqman’ package (Turner 2014) in RStudio®
Results
Genotype-by-environment interactions and trait heritability
FAY15 had the highest mean GY, followed by ROH15, and OKL15, while STU14, STU15, and FAY14 had the lowest. Significant genotype effects were observed for all traits indicating differential performance (Table 1). Genotype-by-environment interaction was highly significant for all traits. Incomplete block treatments as well as incomplete blocks nested within environments did not show a significant effect for measured phenotypic traits. Narrow sense heritability (h 2) estimates ranged from 0.30 to 0.81, with PH the most heritable (h 2 = 0.81), followed by KW (h 2 = 0.71) and HD (h 2 = 0.63). GY was moderately heritable (h 2 = 0.48) while SL was the least heritable trait (h 2 = 0.30).
Principal components analyses (PCA) and phenotypic correlations
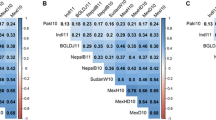
Results from PCA showed PC1 to explain 36.4% of the total variation for phenotypic traits and was positively associated with PL and negatively associated with all other traits (Fig. 1). PC2 contributed 20.1% of the total variation and was in positive correlation with GY and KNS. The PCA biplot was divided into two trait clusters: (1) GY and its components including KNS, KWS, and KW; and (2) HD and agronomic traits including PH, SL, and PL. Pearson correlation coefficients (r) further supported these PCA groupings as GY was strongly correlated with KW (r = 0.48), KNS (r = 0.67) and KWS (r = 0.73) (Table 2). PH was positively correlated with PL (r = 0.49) and HD (r = 0.19). Neither HD nor PH was significantly correlated with GY.
PCA biplots for the a measured traits and b adjusted GY across different site-years for the soft winter wheat AMP. Site-years: FAY14 Fayetteville14; FAY15 Fayetteville15; KEI15 Keiser15; MAR15 Marianna15; OKL15 Oklahoma15; ROH15 Rohwer15; STU14 Stuttgart14; STU15 Stuttgart15. Traits GY grain yield; HD heading date; KNS kernel number spike−1; KW kernel weight; KWS kernel weight spike−1; PH plant height; PL peduncle length; SL spike length
PCA biplot analyses for GY across site-years revealed separation based on year, with the 2014 (FAY14 and STU14) and 2015 (excluding MAR15) clustering separately (Fig. 1). PC1 explained 21.9% of the variation for GY and was positively correlated with MAR15. PC2 contributed 15.2% of variation for GY across environments, was positively correlated with OKL15, STU14, FAY14, and MAR15 and was negatively correlated with STU15, FAY15, ROH15, and KEI15.
Analysis of LD
A total of 74,822 intrachromosomal pairs were in significant LD (p < 0.005) at the whole genome level (Online Resource 1). Average distance of markers in significant LD was ~ 14.40 cM, while markers in complete LD (r 2 = 1.0) had an average distance of 1.71 cM for the whole genome. Genome D had the highest average distance for pairs in complete LD (3.14 cM), followed by Genomes B (1.90 cM) and A (1.34 cM). Average r 2 value for significant pairs across the whole genome was 0.32. Among the subgenomes, genome D also had the highest mean r 2 for all significant pairs (0.37), followed by genomes A (0.32) and B (0.31). LD was estimated to decay at ~7 cM for the whole genome, while genome D had the highest extent of LD among the subgenomes, estimated at ~10 cM, compared to genomes A and B (both estimated at ~7 cM) (Online Resource 2).
Population structure
Genetic structure was evaluated using 5661 genome-wide SNP markers where markers linked to major genes were designated as fixed effects. Inference for the true number of clusters (K) using the Evanno criterion (Evanno et al. 2005) revealed the optimum number of subpopulations for this panel at K = 3 (Online Resource 3). Each entry was assigned to one of three subpopulations based on its largest value for coefficient of membership (Q). Fifty-nine lines were assigned to the first subgroup (Q1), 54 lines were assigned to the second subgroup, Q2, and 126 lines to the third subgroup, Q3 (Online Resource 4). There was no observable clustering based on geographic origin for the lines across the different subgroups. Analysis of molecular variance (AMOVA) further revealed the presence of within population variation, which accounted for 89.1% of the total variance (Online Resource 5). Mean value for Fst was highest for Q1 (0.69), followed by the Q2 (0.43) and Q3 (0.23) subpopulations (Online Resource 6).
Genetic diversity for developmental genes
A total of 207 (87%) lines were semi-dwarfs, having a dwarfing allele in combination with a tall allele for either Rht-B1 or Rht-D1 (Online Resource 7). Two of the lines were double dwarfs, while 26 lines possessed wild-type tall alleles for both loci. Subgroup Q3 had the highest number of semi-dwarf entries for both the Rht-B1a/Rht-D1b and Rht-B1b/Rht-D1a (semi dwarf) allelic combinations (106; 51.2%), in addition to 17 wild-type lines. Majority of lines possessing the photoperiod insensitive Ppd-D1a allele also belonged to the Q3 subpopulation (56; 57.7%). Forty-seven of the entries (19.7%) had a short vernalization allele at the winter vrn-1A locus (vrn-A1b, M_vrn_A1_ex4 locus) with 23 of these lines belonging to subgroup Q3, while 40 of the lines (16.7%) had short vernalization at vrn-B1 (Vrn-B1a, Vrn-B1_AGS2000 locus) (Guedira et al. 2014).
Summary for marker-trait associations (MTA) identified
Predictability for PH (i.e. the most heritable trait) for the ABLUP dataset was highest for K-PC (0.25) under an rrBLUP model; hence this was regarded as the most reliable in identifying significant MTA. K-Q and K only models, performed similarly with prediction values equal to 0.18 and 0.16 (data not shown). GWAS identified 112 loci significantly associated with the eight measured traits at a threshold of p < 0.0005 (Online Resource 8; Online Resource 9).
MTA were detected in all chromosomes except 1D, 3D, 5D, and 6D based on a significance threshold of p < 0.0005. SNPs associated with multiple traits included: SNP wsnp_Ex_c12254_19574891 (1A) associated with HD and KNS; Ppd-D1- ‘Norstar’ allele (2D) associated with both PH and HD (Table 3; Fig. 2). SNP wsnp_Ex_c2500_4671165 (3B) associated with PH and KNS; wsnp_Ex_c13849_21698240 (4B) with GY and KNS, and wsnp_Ex_c48922_53681502 (4B), associated with GY and KWS.
Manhattan plot showing genome-wide SNP loci associated with GY, HD, and PH. Horizontal line represents the significance threshold by which markers were considered associated with a trait (p < 0.0005; ~3.30). a Plot of genome-wide markers associated with GY under a K-PC model, BLUP15; b Plot of genome-wide markers associated with HD under a K–Q model, ABLUP c Plot of genome-wide markers associated with PH under a K–Q model, NBLUP
MTA for GY and yield components
Fifteen markers significant for GY were distributed across eight chromosomes and responsible for 8–28% of the phenotypic variation. Highly significant GY MTA (wsnp_Ex_c259_497455; p = 8.56E−05) in chromosome 2B showed an overall negative allelic effect (−49.35) under a K-Q model. Using Rht-B1 and Vrn-A1 as covariates in a K and K-PC model identified nine SNPs associated with GY in four different datasets. There were 19 markers in 11chromosomes associated with KNS, explaining 6–16% of the phenotypic variation. MTA for KWS (19) were distributed across seven chromosomes and responsible for 8–26% of the phenotypic variance. Markers associated with KW (9) which accounted for 10–29% of the variation were located in four chromosomes (1A, 2B, 3A, 6A).
MTA for agronomic traits
Fourteen trait-specific MTA for HD were detected in four chromosomes with KASP® markers for the alleles of Ppd-D1 ‘Ciano 67’ and Ppd-D1 ‘Norstar’ being highly significant across four datasets. PH had the largest number of detected MTA (24) which included Rht-D1 (4D) detected across all BLUP datasets and responsible for 17–34% of variation. Rht-D1 was highly significant for PH, with p values ranging from 1.90E−08 to 1.80E−05. Spike length had the least number of detected MTA (8), which mapped to chromosomes 1A, 1B, 7B, and 7D. Significant markers for PL (10) were identified in four chromosomes and were responsible for 6–13% of trait variation.
Discussion
Rapid LD decay
Analysis of LD is a prerequisite for evaluating a collection of genotypes and determining adequate marker density for GWAS (Bellucci et al. 2015; Chen et al. 2012; Lopes et al. 2015). LD in the AMP was estimated at ~7 cM across the whole genome, with the low proportion of observed marker pairs in complete LD (3.96%) and significant LD (48.71%) leading to this rapid decay. The mean r 2 value for significant marker pairs was 0.32, comparable to a previous study on eastern US soft winter wheat (Cabrera et al. 2014). Other studies have shown LD in winter wheat to decay at distances from 2 to 5 cM (Chen et al. 2012; Hoffstetter et al. 2016; Tadesse et al. 2015) and up to >10 cM distances (Benson et al. 2012; Zhang et al. 2010). Higher LD in the D compared to the A and B genomes was consistent with previous reports (Chao et al. 2010; Sukumaran et al. 2014) and is a possible consequence of recent introgression and bottleneck accompanying the origin of hexaploid wheat (Chao et al. 2010).
The relatively rapid LD decay implies a higher number of markers required for GWAS, which can result in higher mapping resolution (Abdurakhmonov and Abdukarimov 2008). Next-generation sequencing (NGS) platforms such as genotyping by sequencing (GBS) (Elshire et al. 2011; Poland and Rife 2012) could help in generating a larger number of markers amenable to GWAS, particularly for the D genome where marker coverage was low. This low marker coverage in the genome D could also have led to higher mean r 2 values, average distance of pairs in significant LD, and markers in complete LD. Using a two-tailed t test to compare the average r 2 values and cM distance revealed significant differences between values for genome D and genomes A and B (p < 0.05). Higher average r 2 value for the D genome, nevertheless, indicates that fewer markers are needed for association mapping (Sukumaran et al. 2014).
Moderate genetic stratification
The presence of population structure (PS) can lead to false positive discoveries in GWAS and thus relationships must be accounted for (Sorrells and Yu 2009; Sukumaran and Yu 2014). Moderate genetic stratification for the AMP was supported by a high within group genetic variance (89.1%). This observation was similar with previous results in spring wheat (Edae et al. 2014) and wheat lines from US and Mexico (Chao et al. 2010) and reflects the impact of selection in maintaining allelic diversity in wheat breeding populations (Edae et al. 2014). The lack of clustering of entries from the same geographic origin within a subpopulation in this study further supported this large within group variation. Subgroup Q1 was more genetically similar with Q3, reflected by a lower Gst value between these subgroups (0.13), compared to Q1 and Q2 (0.17). One possible explanation for this is the presence of more entries possessing the Rht-B1b/Rht-D1b allele combinations in the Q1 (7) and the Q3 (17) subgroups, compared to the Q2 (2) subgroup. Q3 was the least differentiated among the subgroups, as reflected by having the lowest value for Fst. In contrast with the current observation, higher levels of population structure had been detected in Chinese wheat cultivars (Zhang et al. 2011), US elite winter wheat (Zhang et al. 2010), and CIMMYT elite spring wheat yield trial lines (Dreisigacker et al. 2012).
Genome location of identified MTA compared to previous studies
GY is a complex trait and its improvement is a primary objective for wheat breeding programs (Ain et al. 2015; Green et al. 2012). The distribution of MTA in multiple chromosomes confirms a complex genetic architecture for yield (Quarrie et al. 2005; Shi et al. 2009). In the present study, significant associations identified for GY and yield component MTA in chromosomes 1A, 2A, 2B, 3B, and 5A agreed with previous reports (Addison et al. 2016; Bennett et al. 2012; Bordes et al. 2014; Lopes et al. 2015). Markers in LD in chromosome 4B associated with GY (wsnp_Ex_c13849_21698240, wsnp_Ex_c48922_53681502, and wsnp_CAP11_c84_120095) were mapped in a region flanking the Rht-B1 locus, which was previously associated with variation for GY in a CIMMYT spring wheat GWAS (Lopes et al. 2015). SNP wsnp_Ex_c259_497455, identified in the SBLUP dataset, coincided with a GY QTL mapped between 9 and 12.5 cM in chromosome 2B by Bordes et al. (2014). Additionally, GY-associated markers wsnp_Ex_c2723_5047696, mapped in ABLUP, BLUP15, and SBLUP datasets under a K-Q model, together with wsnp_Ex_rep_c66331_64502363 and wsnp_Ex_rep_c66331_64502558 co-localized with a QTL previously mapped in chromosome 3BS for yield under irrigated conditions (Bennett et al. 2012). The use of BLUP trait values from combined analyses increased the power in finding significant QTL as BLUPs are robust in identifying significant associations (Mason et al. 2013). Majority of the GY MTA observed in this study showed negative allelic effects with respect to the minor allele, indicating that breeders have been successful in selecting alleles that improve yield and productivity in modern winter wheat cultivars. Validation of yield QTL in CIMMYT’s WAMI panel (Lopes et al. 2015; Sukumaran et al. 2014) also showed that selections were made for the yield “enhancing” major allele (DN Lozada, unpublished data), suggesting that both winter and spring classes have undergone similar selection pressures to achieve optimum yield. Simultaneously capturing these favorable alleles into new germplasm would be beneficial for breeding higher yielding varieties of wheat.
Yield component traits are generally more heritable than GY itself and therefore have potential for genetic improvement. A SNP associated with KNS, wsnp_Ex_c12254_19574891 (1A), was mapped within a 6 cM distance from marker wPt6122, previously associated with grain number and spike number m−2 in a winter wheat core collection (Neumann et al. 2011). The same marker was also located proximal to a KNS QTL (within 1 cM) region previously detected by Edae et al. (2014). SNP wsnp_Ex_c1276_2445537 mapped at 172.32 cM in chromosome 6B coincided with a KWS-associated region reported by Neumann et al. (2011) at 175.9 cM. For KW, wsnp_JD_c5699_6859527 (3A) co-located with a thousand grain weight “enhancing” locus BARC0197_174 in a panel of European winter and spring wheat varieties (Zanke et al. 2015). The positive minor allele effect of this marker and its detection in three BLUP datasets (ABLUP, BLUP15, NBLUP) under a K-Q model indicate that it could be a potential target for improving KW in existing germplasm.
Twenty-four markers distributed across 10 chromosomes were associated with variation in PH. Although influenced by many genes, PH is highly heritable and controlled in large part by Rht-B1 and Rht-D1 (Snape et al. 1977; Würschum et al. 2015; Zanke et al. 2014b). Rht-D1 was highly significant for PH across all BLUP datasets and models used with the dwarfing allele present in 64% of the lines (Online Resource 7). The positive allelic effect for this locus indicates that selection by breeders has favored the “height reducing” major allele, as shorter stature has been shown to reduce lodging and increase harvest index (Rebetzke et al. 2011). Despite this, PH was not correlated with GY, in agreement with a previous study by Sukumaran et al. (2014) and in contrast with Bellucci et al. (2015) where negative correlation between these traits was observed. No PH MTA were detected in chromosome 4B harboring the Rht-B1 gene, consistent with other studies that have shown Rht-D1 to have a larger genetic effect (Bellucci et al. 2015; Neumann et al. 2011; Würschum et al. 2015; Zanke et al. 2014b). It is also worth noting that PH did not share common significant loci with PL and SL, an unexpected result considering a high correlation observed between these traits and in contrast with previous studies (Heidari et al. 2012; Sukumaran et al. 2014).
The timing of anthesis is a critical trait for adaptation of wheat to diverse environments and is primarily affected by genes for vernalization and photoperiod response (Zanke et al. 2014a). In the present study, MTA for HD were identified in four chromosomes and did not include the Ppd-B1 region on 2B. This result is likely due to both the stronger effect of the Ppd-D1a allele for conferring photoperiod insensitivity (Guedira et al. 2016; Kamran et al. 2014) and its higher frequency within the population (54.8%) compared to Ppd-B1a (14.6%) (Online Resource 7). Ppd-D1 markers for ‘Ciano 67’ and ‘Norstar’ alleles were significantly associated with HD across four BLUP datasets and all GWAS models used, similar to previous observations (Zanke et al. 2014a). Major alleles for these loci had negative allelic effects for HD, indicating that insensitivity to photoperiod decreased days to HD, which plays a large role in the adaptation of wheat to the southern US growing areas.
Current and future genetic improvement of southern US winter wheat
The pleiotropic effect of photoperiod insensitivity conferred by Ppd-D1a on plant development has previously been shown (Snape et al. 2001; Zanke et al. 2014b) and has its importance for adaptation of southern US winter wheat (Addison et al. 2016; Guedira et al. 2016). In addition to HD, Ppd-D1 ‘Norstar’ allele was associated with PH, with a positive minor allele effect indicating selection for reduced PH to improve grain yield. Bentley et al. (2014) and Wilhelm et al. (2013) noted a reduction in PH caused by Ppd-D1a among elite European lines and in a worldwide wheat germplasm panel. In this study, 66 of the 100 highest yielding lines possessed the Ppd-D1b allele for the Ppd-D1 ‘Norstar’ allele, which was higher than expected based on allele frequency (Online Resource 11), indicating its importance for yield and adaptation in the current germplasm. The Rht-D1b dwarfing allele was also present in 60 of the 100 highest yielding entries. Taken together, our results showed the interplay of reduced PH and photoperiod to produce higher yielding cultivars of soft winter wheat adapted to the southern US.
Several studies have previously reported multi-trait MTA associated with GY, yield components and agronomic traits using a GWAS approach in spring wheat (Edae et al. 2014; Sukumaran et al. 2014). GY shared common MTA (wsnp_Ex_c13849_21698240 and wsnp_Ex_c48922_53681502 (4B)) with KNS and KWS (Table 3), which explained 10–26% of trait variation (Table 2). To our knowledge, there has not been a report on multi-trait loci related with controlling variation for GY and yield components mapped in chromosome 4B. Edae et al. (2014) previously identified multi-trait markers associated with GY, spikes m−2, KW, and TW in chromosome 5B while Wang et al. (2009) mapped loci in 1B, 2A, and 3B associated with grain filling rate, KWS, and KW. Our results here thus provide additional multi-trait loci associated with yield and yield components which can be targeted for future MAS to improve GY and adaptation in soft winter wheat. The multi-trait markers identified in this study could ultimately be used to accelerate pyramiding of yield and adaptation-related QTL to develop southern US winter wheat varieties with increased GY potential and broader adaptations.
Conclusions
A GWAS for GY, yield components, and agronomic traits in soft winter wheat was conducted using genome-wide SNP markers. Multi-trait MTA in chromosomes 1A, 2D, 3B, and 4B were identified and could be potential targets of selection for marker-assisted breeding to capitalize on variation for GY, yield components, and adaptation traits in winter wheat. QTL validation and development of breeder-friendly assays for these multi-trait loci and their deployment to existing breeding programs could ultimately help accelerate MAS to improve GY and adaptation in soft winter wheat. Results from this study serve as valuable resources for molecular breeding towards varietal improvement of wheat. The utility of association mapping approach for determining genomic regions affecting variation for traits of agricultural and economic importance was demonstrated.
References
Abdurakhmonov IY, Abdukarimov A (2008) Application of association mapping to understanding the genetic diversity of plant germplasm resources. Int J Plant Genom 2008:574927. doi:10.1155/2008/574927
Addison CK, Mason RE, Brown-Guedira G, Guedira M, Hao Y, Miller RG, Subramanian N, Lozada DN, Acuna A, Arguello MN, Johnson JW, Ibrahim AMH, Sutton R, Harrison SA (2016) QTL and major genes influencing grain yield potential in soft red winter wheat adapted to the southern United States. Euphytica:1-13. doi:10.1007/s10681-016-1650-1
Bellucci A, Torp AM, Bruun S, Magid J, Andersen SB, Rasmussen SK (2015) Association mapping in Scandinavian Winter wheat for yield, plant height, and traits important for second-generation bioethanol production. Front Plant Sci 6:1046. doi:10.3389/fpls.2015.01046
Bennett D, Reynolds M, Mullan D, Izanloo A, Kuchel H, Langridge P, Schnurbusch T (2012) Detection of two major grain yield QTL in bread wheat (Triticum aestivum L.) under heat, drought and high yield potential environments. Theor Appl Genet 125(7):1473–1485
Benson J, Brown-Guedira G, Paul Murphy J, Sneller C (2012) Population structure, linkage disequilibrium, and genetic diversity in soft winter wheat enriched for fusarium head blight resistance. Plant Genome 5(2):71–80
Bentley AR, Scutari M, Gosman N, Faure S, Bedford F, Howell P, Cockram J, Rose GA, Barber T, Irigoyen J, Horsnell R, Pumfrey C, Winnie E, Schacht J, Beauchêne K, Praud S, Greenland A, Balding D, Mackay IJ (2014) Applying association mapping and genomic selection to the dissection of key traits in elite European wheat. Theor Appl Genet 127(12):2619–2633. doi:10.1007/s00122-014-2403-y
Bordes J, Goudemand E, Duchalais L, Chevarin L, Oury FX, Heumez E, Lapierre A, Perretant MR, Rolland B, Beghin D (2014) Genome-wide association mapping of three important traits using bread wheat elite breeding populations. Mol Breed 33(4):755–768
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23(19):2633–2635. doi:10.1093/bioinformatics/btm308
Breseghello F, Sorrells ME (2006a) Association analysis as a strategy for improvement of quantitative traits in plants. Crop Sci 46(3):1323–1330. doi:10.2135/cropsci2005.09-0305
Breseghello F, Sorrells ME (2006b) Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172(2):1165–1177
Cabrera A, Souza E, Guttieri M, Sturbaum A, Hoffstetter A, Sneller C (2014) Genetic diversity, linkage disequilibrium, and genome evolution in soft winter wheat. Crop Sci 54(6):2433–2448
Cavanagh C, Chao S, Wang S, Huang B, Stephen S, Kiani S, Forrest K, Saintenacc C, Brown-Guedira G, Akhunova A, See D, Bai G, Pumphrey M, LTomar, Wong D, Kong S, Reynolds M, Silva MLd, Bockelman H, LTalbert, Anderson J, Dreisigacker S, Baenziger S, Carter A, Korzun V, Morrell P, Dubcovsky J, MMorell, Sorrells M, Hayden M, Akhunov E (2013) Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. PNAS 110 (20):8057–8062. doi:www.pnas.org/cgi/doi/10.1073/pnas.1217133110
Chao S, Dubcovsky J, Dvorak J, Luo M-C, Baenziger SP, Matnyazov R, Clark DR, Talbert LE, Anderson JA, Dreisigacker S, Glover K, Chen J, Campbell K, Bruckner PL, Rudd JC, Haley S, Carver BF, Perry S, Sorrells ME, Akhunov ED (2010) Population- and genome-specific patterns of linkage disequilibrium and SNP variation in spring and winter wheat (Triticum aestivum L.). BMC Genomics 11(1):1–17. doi:10.1186/1471-2164-11-727
Chen X, Min D, Yasir TA, Hu Y-G (2012) Genetic diversity, population structure and linkage disequilibrium in elite Chinese winter wheat investigated with SSR markers. PLoS ONE 7(9):e44510
Cleveland WS (1979) Robust locally weighted regression and smoothing scatterplots. J Am Stat Assoc 74(368):829–836
Dreisigacker S, Shewayrga H, Crossa J, Arief VN, DeLacy IH, Singh RP, Dieters MJ, Braun H-J (2012) Genetic structures of the CIMMYT international yield trial targeted to irrigated environments. Mol Breed 29(2):529–541
Earl DA (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conser Genet Resour 4(2):359–361
Edae EA, Byrne PF, Haley SD, Lopes MS, Reynolds MP (2014) Genome-wide association mapping of yield and yield components of spring wheat under contrasting moisture regimes. Theor Appl Genet 127(4):791–807
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6(5):e19379
Endelman JB (2011) Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 4(3):250–255
Endelman JB, Jannink J-L (2012) Shrinkage estimation of the realized relationship matrix. G3 (Bethesda) 2(11):1405–1413
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620. doi:10.1111/j.1365-294X.2005.02553.x
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131(2):479–491
Federer WT, Crossa J (2012) I.4 screening experimental designs for quantitative trait loci, association mapping, genotype-by environment interaction, and other investigations. Front Physiol 3:156. doi:10.3389/fphys.2012.00156
Federer WT, Raghavarao D (1975) Augmented designs. Biometrics 31(1):29–35. doi:10.2307/2529707
Federer WT, Reynolds M, Crossa J (2001) Combining results from augmented designs over sites. Agron J 93(2):389–395
Flint-Garcia SA, Thornsberry JM, Buckler ES (2003) Structure of linkage disequilibrium in plants. Annu Rev Plant Biol 54:357–374. doi:10.1146/annurev.arplant.54.031902.134907
Francis R (2016) Pophelper: an R package and web app to analyse and visualize population structure. Mol Ecol Resour 17(1):27–32. doi:10.1111/1755-0998.12509
Green AJ, Berger G, Griffey CA, Pitman R, Thomason W, Balota M, Ahmed A (2012) Genetic yield improvement in soft red winter wheat in the Eastern United States from 1919 to 2009. Crop Sci 52(5):2097–2108. doi:10.2135/cropsci2012.01.0026
Guedira M, Maloney P, Xiong M, Petersen S, Murphy JP, Marshall D, Johnson J, Harrison S, Brown-Guedira G (2014) Vernalization duration requirement in soft winter wheat is associated with variation at the locus. Crop Sci 54(5):1960–1971
Guedira M, Xiong M, Hao YF, Johnson J, Harrison S, Marshall D, Brown-Guedira G (2016) Heading date QTL in winter wheat (Triticum aestivum) coincide with major developmental genes VERNALIZATION1 and PHOTOPERIOD1. PLoS ONE 11(5):e0154242. doi:10.1371/journal.pone.0154242
Heidari B, Saeidi G, Sayed Tabatabaei BE, Suenaga K (2012) QTLs involved in plant height, peduncle length and heading date of wheat (Triticum aestivum L.). J Agr Sci Technol 14(5):1090–1104
Hoffstetter A, Cabrera A, Sneller C (2016) Identifying quantitative trait loci for economic traits in an elite soft red winter wheat population. Crop Sci 56:1–12
Institute SAS (2011) SAS system options: reference, 2nd edn. SAS Institute, Cary
Kamran A, Iqbal M, Spaner D (2014) Flowering time in wheat (Triticum aestivum L.): a key factor for global adaptability. Euphytica 197(1):1–26. doi:10.1007/s10681-014-1075-7
Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E (2008) Efficient control of population structure in model organism association mapping. Genetics 178(3):1709–1723. doi:10.1534/genetics.107.080101
Laido G, Marone D, Russo MA, Colecchia SA, Mastrangelo AM, De Vita P, Papa R (2014) Linkage disequilibrium and genome-wide association mapping in tetraploid wheat (Triticum turgidum L.). PLoS ONE 9(4):e95211
Lee S, Abecasis GR, Boehnke M, Lin X (2014) Rare-variant association analysis: study designs and statistical tests. Am J of Hum Genet 95(1):5–23. doi:10.1016/j.ajhg.2014.06.009
Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28(18):2397–2399. doi:10.1093/bioinformatics/bts444
Lopes MS, Dreisigacker S, Pena RJ, Sukumaran S, Reynolds MP (2015) Genetic characterization of the wheat association mapping initiative (WAMI) panel for dissection of complex traits in spring wheat. Theor Appl Genet 128(3):453–464. doi:10.1007/s00122-014-2444-2
Mason RE, Hays DB, Mondal S, Ibrahim AMH, Basnet BR (2013) QTL for yield, yield components and canopy temperature depression in wheat under late sown field conditions. Euphytica 194(2):243–259. doi:10.1007/s10681-013-0951-x
Meirmans PG, Van Tienderen PH (2004) GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes 4(4):792–794
Moose SP, Mumm RH (2008) Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiol 147(3):969–977. doi:10.1104/pp.108.118232
Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang Z, Costich DE, Buckler ES (2009) Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell 21(8):2194–2202. doi:10.1105/tpc.109.068437
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Neumann K, Kobiljski B, Denčić S, Varshney RK, Varshney RK, Börner A (2011) Genome-wide association mapping: a case study in bread wheat (Triticum aestivum L.). Mol Breeding 27(1):37–58. doi:10.1007/s11032-010-9411-7
Nielsen NH, Backes G, Stougaard J, Andersen SU, Jahoor A (2014) Genetic diversity and population structure analysis of European Hexaploid bread wheat (Triticum aestivum L.) varieties. PLoS ONE 9(4):e94000. doi:10.1371/journal.pone.0094000
Pallotta M, Warner P, Fox R, Kuchel H, Jefferies S, Langridge P (2003) Marker assisted wheat breeding in the southern region of Australia. In: Proceedings of 10th International Wheat Genetics Symposium, Paestum, Italy, pp 1–6
Patel D, Zander M, Dalton-Morgan J, Batley J (2015) Advances in plant genotyping: Where the future will take us. In: Batley J (ed) Plant genotyping, vol 1245. Methods Mol Biol. Springer, New York, pp 1–11. doi:10.1007/978-1-4939-1966-6_1
Poland JA, Rife TW (2012) Genotyping-by-sequencing for plant breeding and genetics. Plant Genome 5(3):92–102
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Q-u Ain, Rasheed A, Anwar A, Mahmood T, Imtiaz M, Mahmood T, Xia X, He Z, Quraishi UM (2015) Genome-wide association for grain yield under rainfed conditions in historical wheat cultivars from Pakistan. Front Plant Sci 6:743. doi:10.3389/fpls.2015.00743
Quarrie SA, Steed A, Calestani C, Semikhodskii A, Lebreton C, Chinoy C, Steele N, Pljevljakusić D, Waterman E, Weyen J, Schondelmaier J, Habash DZ, Farmer P, Saker L, Clarkson DT, Abugalieva A, Yessimbekova M, Turuspekov Y, Abugalieva S, Tuberosa R, Sanguineti M-C, Hollington PA, Aragués R, Royo A, Dodig D (2005) A high-density genetic map of hexaploid wheat (Triticum aestivum L.) from the cross Chinese Spring × SQ1 and its use to compare QTLs for grain yield across a range of environments. Theor Appl Genet 110(5):865–880. doi:10.1007/s00122-004-1902-7
Rebetzke GJ, Ellis MH, Bonnett DG, Condon AG, Falk D, Richards RA (2011) The Rht13 dwarfing gene reduces peduncle length and plant height to increase grain number and yield of wheat. Field Crops Res 124(3):323–331. doi:10.1016/j.fcr.2011.06.022
Ruggieri V, Francese G, Sacco A, D’Alessandro A, Rigano MM, Parisi M, Milone M, Cardi T, Mennella G, Barone A (2014) An association mapping approach to identify favourable alleles for tomato fruit quality breeding. BMC Plant Biol. doi:10.1186/s12870-014-0337-9
Sehgal D, Autrique E, Singh R, Ellis M, Singh S, Dreisigacker S (2017) Identification of genomic regions for grain yield and yield stability and their epistatic interactions. Sci Rep. 7: 41578. doi:10.1038/srep41578. https://www.nature.com/articles/srep41578#supplementary-information
Shi J, Li R, Qiu D, Jiang C, Long Y, Morgan C, Bancroft I, Zhao J, Meng J (2009) Unraveling the complex trait of crop yield With quantitative trait loci mapping in Brassica napus. Genetics 182(3):851–861. doi:10.1534/genetics.109.101642
Snape JW, Law CN, Worland AJ (1977) Whole chromosome analysis of height in wheat. Heredity 38(1):25–36
Snape JW, Butterworth K, Whitechurch E, Worland AJ (2001) Waiting for fine times: genetics of flowering time in wheat. Euphytica 119(1–2):185–190. doi:10.1023/a:1017594422176
Sorrells ME, Yu J (2009) Linkage disequilibrium and association mapping in the Triticeae. In: Genetics and genomics of the Triticeae. Springer: New York, pp 655–683
Sukumaran S, Yu J (2014) Association Mapping of genetic resources: Achievements and future perspectives. In: Tuberosa R, Graner A, Frison E (eds) Genomics of plant genetic resources: Volume 1. Managing, sequencing and mining genetic resources. Springer Netherlands, Dordrecht, pp 207–235. doi:10.1007/978-94-007-7572-5_9
Sukumaran S, Dreisigacker S, Lopes M, Chavez P, Reynolds M (2014) Genome-wide association study for grain yield and related traits in an elite spring wheat population grown in temperate irrigated environments. Theor Appl Genet:1–11. doi:10.1007/s00122-014-2435-3
Tadesse W, Ogbonnaya FC, Jighly A, Sanchez-Garcia M, Sohail Q, Rajaram S, Baum M (2015) Genome-wide association mapping of yield and grain quality traits in winter wheat genotypes. PLOS ONE. doi:10.1371/journal.pone.0141339
R Development Core Team (2010) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/
Thomson M (2014) High-throughput SNP genotyping to accelerate crop improvement. Plant Breed Biotech 2(3):195–212
Turner SD (2014) qqman: an R package for visualizing GWAS results using QQ and manhattan plots. bioRxiv:005165
Wang RX, Hai L, Zhang XY, You GX, Yan CS, Xiao SH (2009) QTL mapping for grain filling rate and yield-related traits in RILs of the Chinese winter wheat population Heshangmai × Yu8679. Theor Appl Genet 118(2):313–325. doi:10.1007/s00122-008-0901-5
Wang G, Leonard JM, von Zitzewitz J, Peterson CJ, Ross AS, Riera-Lizarazu O (2014a) Marker-trait association analysis of kernel hardness and related agronomic traits in a core collection of wheat lines. Mol Breeding 34(1):177–184. doi:10.1007/s11032-014-0028-0
Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L, Mastrangelo AM, Whan A, Stephen S, Barker G, Wieseke R, Plieske J, Lillemo M, Mather D, Appels R, Dolferus R, Brown-Guedira G, Korol A, Akhunova AR, Feuillet C, Salse J, Morgante M, Pozniak C, Luo M-C, Dvorak J, Morell M, Dubcovsky J, Ganal M, Tuberosa R, Lawley C, Mikoulitch I, Cavanagh C, Edwards KJ, Hayden M, Akhunov E, Int Wheat Genome S (2014b) Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol J 12(6):787–796. doi:10.1111/pbi.12183
Weir BS, Cockerham C (1996) Genetic data analysis II: Methods for discrete population genetic data. Sinauer Assoc, Inc, Sunderland, MA, USA
Wilhelm EP, Boulton MI, Al-Kaff N, Balfourier F, Bordes J, Greenland AJ, Powell W, Mackay IJ (2013) Rht-1 and Ppd-D1 associations with height, GA sensitivity, and days to heading in a worldwide bread wheat collection. Theor Appl Genet 126(9):2233–2243. doi:10.1007/s00122-013-2130-9
Wolfinger R, Federer WT, Cordero-Brana O (1997) Recovering information in augmented designs, using SAS PROC GLM and PROC MIXED. Agron J 89(6):856–859
Würschum T, Langer SM, Longin CFH (2015) Genetic control of plant height in European winter wheat cultivars. Theor Appl Genet 128(5):865–874. doi:10.1007/s00122-015-2476-2
Zanke C, Ling J, Plieske J, Kollers S, Ebmeyer E, Korzun V, Argillier O, Stiewe G, Hinze M, Beier S, Ganal MW, Röder MS (2014a) Genetic architecture of main effect QTL for heading date in European winter wheat. Front Plant Sci 5:217. doi:10.3389/fpls.2014.00217
Zanke CD, Ling J, Plieske J, Kollers S, Ebmeyer E, Korzun V, Argillier O, Stiewe G, Hinze M, Neumann K (2014b) Whole genome association mapping of plant height in winter wheat (Triticum aestivum L.). PLoS ONE 9(11):e113287
Zanke CD, Ling J, Plieske J, Kollers S, Ebmeyer E, Korzun V, Argillier O, Stiewe G, Hinze M, Neumann F, Eichhorn A, Polley A, Jaenecke C, Ganal MW, Röder MS (2015) Analysis of main effect QTL for thousand grain weight in European winter wheat (Triticum aestivum L.) by genome-wide association mapping. Front. Plant Sci 6:644. doi:10.3389/fpls.2015.00644
Zhang D, Bai G, Zhu C, Yu J, Carver BF (2010) Genetic diversity, population structure, and linkage disequilibrium in US elite winter wheat. Plant Genome 3(2):117–127
Zhang L, Liu D, Guo X, Yang W, Sun J, Wang D, Sourdille P, Zhang A (2011) Investigation of genetic diversity and population structure of common wheat cultivars in northern China using DArT markers. BMC Genet 12(1):42
Zhao K, Aranzana MJ, Kim S, Lister C, Shindo C, Tang C, Toomajian C, Zheng H, Dean C, Marjoram P, Nordborg M (2007) An Arabidopsis example of association mapping in structured samples. PLOS Genet. doi:10.1371/journal.pgen.0030004
Zhao K, Tung C-W, Eizenga GC, Wright MH, Ali ML, Price AH, Norton GJ, Islam MR, Reynolds A, Mezey J, McClung AM, Bustamante CD, McCouch SR (2011) Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun 2:467. http://www.nature.com/ncomms/journal/v2/n9/suppinfo/ncomms1467_S1.html
Zhu C, Gore M, Buckler ES, Yu J (2008) Status and prospects of association mapping in plants. Plant Genome 1(1):5–20. doi:10.3835/plantgenome2008.02.0089
Acknowledgments
This research was supported by the Agriculture and Food Research Initiative (AFRI) of the US Department of Agriculture’s National Institute of Food and Agriculture (USDA-NIFA) Grant 2012-67013-19436, the Monsanto Beachell-Borlaug International Scholars Program (MBBISP), and the National Research Initiative Competitive Grants 2011-68002-30029 and 2017-67007-25939 from USDA-NIFA.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lozada, D.N., Mason, R.E., Babar, M.A. et al. Association mapping reveals loci associated with multiple traits that affect grain yield and adaptation in soft winter wheat. Euphytica 213, 222 (2017). https://doi.org/10.1007/s10681-017-2005-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-017-2005-2