Abstract
A survey of bacterial and archaeal community structure was carried out in 10 shallow tube wells in a high arsenic groundwater system located in Hetao Basin, Inner Mongolia by 16S rRNA gene based two-step nested PCR-DGGE, clone libraries and 454 pyrosequencing. 12 bacterial and 18 archaeal DGGE bands and 26–136 species-level OTUs were detected for all the samples. 299 bacterial and 283 archaeal 16S rRNA gene clones for two typical samples were identified by phylogenetic analysis. Most of the results from these different methods were consistent with the dominant bacterial populations. But the proportions of the microbial populations were mostly different and the bacterial communities in most of these samples from pyrosequencing were both more abundant and more diverse than those from the traditional methods. Even after quality filtering, pyrosequencing revealed some populations including Alishewanella, Sulfuricurvum, Arthrobacter, Sporosarcina and Algoriphagus which were not detected with traditional techniques. The most dominant bacterial populations in these samples identified as some arsenic, iron, nitrogen and sulfur reducing and oxidizing related populations including Acinetobacter, Pseudomonas, Flavobacterium, Brevundimonas, Massilia, Planococcus, and Aquabacterium and archaeal communities Nitrosophaera and Methanosaeta. Acinetobacter and Pseudomonas were distinctly abundant in most of these samples. Methanogens were found as the dominant archeal population with three methods. From the results of traditional methods, the dominant archaeal populations apparently changed from phylum Thaumarchaeota to Euryarchaeota with the arsenic concentrations increasing. But this structure dynamic change was not revealed with pyrosequencing. Our results imply that an integrated approach combining the traditional methods and next generation sequencing approaches to characterize the microbial communities in high arsenic groundwater is recommended.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Arsenic (As) in groundwater is a serious environmental issue that affects the health of millions of people in parts of Bangladesh, West Bengal, Cambodia, China, Japan, Argentina, Chile and USA (Islam et al. 2004; Sutton et al. 2009; Polya and Charlet 2009; Liao et al. 2011; Farooq et al. 2012; Jiang et al. 2014; Rodríguez-Lado et al. 2013). Common chronic problems include skin diseases (pigmentation, dermal hyperkeratosis, skin cancer) and many cardiovascular, neurological, hematological, renal and respiratory diseases, as well as lung, bladder, liver, kidney and prostate cancers (Smith et al. 2002; Herbert and Snow 2012). Hetao Basin (the Great Bend of Yellow River) of Inner Mongolia located in the arid-semiarid region in northwestern China is typical of an area affected by As poisoning, with >300,000 victims of arseniasis (He et al. 2009). Most groundwater from this region contains high As with concentrations varying from 0.35 to 1.74 mg/L, which exceeds the maximum contaminant level (10 μg/L) by more than 100 times (Deng 2008).
In the last decade, there have been many studies about As release and transformation mechanism, with a focus on geochemical controls and microbial activity (Nordstrom 2002; Smedley and Kinniburgh 2002; Berg et al. 2007; Guo et al. 2008; Deng et al. 2009; Sutton et al. 2009; Mukherjee et al. 2009; Kocar et al. 2010; Neidhardt et al. 2012; Rodríguez-Lado et al. 2013). These previous studies showed that As mobilization is a result of complex interplay of microbial mediated reactions and geochemical processes that are sensitive to site-specific hydrology and sediment composition. To date, many microorganisms, such as Shewanella sp. strain ANA-3, Desulfosporosinus sp. strain Y5, Bacillus sp. UWC have been linked with As mobilization at different aquifers and sediment depths (Malasarn et al. 2004; Pe′rez-Jime′nez et al. 2005; Liao et al. 2011). However, previous research supporting the biogeochemical basis for As mobilization at Hetao Basin has so far failed to identify the native microbial populations responsible for high arsenic concentration in these aquifers. In recently studies, we have successfully characterized the in situ microbial communities in the high arsenic aquifers with traditional Sanger sequencing methods (Li et al. 2013; Jiang et al. 2014). Although some useful information have been found, the disadvantages of the methods are prominent due to its the limit amounts of sequence data and the relatively high sequencing cost. With recent advancement in sequencing technologies, the next generation sequencing, such as Tag-encoded FLX amplicon pyrosequencing (Roche Titanium 454 FLX pyrosequencing) is increasingly being used to characterize microbial community composition especially for the study of uncultured microbial populations in environmental samples (Hou et al. 2013; Bouétard et al. 2012). But more recently studies showed this sequencing technology still its defects such as underestimate on the abundant and diverse of archaeal communities (Sundberg et al. 2013).
In this study, we employed an integrated approach, including 16S rRNA gene based denaturing gradient gel electrophoresis (DGGE), clone libraries and 454 pyrosequencing to investigate the structure and diversity of the in situ microbial communities in the representative arsenic-rich shallow aquifers by comparing the results of the traditional and new generation sequencing methods.
Materials and methods
Sample collection and laboratory analytical methods
Groundwater samples were collected in May 2010 from tube wells from ten villages in Hangjihouqi, Hetao Basin, Inner Mongolia (Fig. 1): three located in Shahai district (samples IMCUGW8, 9, 10) and three in Taiyangmiao (samples IMCUGW4, 5, 7), two in Manhui Town (samples IMCUGW1, 3), one in Sandaoqiao (sample IMCUGW6) and one in Erdaoqiao (sample IMCUGW2). Microbial samples were collected by on-line filtering of 5–10 L water through 0.2-μm filters (Millipore), and the filters were immediately frozen in dry ice. All samples were stored with dry ice in the field and during transportation, and then at −80 °C in laboratory until further analysis. Methods for anion and As analysis were from Deng et al. (2009). The geochemical parameters of water samples were measured in situ using a multiple parameter water quality meter (Horiba, Japan). Methane in water samples was analyzed at Lanzhou Institute of Geology, Chinese Academy of Sciences analyzed using a MAT-271 mass spectrometry. Water samples used As concentration and elemental species analysis were filtered and acidified with HCl to pH 1.0 and then analyzed with IC-HG-AFS (Deng et al. 2009). Anions including NO3 −, SO4 2− were determined by ion chromatography (DX-120, Dionex, USA). Fe(II, III) concentration was determined by the Ferrozine-based assay (Riemer et al. 2004).
DNA extraction, PCR-DGGE analysis and clone library construction
DNA of the collected microbial samples was extracted using FastDNA SPIN Kit for soil (Qbiogene, Inc. CA) according to the manufacturer’s protocols. DGGE was performed using a D-Code Universal Mutation Detection System (Bio-Rad) as previously described (Webster et al. 2006; O’Sullivan et al. 2008). A nested PCR approach for amplification of 16S rRNA gene fragments suitable for DGGE analysis was used with the standard bacterial DGGE primers (F27 5′-AGA GTT TGA TCM TGG CTC AG and R907 5′-CCG TCA ATT CCT TTG AGT TT) (Marchesi et al. 1998), or archaeal DGGE primers (F21 5′-TTC CGG TTG ATC CYG CCG GA and R958 5′-YCC GGC GTT GAM TCC AAT T) for the first PCR (DeLong 1992) for the first PCR. The amplicons obtained from the first PCR were then diluted 1:20 and were used as templates for the second PCR with bacterial DGGE primers F357-GC (5′ GC-Clamp (CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG G)-CCT ACG GGA GGC AGC AG-) and R518 (5′-ATT ACC GCG GCT GCT GG). For the second PCR of archaeal DGGE, primers were SAf (5′ GC-Clamp-CCT AYG GGG CGC AGM AGG) and PARCH519R (5′-GTA AAA CGA CGG CCA G) (Nicol et al. 2003). Bands showing intensity under a certain value (15 % of the highest peak within a lane) were omitted from further analyses. DGGE band densities were analyzed by Quantity One 4.6.2 (Bio-rad, USA). At least one band per banding position was excised and sequenced. At least 12 bacterial and 18 archaeal major DGGE bands designated as IMCUGWBD-Band1–12 and IMCUGWAD-Band1–18 respectively were visible and excised from the gel, re-amplified, purified and sequenced. The affiliations of the 16S rRNA gene sequence were determined by comparing with the GenBank database. Fragments of the 16S rRNA gene for clone libraries were amplified using the same sets of primers with the first run of the nested PCR for DGGE, and ligated into pGEM-T Easy vector using the manufacturer’s protocol (Promega). The recombinant plasmids were used to transform competent Escherichia coli JM109 cells. Clone library construction was referenced from previously study (Li et al. 2013). Randomly selected white clones were screened by colony PCR amplification of the 16S rRNA gene inserts using M13 primers. Pyrosequencing analyses were performed by detecting bacterial and archaeal V4–V8 variable regions of the 16S rRNA genes with the primers (515F 5′ GTG CCA GCM GCC GCG GTA A and 1391R 5′ GAC GGG CGG TGT GTR CA). PCRs were performed with the following thermocycler program: denaturation at 95 °C for 5 min, 30 cycles of denaturation at 94 °C for 1 min, annealing at 56 and 52 °C (for the first and second PCRs, respectively) for 1 min, and extension at 72 °C for 1 min, and final extension at 72 °C for 10 min.
Sequencing and data analysis
Pyrosequencing of the V4 region of the 16S rRNA gene was carried out from the 515F-end of the amplicons with Roche (454) genome sequencer FLX+system (454 Life Sciences, USA) at SeqWright Inc (Houston, USA). Low quality reads were removed in Mothur for quality screening (Schloss et al. 2009). The following analysis was accomplished with QIIME software package. All the remaining reads were de-noised and trimmed to a uniform length of 238 bp, clustering analysis at the similarity levels of 80, 90, 95 and 97 % using UCLUST (Edgar 2010). These OTU (operational taxonomic unit) levels approximately correspond to the phylum, order, genus, and species level, respectively (Schloss and Handelsman 2004). The most abundant sequences from each cluster were chosen as representatives and these representative sequences were aligned with the PyNAST method (Knight et al. 2010). The aligned sequences were sent for chimera-checking and taxonomic assignment through ChimeraSlayer (Costa et al. 2009) and BLAST (Vick et al. 2010) online using 16S.gold.NAST_ALIGNED.fasta and gg_97_otus_4feb2011.fasta (http://greengenes.lbl.gov) respectively. All chimeric sequences were discarded.
For clone library construction, positive clones were sequenced using primers M13 on an ABI 3100 automated sequencer. The raw sequences were trimmed by using DNAman 6.0. and classified by RDP online (http://rdp.cme.msu.edu/). Potential chimeric sequences were identified with Bellerophon (http://foo.maths.uq.edu.au/~huber/bellerophon.pl). Phylogenetic and statistical analyses were performed with Bioedit 7.0.9, MEGA 4.0 and DOTUR 1.53 (Song et al. 2010). Sequences with 97 % identity were defined as one OTU. The most similar 16S rRNA sequences in each OTU from the GenBank databases were chosen to construct phylogenetic trees. Rarefaction analysis was used to evaluate the saturation of the sampled clones, and was performed with Shannon diversity index and clone numbers. Coverage (C) value was calculated to evaluate the representativeness of the analyzed clones for species diversity in samples according to the formula: \( {\text{C}} = \left( { 1- {\text{n}}/{\text{N}}} \right) \times 100 \% \), where n is the number of 16S rDNA types appearing only once in the library and N is the total number of positive clones detected (Li et al. 2010). Shannon indices were calculated by using the DOTUR program.
Nucleotide sequence accession numbers
The sequences obtained in this study have been deposited in the GenBank database. The 16S rRNA genes derived by DGGE named IMCUGWBD-Band1–12 and IMCUGWAD-Band1–18 were given the accession numbers AB622794–AB622805 and AB622753–AB622770 respectively and the 16S rRNA gene clones were given the accession numbers as AB622771–AB622793, AB622731–AB622752, AB623001–AB623006, JX196251–JX196266 and JX196317–JX196333.
Results
Sample characteristics and chemical composition
Groundwater samples were analyzed for chemical constituents to assess the geochemical conditions at each site. Arsenic concentrations were between 23 and 1,001 μg/L. The concentrations of NO3 −, SO4 2−, irons and methane indicate a difference in geochemical conditions between these tube wells. Four samples with As concentration more than 917 μg/L were distinctly characterized with high concentrations of methane. Most of the high As groundwater samples contained low concentrations of SO4 2− and NO3. Sample IMCUGW1 is characterized with low concentration of As, low ratios of As(III)/As(V) and Fe(II)/Fe(III), and high concentration of NO3 −, and SO4 2− (Table 1).
Microbial communities by DGGE profiling
At least 12 bacterial and 18 archaeal major DGGE bands designated as IMCUGWBD-Band1–12 and IMCUGWAD-Band1–18 respectively. Of the 12 bacterial sequences, five sequences belonged to the Betaproteobacteria, four to Gammaproteobacteria, one to Alphaproteobacteria, and the remaining two were Bacteroidetes and Firmicutes (Table 2; Fig. 2). Of the 18 archeaal sequences, twelve belonged to the Euryarchaeota and six to Thaumarchaeota (Table 3; Fig. 3).
a PCR-based DGGE fingerprints of bacterial 16S rRNA gene fragments. Lanes 1–10 corresponded to samples IMCUGW1–10 respectively. Sequence from band 1 belonged to Bacteroidetes; Sequences from bands 2, 5, 9, 10, 11 belonged to Betaproteobacteria; Sequences from bands 3, 4, 6, 8 belonged to Gammaproteobacteria; Sequence from band 7 belonged to Firmicutes; Sequence from band 12 belonged to Alphaproteobacteria. b Bacterial community composition on genus level based on relative DGGE band intensities. Bands showing intensity under 15 % of the highest peak within a lane were omitted. Samples umbers S1–S10 refer to samples IMCUGW1–10
a PCR-based DGGE fingerprints of archaeal 16S rRNA gene fragments. Lanes 1–10 corresponded to samples IMCUGW1–10 respectively. Sequences from bands 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18 belonged to Euryarchaeote; Sequences from bands 1, 2, 3, 4, 5 belonged to Thaumarchaeota. b Archaeal community composition on family or genus level based on relative DGGE band intensities. Bands showing intensity under 15 % of the highest peak within a lane were omitted. Samples umbers S1–S10 refer to samples IMCUGW1–10
DGGE band patterns of bacteria and archaea showed great bacterial complexity. Several major bacterial bands 3, 4, 6, 8 and 12 which showed a high similarity with genus Acinetobacter, Psuedomonas, Brevundimonas and Massilia respectively, were dominated in most of the samples (Table 2; Fig. 2). The dominant archaeal populations changed from phylum Thaumarchaeota to Euryarchaeote as the As concentration became higher than 335 µg/L. Band 1, 2, 3, 4 and 5 were found highly similar with the sequences identified as Candidatus Nitrosopumilus. This population is distinctly dominated in those samples with relatively low arsenic including IMCUGW1-4. Band 7, 8 and 10 was identified as methanogens which were dominant in sample IMCUGW7, 8, 9 and 10. This result was consistent with the increased concentration of methane in water samples (Table 3; Fig. 3). Band 15 was similar to family Halobacteriaceae which was found to be dominant in alkaline-saline soil (Keshri et al. 2013).
Microbial community by cloning
Sample IMCUGW1 and IMCUGW9 were used for clone library construction of 16S rRNA gene sequences. In total 299 bacterial and 283 archaeal randomly selected white clones were sequenced. 28 and 24 bacterial and 30 and 26 achaeal OTUs were defined at >97 % similarity level, respectively. The coverage of the clone libraries were 93.9 and 94.0 % for bacteria and, 93.6 and 92.1 % for archaea, respectively. The shannon index of the clone libraries were 2.29 and 2.56 for bacteria and, 2.70 and 2.78 for archaea, respectively. Phylogenetic analysis showed that the results of bacterial 16S rRNA gene clone libraries were mostly consistent with those of DGGE which were mainly composed of proteobacterial sequences, with the relative abundances being 81.4 and 89.6 % for sample IMCUGW1 and IMCUGW9, respectively. The proteobacterial clone sequences were respectively affiliated with Gammaproteobacteria (62.9 and 68.6 %), Betaproteobacteria (9.5 and 17.0 %), Alphaproteobacteria (7.5 and 4.0 %). The class Gammaproteobacteria in these two samples was mainly related to genus the Pseudomonas (30.4 and 29.4 %) and Acinetobacter (29.7 and 28.8 %), respectively. Aquabacterium dominated in both of these two samples with the proportions of 5.4.0 and 4.6 %. The other major groups in sample IMCUGW1 included Arthrobacter and Rhizobium (3.4 and 2.7 %, respectively). Sample IMCUGW9 consisted of three other major genera including Psychrobacte, Brevundimonas, and Flavobacterium (4.6, 3.2 and 4.6 % respectively) (Fig. 4).
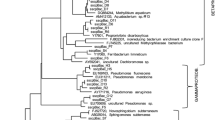
Phylogenetic relationships between bacterial the 16S rRNA (partial sequences, ~900 bp) gene sequences of sample IMCUGW1 and 9 and closely related sequences from the GenBank database. Sequence accession numbers are shown in front of the samples numbers. One representative clone type within each operational taxonomic unit is shown, and the number of clones within each phylotype is shown in the parentheses. Clone sequences from this study are coded as follows with IMCUGW1BD12(5) as an example: bacterial 16S rRNA gene clone number 5 from the IMCUGW1 sample. Scale bars indicate the Jukes-Cantor distances. Bootstrap values of >50 % (for 1000 iterations) are shown. Aquifex pyrophilius is used as outgroup
The obtained archaeal 16S rRNA gene clone sequences fell into Thaumarchaeota, Crenarcheota and Euryarchaeota. The major archaeal groups were significantly different between these two samples. Consistent with the DGGE results, Nitrosophaera was dominant in relative abundance of 78.8 % in sample IMCUGW1. Euryarchaeota in sample IMCUGW9 comprised 21.9 % of the archaeal community, and Methanosaeta belonging to this phylum was the only dominant population in sample IMCUGW9 (Fig. 5).
Phylogenetic relationships between archaeal the 16S rRNA (partial sequences, ~900 bp) gene sequences of sample IMCUGW1 and 9 and closely related sequences from the GenBank database. Sequence accession numbers are shown in front of the samples numbers. One representative clone type within each operational taxonomic unit is shown, and the number of clones within each phylotype is shown in the parentheses. Clone sequences from this study are coded as follows with IMCUGW9AC7(4) as an example: archeal 16S rRNA gene clone number 4 from the IMCUGW9 sample. Scale bars indicate the Jukes-Cantor distances. Bootstrap values of >50 % (for 1,000 iterations) are shown
Microbial community by pyrosequencing
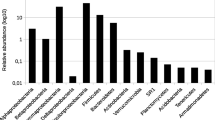
From the result of pyrosequencing, 49, 283 microbial 16S rRNA reads from ten samples were yielded. A variety of taxa were observed at the 97 % OTU level, with 26–136 observed and 43–267 predicted OTUs (based on Chao1) and coverage values ranging from 39.4 to 77.5 %. Bacterial populations are the main groups in all samples. The obtained bacterial 16S rRNA gene clone sequences could be grouped into Alpha-, Beta- Delta-, Epsilon-, and Gammaproteobacteria, Bacteroidetes, Firmicutes, Chloroflexi, Actinobacteria, Firmicutes, and Elusimicrobia (Fig. 6). Consistent with the result of DGGE, the bacterial communities were mainly composed of proteobacterial sequences, with the relative abundances 42.4–98.8 %. The proteobacterial sequences were mainly affiliated with Alpha-, Beta- and Gammaproteobacteria, and the latter class was the distinctly dominant group with the proportions from 42.2 to 98.5 %. The major groups on genus level mainly included Acinetobacter, Pseudomonas, Flavobacterium, Brevundimonas, Aquabacterium, Psychrobacter, Alkanindiges, Thiobacillus, Arthrobacter, Alishewanella, and Sulfuricurvum, Sporosarcina, Planomicrobium, with Pseudomonas and Acinetobacter distinctly being dominant with the percentages of 0–82.7 and 1.0–96.7, respectively (Fig. 6).
Average archaeal abundance was very low with the highest two value 1.0 % (IMCUGW07) and 4.6 % (IMCUGW10). The mainly archaeal groups of these samples are Euryarchaeota, Thaumarchaeota and Crenarchaeota with the former phylum is relatively high (the highest is IMCUGW10, 3.67 %). The main populations in Euryarchaeota are related to methanogens. In the results of pyrosequencing, a few sequences related to Halobacteriaceae or Candidatus Nitrosopumilus were also found, which is accordance with the results of DGGE and clone libraries.
From the results of the three different methods, some arsenic, iron, nitrogen and sulfur reducing and oxidizing related populations have been found. Acinetobacter was documented to distinctly dominate the microbial population as observed in most samples. This bacterial genus is more likely to be more As resistant than function other species reported in previously studies (Sutton et al. 2009), and some of them can even oxidize or reduce arsenic (Fan et al. 2008). Recently a partial sequence of the arsenic reductase (arsC) gene from strain Acinetobacter sp. VKPM14 (accession No. ABQ28709) has been submitted to the Genbank of EMBL. The other dominant population was identified as genus Pseudomonas which also possesses the function of arsenate reduction and tolerance (Freikowski et al. 2010; Srivastava et al. 2010; Sutton et al. 2009). In addition, some possible As-related microorganisms include Acidovorax, Aquabacterium, Hydrogenophaga and Methylotenera, Planococcus, Flavobacterium, Brevundimonas, Thiobacillus and Arthrobacter were also detected to dominate the microbial population in As contaminated environment which have been previously found in association with As-oxidation, As resistance, denitrification, sulfide oxidation and Fe(II)-oxidation (Sutton et al. 2009; Fisher et al. 2008; Hohmann et al. 2011). The genus Psychrobacter was firstly reported as an arsenate-reducing bacterium by Liao et al. (2011), was also predominant in some of our high arsenic samples. The genus Alkaliphilus was reported as the type species from alkaline sediment to reduce metals such as Fe(III), Co(III), and Cr(VI) (Jiang et al. 2006).
Discussion
With the rapid development of high-throughput sequencing technology, the limitation of traditional Sanger sequencing technology have become increasingly prominent. The microbial biosphere is considerably larger and more diverse than previous studies using traditional Sanger sequencing. Reliably assessing microbial abundance and diversity is increasingly required. The next-generation sequencing approaches such as 454 pyrosequencing are facilitating this endeavor. Pyrosequencing showed much greater detection capability than the traditional sequencing methods (Kautz et al. 2013). The deep sequencing to explore the microbial community diversity that has been undetected by the traditional methods due to methodological constraints is the key advantage that makes this new sequencing technology more attractive (Pinto and Raskin 2012). However, there are still some inadequacies for the current technology. For instance, the entire length of the 16S rRNA gene cannot be sequenced; PCR based methods introduce biases such as the amplification efficiency in multi-template PCR reactions and primer mismatches could affect the results of microbial community structure analyses (Pinto and Raskin 2012).
In our study, the microbial communities in ten high arsenic groundwater samples were comparatively detected among the utility of 16S rRNA amplicon 454 pyrosequencing, Sanger-sequenced clone libraries, and DGGE. Most of the results from these three different methods were consistent with the dominant bacterial populations (Figs. 7 and 8). The most dominant bacterial populations in these high arsenic samples detected by these three methods were Acinetobacter, Pseudomonas, Flavobacterium, Brevundimonas, Massilia, Planococcus, and Aquabacterium. Acinetobacter and Pseudomonas were distinctly abundant in most of these samples. However the proportions of these bacterial populations were mostly different (Figs. 7 and 8). The bacterial communities in most of these samples from pyrosequencing were both more abundant and more diverse than those from the traditional methods. Even after quality filtering, pyrosequencing revealed some other populations including Alishewanella, Sulfuricurvum, Arthrobacter, Sporosarcina and Algoriphagus which were not detected with traditional techniques. Moreover, the proportions of most of the predominant populations in these ten samples were distinctly different with those of traditional methods. These differences are to be expected and could be explained by the detection of rare microbes, but they might in part also be due to a higher error rate of 454 sequencing (Kautz et al. 2013).
Comparison of major bacterial phyla (>1 %) from high arsenic groundwater samples analyzed by pyrosequencing (denoted as P), PCR-DGGE (denoted as D) and cloning (denoted as C) methods. Samples numbers 1–10 refer to samples IMCUGW1–10. Samples number from this study is coded as follows with C9 as an example: bacterial 16S rRNA gene sequence frequencies of IMCUGW9 sample analyzed by 16S rRNA gene clone library method. The phylum Proteobacteria is shown at class level resolution
Comparison of known major bacterial genera or family (>1 %) from high arsenic groundwater samples analyzed by pyrosequencing (denoted as P), PCR-DGGE (denoted as D) and cloning (denoted as C) methods. Samples numbers 1–10 refer to samples IMCUGW1–10. Samples number from this study is coded as follows with C9 as an example: bacterial 16S rRNA gene sequence frequencies of IMCUGW9 sample analyzed by 16S rRNA gene clone library method
The bacterial communities of these samples were always distinctly both more abundant and more diverse than those of archaea by 454 pyrosequencing. Similar result has been found in very recent study (Sundberg et al. 2013). From sanger-sequenced clone libraries and DGGE results (Figs. 3 and 5), the dominant archaeal populations apparently changed from pylum Thaumarchaeota (distinctly dominated by Candidatus Nitrosopumilus) to Euryarchaeota (mainly dominant with methanogens) as the arsenic concentrations increasing. Methanogens were found as the dominant population in four samples with extremely high arsenic concentrations. While from the result of pyrosequencing, average archaeal abundance was very low. Only two samples (IMCUGW07 and IMCUGW10) have been detected with the archaeal percentages higher than 1.0. The relatively abundant populations are methanogens, which was accordance with the results of DGGE and clone libraries. However, none of the other dominant populations including Candidatus Nitrosopumilus and Halobacteriaceae which were both detected in DGGE and cloning, were revealed with pyrosequencing. The possible reason of the difference could be the primers used in our 454 pyrosequencing might have low number of mismatches (Hou et al. 2013; Dodsworth et al. 2011; Zhou et al. 2011). Modified sets of primers with high number of mismatches for archaeal populations might improve the results. Therefore, based on the direct comparison of these traditional sequencing techniques, an integrative approach combining the traditional methods and next generation sequencing approaches to characterize the microbial communities in high arsenic groundwater is recommended.
References
Berg M, Stengel C, Trang PTK, Viet PH, Sampson ML, Leng M, Samreth S, Fredericks D (2007) Magnitude of arsenic pollution in the Mekong and Red River Deltas-Combodia and Vietnam. Sci Total Environ 372:413–425
Bouétard A, Noirot C, Besnard A-L, Bouchez O, Choisne D, Robe E, Klopp C, Lagadic C, Coutellec M-A (2012) Pyrosequencing-based transcriptomic resources in the pond snail Lymnaea stagnalis, with a focus on genes involved in molecular response to diquat-induced stress. Ecotoxicology 21:2222–2234
Costa KC, Navarro JB, Shock EL, Zhang CL, Soukup D (2009) Microbiology and geochemistry of great boiling and mud hot springs in the United States Great Basin. Extremophiles 13:447–459
DeLong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci USA 89:5685–5689
Deng YM (2008) Geochemical proeesses of high arsenic groundwate system at Western Hetao Basin. Dissertation, China University of Geoscience
Deng YM, Wang YX, Ma T (2009) Speciation and enrichment of arsenic in strongly reducing shallow aquifers at western Hetao Plain, northern China. Environ Geol 56:1467–1477
Dodsworth JA, Hungate BA, Hedlund BP (2011) Ammonia oxidation, denitrification and dissimilatory nitrate reduction to ammonium in two US Great Basin hot springs with abundant ammonia-oxidizing archaea. Environ Microb 13:2371–2386
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Fan H, Su C, Wang Y, Yao J, Zhao K, Wang G (2008) Sedimentary arsenite-oxidizing and arsenate-reducing bacteria associated with high arsenic groundwater from Shanyin, Northwestern China. J Appl Microb 105:529–539
Farooq SH, Chandrasekharam D, Abbt-Braun G, Berner Z, Norra S, Stueben D (2012) Dissolved organic carbon from the traditional jute processing technique and its potential influence on arsenic enrichment in the Bengal Delta. Appl Geochem 27:292–303
Fisher OE, Dawson AM, Polshyna G, Ann NY (2008) Transformation of inorganic and organic arsenic by Alkaliphilus oremlandii sp. nov. Strain Acad Sci 1125:230–241
Freikowski D, Winter J, Gallert C (2010) Hydrogen formation by an arsenate-reducing Pseudomonas putida, isolated from arsenic-contaminated groundwater in West Bengal, India. Appl Microbiol Biotechnol 88:1363–1371
Guo H, Tang X, Yang S, Shen Z (2008) Effect of indigenous bacteria on geochemical behavior of arsenic in aquifer sediments from the Hetao Basin, Inner Mongolia: evidence from sediment incubations. Appl Geochem 23:3267–3277
He J, Ma T, Deng Y, Yang H, Wang Y (2009) Environmental geochemistry of high arsenic groundwater at western Hetao plain, Inner Mongolia. Front Earth Sci China 3:63–72
Herbert KJ, Snow ET (2012) Modulation of arsenic-induced epidermal growth factor receptor pathway signalling by resveratrol. Chem-Biol Interact 198:38–48
Hohmann C, Morin G, Ona-Nguema G, Guigner J-M, Brown GE Jr, Kappler A (2011) Molecular-level modes of As binding to Fe(III) (oxyhydr)oxides precipitated by the anaerobic nitrate-reducing Fe(II)-oxidizing Acidovorax sp. strain BoFeN1. Geochim Cosmochim Acta 75:4699–4712
Hou W, Wang S, Dong H, Jiang H, Briggs BR, Peacock JP, Huang Q, Huang L, Wu G, Zhi X (2013) A comprehensive census of microbial diversity in hot springs of Tengchong, Yunnan Province China using 16S rRNA gene pyrosequencing. PLoS One 8(1):e53350
Islam FS, Gault AG, Boothman C, Polya DA, Charnock JM, Chatterjee D, Lloyd JR (2004) Role of metal-reducing bacteria in arsenic release from Bengal delta sediments. Nature 430:68–71
Jiang H, Dong H, Zhang G, Yu B, Chapman LR, Fields MW (2006) Microbial diversity in water and sediment of Lake Chaka, an athalassohaline lake in Northwestern China. Appl Environ Microbiol 72:7430–7431
Jiang Z, Li P, Wang Y, Li B, Deng Y, Wang Y (2014) Vertical distribution of bacterial populations associated with arsenic mobilization in aquifer sedimentsfrom the Hetao plain, Inner Mongolia. Environ Earth Sci 71:311–318
Kautz S, Rubin BE, Russell JA, Moreau CS (2013) Surveying the microbiome of ants: comparing 454 pyrosequencing with traditional methods to uncover bacterial diversity. Appl Environ Microbiol 79:525–534
Keshri J, Mishra A, Jha B (2013) Microbial population index and community structure in saline–alkaline soil using gene targeted metagenomics. Microbiol Res 168:165–173
Knight R, Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ (2010) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267
Kocar BD, Borch T, Fendorf S (2010) Arsenic repartitioning during biogenic sulfidization and transformation of ferrihydrite. Geochim Cosmochim Acta 74:980–994
Li P, Wang Y, Wang Y, Liu K, Tong L (2010) Bacterial community structure and diversity during establishment of an anaerobic bioreactor to treat swine wastewater. Water Sci Technol 61:243–252
Li P, Wang Y, Jiang Z, Jiang H, Li B, Dong H, Wang Y (2013) Microbial diversity in high arsenic groundwater in Hetao Basin of Inner Mongolia, China. Geomicrobiol J 30:897–909
Liao VH, Chu YJ, Su YC, Hsiao SY, Wei CC, Liu CW, Liao CM, Shen WC, Chang FJ (2011) Arsenite-oxidizing and arsenate-reducing bacteria associated with arsenic-rich groundwater in Taiwan. J Contam Hydrol 123:20–29
Malasarn D, Saltikov CW, Campbell KM, Santini JM, Hering JG, Newman DK (2004) arrA is a reliable marker for As(V) respiration. Science 306:455
Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Wade WG (1998) Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol 64:795–799
Mukherjee A, Bhattacharya P, Shi F (2009) Chemical evolution in the high arsenic groundwater of the Huhhot basin(Inner Mongolia, PR China) and its difference from the western Bengal basin (India). Appl Geochem 24:1835–1851
Neidhardt H, Norra S, Tang X, Guo H, Stüben D (2012) Impact of irrigation with high arsenic burdened groundwater on the soil–plant system: results from a case study in the Inner Mongolia, China. Environ Pollut 163:8–13
Nicol GW, Glover LA, Prosser JI (2003) The impact of grassland management on archaeal community structure in upland pasture rhizosphere soil. Environ Microbiol 5:152–162
Nordstrom DK (2002) Worldwide occurrences of arsenic in ground water. Science 296:2143–2145
O’Sullivan LA, Webster G, Fry JC, Parkes RJ, Weightman AJ (2008) Modified linker-PCR primers facilitate complete sequencing of DGGE DNA fragments. J Microbiol Methods 75:579–581
Pe′rez-Jime′nez JR, DeFraia C, Young LY (2005) Arsenate respiratory reductase gene (arrA) for Desulfosporosinus sp. strain Y5. Biochem Biophys Res Commun 338:825–829
Pinto AJ, Raskin L (2012) PCR biases distort bacterial and archaeal community structure in pyrosequencing datasets. PLoS One 7:e43093
Polya D, Charlet L (2009) Environmental science: Rising arsenic risk? Nat Geosci 2:383–384
Riemer J, Hoepken HH, Czerwinska H, Robinson SR, Dringen R (2004) Colorimetric ferrozine-based assay for the quantitation of iron in cultured cells. Anal Biochem 331:370–375
Rodríguez-Lado L, Sun G, Berg M, Zhang Q, Xue H, Zheng Q, Johnson CA (2013) Groundwater arsenic contamination throughout China. Science 341:866–868
Schloss PD, Handelsman J (2004) Status of the microbial census. Microbiol Mol Biol Rev 68:686–691
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Micobiol 75:7537
Smedley PL, Kinniburgh DG (2002) A review of the source, behaviour and distribution of arsenic in natural waters. Appl Geochem 17:517–568
Smith AH, Lopipero PA, Bates MN, Steinmaus CM (2002) Arsenic epidemiology and drinking water standards. Science 296:2145–2146
Song ZQ, Chen JQ, Jiang HC, Zhou EM (2010) Diversity of Crenarchaeota in terrestrial hot springs in Tengchong, China. Extremophiles 14:287–296
Srivastava D, Madamwar D, Subramanian RB (2010) Pentavalent arsenate reductase activity in cytosolic fractions of Pseudomonas sp., isolated from arsenic-contaminated sites of Tezpur, Assam. Appl Biochem Biotechnol 162:3766–3779
Sundberg C, Al-Soud WA, Larsson M, Alm E, Yekta SS, Svensson BH, Sørensen SJ, Karlsson A (2013) 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol Ecol 85:612–626
Sutton NB, van der Kraan GM, van Loosdrecht MCM, Muyzer G, Bruining J, Schotting RJ (2009) Characterization of geochemical constituents and bacterial populations associated with As mobilization in deep and shallow tube wells in Bangladesh. Water Res 43:1720–1730
Vick TJ, Dodsworth JA, Costa KC, Shock EL, Hedlund BP (2010) Microbiology and geochemistry of Little Hot Creek, a hot spring environment in the Long Valley Caldera. Geobiology 8:140–154
Webster G, Parkes RJ, Cragg BA, Newberry CJ, Weightman AJ, Fry JC (2006) Prokaryotic community composition and biogeochemical processes in deep subseafloor sediments from the Peru Margin. FEMS Microbiol Ecol 58:65–85
Zhou J, Wu L, Deng Y, Zhi X, Jiang Y-H, Tu Q, Xie J, Van Nostrand JD, He Z, Yang Y (2011) Reproducibility and quantitation of amplicon sequencing-based detection. ISME J 5:1303–1313
Acknowledgements
This research was financially supported by National Natural Science Foundation of China (Grant No. 41372348, 41120124003), National Science Foundation for Post-doctoral Scientists of China (Grant No. 2012M521491, 2013T60757) and the Fundamental Research Funds for the Central Universities, China University of Geosciences (No. CUG140505, GBL11204)
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, P., Jiang, D., Li, B. et al. Comparative survey of bacterial and archaeal communities in high arsenic shallow aquifers using 454 pyrosequencing and traditional methods. Ecotoxicology 23, 1878–1889 (2014). https://doi.org/10.1007/s10646-014-1316-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10646-014-1316-5