Abstract
Pentavalent arsenate reductase activity was localized and characterized in vitro in the cytosolic fraction of a newly isolated bacterial strain from arsenic-contaminated sites. The bacterium was gram negative, rod-shaped, nonmotile, non-spore-forming, and noncapsulated, and the strain was identified as Pseudomonas sp. DRBS1 following biochemical and molecular approaches. The strain Pseudomonas sp. DRBS1 exhibited enzymatic machinery for reduction of arsenate(V) to arsenite(III). The suspended culture of the bacterium reduced more than 97% of As(V) (40–100 mM) to As(III) in 48 h. The growth rate and total cellular yield decreased in the presence of higher concentration of arsenate. The suspended culture repeatedly reduced 10 mM As(V) within 5 h up to five consecutive inputs. The cell-free extracts reduced 86% of 100 µM As(V) in 40 min. The specific activity of arsenate reductase enzyme in the presence of 100 µM arsenate is 6.68 µmol/min per milligram protein. The arsenate reductase activity is maximum at 30 °C and at pH 5.2. The arsenate reductase activity increased in the presence of electron donors like citrate, glucose, and galactose and metal ions like Cd+2, Cu+2, Ca+2, and Fe+2. Selenate as an electron donor also supports the growth of strain DRBS1 and significantly increased the arsenate reduction.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Arsenic (As) is found in the earth’s crust at an average content of 2 to 5 mg kg−1 of soil sample [1] and widely distributed in the natural environment as a consequence of natural phenomenon (e.g., weathering and volcanic activity) and anthropogenic activity [2]. Global and regional cycling of arsenic, with industrial activity in particular, accounts for 54% of its mobilization in the biosphere [3] which causes environmental pollution. Arsenic is used in pesticides, herbicides, wood preservatives, and dye stuffs and in smelting and mining operations which leads to the production of arsenic-containing wastes [4]. In arsenic-enriched environments, a major concern is the potential for mobilization and transport of this toxic element to groundwater and drinking water supplies.
In soils, arsenic is mainly in two inorganic forms, arsenate (oxidized pentavalent state; As(V)) and arsenite (reduced trivalent state; As(III)), both forms are toxic. Arsenite disrupts sulfhydryl groups of proteins and interferes with enzyme function, whereas arsenate acts as a phosphate analog and can interfere with phosphate uptake and transport [5]. Environmental cleanup strategies for As(V) removal involve physicochemical or biological detoxification. Major limitations of physicochemical processes are the high energy inputs, different chemical treatments, and generation of unnecessary sludge, with reactive chemical species as secondary wastes. These problems can be overcome by biological As(V) detoxification which is more ecofriendly and an economically feasible technology [6].
Bacteria have evolved a number of mechanisms to cope with or benefit from arsenic exposure [7]. One of the mechanisms followed by arsenate-reducing bacteria is that they achieve growth by respiratory reduction of arsenate to arsenite. Arsenate reduction is also achieved by various bacteria possessing cytoplasmic arsenate reductase (Ars C) which is a part of an arsenic resistance system [8–11]. Arsenic detoxification has been reported in number of bacterial species such as Escherichia coli, Bacillus sp., Staphylococcus aureus, Staphylococcus xylosus, Chromobacterium violaceum, and Pseudomonas sp. [12–14].
In particular, direct arsenite release mediated by arsenate-reducing bacteria seems to pose more serious effects due to the higher toxicity of arsenite than arsenate. Under oxidizing conditions, arsenic solubility is low because arsenate is the predominant species and strongly absorbs onto rocks or forms minerals with iron (hydrous), manganese, and aluminum, and generally arsenate is found to be a major species in arsenic-contaminated soils [15–17]. It is known that reducing or anaerobic conditions can lead to the mobilization of arsenic as arsenite into the liquid phase such as groundwater [15]. In terms of removing arsenic from contaminated soils, however, microbial arsenic mobilization has important role in arsenic bioremediation [18, 19], and arsenite is desirable because it is more mobile than arsenate. This is the reason we have given importance to arsenate reduction; a potential arsenic hyperresistant gram-negative bacterium was isolated from contaminated sites of Tezpur, Assam, and the resistance mechanism was supported by arsenate reduction into arsenite which can be used as an effective model for bioremediation of arsenic-laden soils.
Materials and Methods
Pentavalent arsenic-resistant and As(V)-reducing bacterial strain was isolated from site contaminated from petroleum wastes of Tezpur, Assam, India. Soil sample of this site was contaminated with an arsenic level of 9,038 mg/kg as estimated by inductively coupled plasma atomic emission spectroscopy (ICP-AES, Optima-3300RL, Perkin Elmer, Norwalk, CT, USA). The bacterial strain was screened from serial dilutions (10−1 to 10−7) of the soil sample plated on nutrient agar plates containing arsenate (0–100 mM). Isolates were repeatedly streaked and maintained on 100 mM As(V) containing nutrient agar plates.
Phylogenetic Identification by 16S rRNA Gene Sequencing and Biochemical Characterization
Bacterial genomic DNA was isolated following a previously described method [12]. The genomic DNA was diluted approximately to 20–50 ng and used as template in (30 µl) polymerase chain reactions (PCR) consisting of 1× Gene Taq buffer B (150 mM Tris–HCl, pH 8.0, 500 mM KCl, and 25 mM MgCl2), 50 µM Gene dNTPs, and 1.5U of Gene Taq polymerase with 20 pmol each of universal eubacteria primers each of 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1497R (5′- GGTTACCTTGTTACGACTT-3′) custom synthesized (MWG Biotech, Edersberg, Germany). Amplification cycles (35 cycles in total) consisted of initial denaturation step at 94 °C for 3 min, followed by a 1-min denaturation step at 94 °C, a 1-min annealing step at 55 °C, and an elongation step of 1 min at 72 °C, and a final extension step of 15 min was performed using Eppendorf Mastercycler gradient thermocycler. The purified PCR product was subjected to sequencing by automated DNA Analyzer 3730 using ABI Prism Big Dye Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA). The partial sequence of bacterial 16S rRNA (800 bp) was submitted to GenBank at NCBI with accession no. FJ655884. BLAST(n) program at NCBI server [20] was used to identify and download the nearest neighbor sequences from the NCBI database. All the sequences were aligned using ClustalW 2.0 program at (http://www.ebi.ac.uk/clustalw), the alignment file thus obtained was analyzed and edited by the Data Analysis in Molecular Biology and Evolution software package [21]. The phylogenetic tree was constructed using aligned sequences by the neighbor-joining algorithm using Kimura two-parameter distance and more than 1,000 replicates in Molecular Evolutionary Genetics Analysis 4.1 software [22]. The strain was biochemically identified by using methods given in Bergey’s Manual of Systematic Bacteriology.
Time Course of Arsenate Reduction and the Growth of Pseudomonas sp. DRBS1
The strain was also grown in nutrient broth (NB; Hi media) supplemented with different concentrations of sodium arsenate (0–100 mM; Loba Chemie). It was also grown with different concentrations of sodium selenate (0–10 mM) in combination with 100 mM arsenate in medium.
Time course of As(V) reduction was monitored in 100-ml sterile NB. Bacterial culture of DRBS1 was grown for overnight to an A600 of 0.3–0.5 in sterile nutrient broth. Four milliliters of suspended culture was inoculated into 100 ml of Nutrient broth, pH 7.0, in a 250-ml conical flask amended with different concentrations of sodium arsenate(V) and incubated at 30 °C, 150 rpm, on an orbital shaker. Reduction experiments were followed from 0 to 48 h under shaking condition (150 rpm) at 30 °C. Samples (2 ml) were removed after every 6 to 48 h and subjected to centrifugation (4,000×g for 10 min), and supernatants were used to measure the concentration of arsenate and arsenite following the method given by Johnson and Pilson [23]. New plastic ware was used to avoid contamination from excess phosphate from detergent. For the assay, 1 ml of each sample was pipetted into each of six tubes. Two tubes were oxidized; two were untreated, and two were reduced. The absorbance was measured at 865 nm. Standard curves were prepared for concentrations of 0–100 µM for both arsenate and arsenite. The growth rate constant (k) for the log phase of growth was determined by plotting the log10 of A600 against time [24], and final cellular yield was expressed in total cellular protein which was determined by the method of Lowry et al. [25]. All experiments were performed in triplicate.
Repeated Detoxification of As(V) by Pseudomonas sp. DRBS1
Bacterial culture grown overnight to an A600 of 0.3–0.5 in 100-ml sterile NB broth was amended with 10 mM As(V) as final concentration and incubated at 30 °C under shaking at 150 rpm. After every 5 h of the incubation time, 2 ml of culture suspension was withdrawn to measure As(V) concentration following a method as described above, and the increments of 10 mM As(V) were repeatedly added in culture flasks until saturation in As(V) reduction was observed.
As(V) Reduction by Resting Cells of Pseudomonas sp. DRBS1
Culture suspensions of Pseudomonas sp. DRBS1 was grown overnight in 100-ml NB (pH 7.0) and harvested by centrifugation at 4,000×g at 4 °C. Cell pellets (2 ml) obtained on centrifugation were washed twice with 50 mM Tris–HCl (pH 7.0) and resuspended in the same buffer. Triplicates of these suspended cell pellets were treated with As(V) concentrations of 1–50 mM, vortexed for 5 min, and incubated at 30 °C for 6 h. Heat-killed (2 ml) culture pellets were used as control. After incubation for 6 h, the tubes were centrifuged, and 200-µl aliquots were withdrawn from each sample to estimate the remaining As(V) by the above-mentioned method.
As(V) Reduction by Permeabilized Cells of Pseudomonas sp. DRBS1
Bacterial culture of Pseudomonas sp. DRBS1 was grown overnight, harvested, and washed with Tris–HCl (pH 7.0) as described above. The suspended culture pellets were treated with 0.2% (v/v) Tween 80 and 0.2% (v/v) Triton X-100 and vortexed for 20 min to achieve cell permeabilization. Permeabilized cell suspensions (1 ml) were then added with 1, 10, 20, 30, 40, 50 mM As(V) as final concentrations and incubated for 6 h at 30 °C. Experiments were performed in triplicates.
Preparation of Cell-Free Extracts
Bacterial suspensions grown for overnight in 100 ml NB broth were harvested at 6,000×g at 4 °C for 10 min, washed, and resuspended in 50 mM Tris–HCl (pH 7.0). The culture pellets thus obtained were resuspended in the 5% (v/v) of the original culture volume in 50 mM Tris–HCl (pH 7.0). These cell suspensions were placed on ice bath and disrupted using an Ultrasonic Probe (Sonics Vibra Cell 500, USA) with amplitude of 35% at 50 W with 9-s pulses and 1-s off mode for 35 min. The sonicate thus obtained was then centrifuged at 32,000×g for 40 min at 4 °C. The supernatant or the cytosolic fraction thus obtained was then filtered through 0.22-µm filters (Millipore, Bedford, USA) to give cell-free extracts (CFE) devoid of membrane fractions. The sonicated cell pellets were accordingly resuspended in the same volume of Tris buffer (pH 7.0) and used for subsequent assays.
Localization and Characterization of Arsenate Reductase Activity
Cell-free extracts or cytosolic fractions and sonicated pellet or membrane fractions were used as aliquots (200 µl) for arsenate reductase assay in order to localize the arsenate reductase activity.
Ars C Enzyme Assay
Ars C enzyme activity was assayed using the coupled enzymatic reaction given by Anderson and Cook [26]. In this assay, arsenate reductase activity is measured by observing NADPH oxidation. The reaction system (1.0 ml) was made up of varying arsenate(V) final concentrations (100 and 200 µM) in 0.8 ml of 100 mM Tris–HCl (pH 7.0) and 50 µl of 3 mM NADPH (final concentration 0.15 mM) added with 0.2-ml aliquots of cell-free extracts for arsenate reduction. The system volume of 1.0 ml was kept constant for all experiments. Assay conditions were kept constant with a reaction time of 30 min and 30 °C unless stated for assays at differential time intervals and temperature conditions. Unit enzyme activity for arsenate reductase was derived as amount of enzyme that reduces 1 µmol of As(V) per minute at 30 °C. Specific activity was defined as unit arsenate reductase activity per milligram protein concentration in the CFE. Measurements were recorded at 340 nm, where 0.15 mM NADPH has an absorbance of approximately 1.0. Absorbance decreases as NADPH is oxidized coupled to arsenate reduction to arsenite. Abiotic control reaction mixtures consisted of 100 mM Tris–HCl buffer and respective As(V) concentrations without the addition of CFE and subjected to the same assay conditions as those followed for experimental reactions. Endogenous NADPH oxidation rates were subtracted from arsenate-induced NADPH oxidation.
As(V) Reduction by CFE as Function of Time
As(V) reductase activity in the CFE was measured at initial As(V) concentrations of 100 and 200 µM with increasing incubation time intervals (0–40 min).
Effect of Temperature on As(V) Reduction of CFE
As(V) reductase activity in the CFE was measured at initial As(V) concentrations of 100 µM with 30-min incubation time at different temperatures (20–70 °C). Nonenzymatic As(V) reduction was checked following inactivation of CFE at 100 °C for 5 min.
Effect of pH on As(V) Reduction by CFE
The cell-free extracts were prepared in buffers of different pH range, and the same buffers were used for As(V) reductase assays. Buffers used for assay were citrate phosphate (pH 4.0–6.0) and Tris–HCl buffers (pH 6.2–7.5).
Effect of Metal Cations, Electron Donors, and Inhibitors on As(V) Reduction by CFE
Cell-free extracts of Pseudomonas sp. DRBS1 were characterized for differential specific activity following As(V) reductase assays in the presence of (1 mM each) metal ions (Pb2+, Cd+2, Cu+2, Ca+2,Fe+2, Ag+, Hg+, Ni2+, and Cs+) and electron donors (citrate, acetate, glucose, galactose, citrate, fructose, carbonate, and lactose).
Results and Discussion
Isolation of Arsenic-Resistant Bacterium
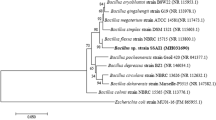
In the present study, isolation and screening for arsenic-resistant bacteria were done from soil samples collected from three arsenic-contaminated sites, namely, Dumping yard of VIP road, Hindustan petroleum loading point, and Prasanti path, Tezpur, Assam. These sites are located in the vicinity of industrial units where arsenic is used in insecticide, fungicide, and weedicide production. Arsenic level in soil samples collected from different sites ranges from 7,120 to 9,038 mg/kg of soil as estimated by ICP-AES Optima-3300RL (Perkin Elmer, Norwalk, CT, USA). The method employed in the present study measures only inorganic soluble arsenic and does not detect any organoarsenicals. Inorganic form of arsenic is likely to be most available to microorganisms and thus is probably most relevant to the study. A total of 25 bacterial strains were isolated from soil samples collected from different sites. Isolates were selected on the basis of the colony characteristics on nutrient agar plates amended with different concentrations of As(V). All isolates could resist up to 50 mM As(V) (data not shown), except DRBS1 bacterial strain which resists up to 100 mM As(V) [significant growth] and tolerate up to 200 mM As(V) [nonsignificant growth]. Microbial resistance to arsenic species is widespread in nature, but resistance to concentrations of As(V) higher than 100 mM is considered as very significant [27]. Resistance and tolerance to arsenate were determined by visible growth after 7 days in nutrient broth amended with varying concentration of arsenate. Microscopic characterization showed the isolate to be a noncapsulated, non-spore-forming, and gram-negative short-rod bacterium. Biochemical characterization was also performed by following the methods given in Bergey’s Manual of Systemic Bacteriology. The strain showed a positive reaction for growth on triple sugar agar, catalase, indole production, nitrate reduction, urease, and liquefaction of gelatin. The morphological and biochemical characteristics were summarized in Table 1. The DNA sequencing and BLAST analysis of partial 16S rRNA gene sequence (800 bp) of strain DRBS1 showed maximum sequence identity (100%) with the sequence of Pseudomonas stutzeri 16S rRNA gene (AY905607). Phylogenetic analysis indicated that the newly isolated strain DRBS1 is affiliated to Pseudomonas sp. (Fig. 1). In other studies, also, Pseudomonas bacteria were isolated from different arsenic-contaminated environments [12, 28–30]. Pseudomonas arsenitoxidans, isolated and described by Ilyaletdinov and Abdrashitova [31], is the only known chemolithotrophic arsenite-oxidizing Pseudomonas species. The genus Pseudomonas has been reported to exhibit the arsenate reductase machinery for arsenate bioreduction in previous studies [12, 32, 33].
Phylogenetic tree derived from 16S rRNA gene sequence of isolate DRBS1. Sequences of closest phylogenetic neighbors obtained by NCBI BLAST(n) analysis; numbers in the parenthesis indicate accession numbers of corresponding sequences. E. coli K12 has been taken as an out-group. The NJ tree was constructed using neighbor-joining algorithm with Kimura two-parameter distances in MEGA 4.0 software. Numbers at nodes indicate percent bootstrap values. The culture shows 100% identity with P. stutzeri upon NCBI BLAST analysis
Salam et al. [34] reported that six morphologically distinct arsenic-resistant bacteria belong to genus Bacillus, Pseudomonas, Listeria, Moraxella, and Planococcus which were isolated from different contaminated sites of Khulna, Bangladesh. Anderson and Cook [26] also reported 17 morphologically distinct arsenic-resistant heterotrophic bacteria. They are the members of genera Exiguobacterium, Aeromonas, Bacillus, Pseudomonas, Escherichia, and Acinetobacter. The total of 22 arsenic-tolerant bacteria was isolated from arsenic-containing rock biofilms on the walls of the Gertruda Adit in the Zloty Stok gold mine. These strains belong to γ-Proteobacteria, Actinobacteria, and Flavobacteria group of bacteria [35]. A group of nine strains were isolated previously from Zloty Stok gold mine and Lubin copper mine [36, 37]. All strains have evolved strong resistance mechanisms supported by arsenate reduction, but none could utilize arsenite or arsenate in respiratory processes [38].
As(V) Reduction and Growth in Culture Flasks
The arsenic resistance mechanism in strain was supported by arsenate reduction, but it did not utilize in respiratory processes. The suspended culture of the Pseudomonas sp. strain DRBS1 was efficient in As(V) reduction even at higher initial arsenate concentrations. Pseudomonas sp. DRBS1, when incubated for 48 h at 30 °C with shaking conditions (150 rpm), could achieve exponential growth and efficiently reduce more than 97% of As(V) at the initial concentrations of 60, 80, and 100 mM and more than 90% As(V) at the initial concentrations of 40 and 20 mM (Table 2). An exponential growth was observed even at higher initial As(V) concentrations of 80 and 100 mM, after 18 h of incubation an increase in cell density was observed, followed by a decrease in cell density after 42 h of incubation. The growth rate of strain DRBS1, in the absence of arsenate, was 0.187 h−1 ± 0.20 (doubling time of 3.7 h), whereas in the presence of 80 mM arsenate it was 0.058 h−1 ± 0.02 (doubling time of 11.8 h; data not shown).The reason for retardation of total growth of As(V)-reducing bacterium may be the toxicity and stress exerted by arsenate. Repeated detoxification of 10 mM arsenate As(V) could be achieved up to five consecutive inputs as shown in Fig. 2, suggesting the utility of bacterium in continuous As(V) bioreduction process.
Effect of Selenate on Arsenate Reduction by Strain DRBS1
Selenate, an oxyanion, coexists with arsenate in a contaminated soil environment and that is why we have investigated the effect of selenate on arsenate reduction in growth experiment. The inoculated culture was amended with different concentration of selenate (0–10 mM) with 100 mM arsenate, incubated at 30 °C under shaking condition for 60 h. An orange- to red-colored precipitate formed in the culture as a result of the formation of insoluble elemental selenium. Selenate did not show any inhibitory effect on the arsenate reduction by Pseudomonas sp. strain DRBS1 while the reduction increased in the presence of all concentrations of selenate. The role of selenate as electron acceptor has been well demonstrated by Yamamura et al. [39] and it did not show any inhibitory effect on arsenate reduction by Bacillus sp. SF-1, a facultative anaerobe. The growth rate of strain DRBS1, in the presence of selenate (10 mM) with arsenate (100 mM) was 0.11 h−1 ± 0.02 (doubling time of 6.3 h) while in the presence of 100 mM arsenate alone it was 0.053 h−1 ± 0.02 (doubling time of 11.8 h; data not shown). In the presence of lower concentrations of selenate (2 and 4 mM) with 100 mM arsenate, the lag phase of bacterial strain DRBS1 did extended but the growth increased in the presence of all concentrations of selenate (0–10 mM) in comparison to 100 mM arsenate alone (Figs. 2 & 3). In recent years, several bacteria capable of reducing selenate to elemental selenium have been isolated from different environments. These isolates include Bacillus sp. SF-1 from selenium-polluted sediment [40], Bacillus selenitireducens MLS10T from alkaline lake sediments [41], Sulfurospirillum barnesii SES-3 T from freshwater sediments [42], Selenihalanaerobacter shriftii DSSe-1 T from deep-sea sediments [43], Salana multivorans Se-3111 T from an aerobic bioreactor [44], and Citrobacter freundii Iso-Z7 from selenium-contaminated sediment [45].
As(V) Reductase Activity in Pseudomonas sp. DRBS1
The resting and permeabilized cells of the bacterium were expedient by reducing 1–50 mM As(V) concentrations in 6 h as shown in Fig. 4. The permeabilization significantly increased the As(V) reduction by the resting cells, as the Tween-80-permeabilized cells could reduce greater than 95% of 50, 40, and 30 mM of As(V) within 6 h, suggesting an efficient intracellular mechanism of arsenate reduction. For localization of As(V) reductase activity, the assay was performed by using the ultrasonicated subcellular fractions. The As(V) reductase activity in the cytosolic fractions (CFE) of cells grown in the absence of arsenate was 0.56 ± 0.04 and in the presence of 100 µM As(V) and 200 µM As(V) it was 6.68 ± 0.01 and 12.15 ± 0.02 µmol/min per milligram protein, respectively, while no activity was detected in membrane fraction (sonicated cell pellet) in the presence and absence of 100 µM As(V) and 200 µM As(V), respectively. The results indicate that the As(V) reductase activity was associated with cytosolic fractions and not with membrane fractions. A cytoplasmic arsenate reductase catalyzes reduction of the less toxic arsenate to more toxic arsenite and is the most well-known mechanism of arsenic resistance, described in many different bacteria [46, 47].
The As(V) reduction by the cell-free extracts as a function of time is shown in Fig. 5. The cell-free extracts could reduce more than 86% of 100 and 200 µM As(V) in 40 min, suggesting the efficient arsenate reductase activity in cytosolic fraction of bacterium. In the presence of 0.2 mM NADPH with initial As(V) concentration of 100 and 200 µM, a twofold increase in arsenate reductase activity was observed. The increase in cytosolic arsenate reductase activity is due to the oxidation of NADH into NAD+ which enhanced the arsenate reduction. Pyridine nucleotides (such as NAD+ or NADP+) increase the As(III) formation; specifically, in the presence of NAD+, the production of As(III) was doubled [48]. There is no effect of 300 µM dithiothreitol (DTT) on the arsenate reductase activity. The enzymatic progress curves of specific As(V) reductase activity in the CFE exhibit the marked increase in the presence of 0.2 mM NADPH with increasing initial concentration as seen in Fig. 6.
Effects of Different In Vitro Conditions on the Arsenate Reductase Activity
The functioning of the arsenate reductase of Pseudomonas sp. DRBS1 was characterized in various in vitro conditions. To define the optimal pH, the As(V) reductase assays were carried out in citrate phosphate, potassium phosphate, and Tris–HCl buffers in various pH ranges. A characteristic pH curve for the enzymatic activity in the presence of 100 µM at pH 5–7 is depicted in Fig. 7. The optimal temperature for the As(V) reductase activity was 30 °C, whereas the reductase activity in presence of 100 and 200 µM was decreased at 37, 40, 50, and 70 °C as shown in Fig. 8. The heat-killed CFE treated for 5 min at 100 °C did not show any As(V) reductase activity.
The effect of different metal cations and electron donors on the arsenate reductase activity of Pseudomonas sp. DRBS1 is shown in Figs. 9 and 10. Among the metal ions tested Cd+2, Cu+2, Ca+2, and Fe+2 stimulated the As(V) reductase activity of the CFE by 6%, 15%, 11%, and 15%, respectively. However, the other divalent cations did not exhibit any significant effect on the reductase activity. Similarly, the reductase activity increased on supplementation in the reaction mixtures with electron donors such as acetate, glucose, citrate, and fructose by more than 25%, respectively.
Conclusions
The present study elucidated the arsenate-reducing potential, localization, and characterization of a very efficient As(V) reductase of Pseudomonas sp. DRBS1. Arsenate reductase assays of the CFE have shown a high As(V) reductase activity, implicating the localization of enzyme in the cytosolic fraction. The As(V) reduction potential of the resting cells was increased by cell permeabilization. The CFE could rapidly reduce more than 86% of 100 and 200 µM of As(V) in 40 min. Optimum temperature and pH of arsenate reductase activity of the bacterium were found to be 30 °C and 5.2, respectively, and activity was enhanced in the presence of 0.2 mM NADH and 1 mM of metal ions like Cu+2, Ca+2, and Fe+2. The As(V) reductase activity was stable in the presence of other metal ions which were tested. The bacterium could resist and reduce higher arsenate concentrations in suspended cultures and repeatedly reduce subsequent doses of 10 mM As(V). The results of higher rates of As(V) reductase at ambient temperatures, pH, and in the presence of polymetals indicate a potential application of Pseudomonas sp. DRBS1 for As(V) bioremediation.
References
Tamaki, S., & Frankenberger, W. T. (1992). Environmental biochemistry of arsenic. Reviews of Environmental Contamination and Toxicology, 124, 79–110.
Cullen, R. W., & Reimer, K. J. (1989). Arsenic speciation in the environment. Chemical Reviews, 89, 713–764.
Nriagu, J. O. (1990). Global metal pollution. Environment, 32(7–11), 28–33.
National Research Council. (1977). Arsenic. Washington, DC: National Academy of Sciences.
Jackson, C. R., Jackson, E. F., Dugas, S. L., Gamble, K., & Williams, S. E. (2003). Microbial transformations of arsenite and arsenate in natural environments. Recent Research Developments in Microbiology, 7, 103–118.
Clausen, C. A. (2000). Isolating metal-tolerant bacteria capable of removing copper, chromium and arsenic from treated wood. Waste Management Research, 18, 264–268.
Cervantes, C., Ji, G., Ramirez, J. L., & Silver, S. (1994). Resistance to arsenic compounds in microorganisms. FEMS Microbiology Reviews, 15, 355–367.
Dowdle, P. R., Laverman, A. M., & Oremland, R. S. (1996). Bacterial dissimilatory reduction of arsenic (V) to arsenic (III) in anoxic sediments. Applied and Environmental Microbiology, 62, 1664–1669.
Oremland, R. S., & Stoltz, J. F. (2003). The ecology of arsenic. Science, 300, 939–944.
Silver, S. (1998). Genes for all metals—a bacterial view of the periodic table. Journal of Industrial Microbiology & Biotechnology, 20, 1–12.
Xu, C., Zhou, T., Kuroda, M., & Rosen, B. P. (1998). Metalloid resistance mechanisms in prokaryotes. Journal of Biochemistry, 123, 16–23.
Patel, P. C., Goulhen, F., Boothman, C., Gault, A. G., et al. (2007). Arsenate detoxification in a Pseudomonad hypertolerant to arsenic. Archives of Microbiology, 187(3), 171–183.
Mateos, L. M., Ordonez, E., Letek, M., & Gil, J. A. (2006). Corynebacterium glutamicum as a model bacterium for the bioremediation of arsenic. International Microbiology, 9, 207–215.
Rosen, B. P. (2002). Biochemistry of arsenic detoxification. FEBS Letters, 529, 86–92.
Mok, M. W., & Wai, C. M. (1994). Mobilization of arsenic in contaminated river waters. In J. O. Nriagu (Ed.), Arsenic in the environment. Part I: cycling and characterization. New York: Wiley.
Garcia-Manyes, S., Jimenez, G., Padro, A., Rubio, R., & Rauret, G. (2002). Arsenic speciation in contaminated soils. Talanta, 58, 97–109.
Bissen, M., & Frimmet, H. F. (2000). Speciation of As (III), As (V), MMA and DMA in contaminated soil extracts by HPLC-ICP/MS. Fresenius’ Journal of Analytical Chemistry, 367, 51–55.
Lovley, D. R., & Coates, J. D. (1997). Bioremediation of metal contamination. Current Opinion in Biotechnology, 8, 285–289.
Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., et al. (1997). Current protocols in molecular biology, unit 24. New York: Wiley.
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research, 25, 3389–3402.
Xia, X., & Xie, Z. (2001). DAMBE: software package for data analysis in molecular biology and evolution. Journal of Heredity, 92, 371–373.
Kumar, S., Tamura, K., Jakobsen, I. B., & Nei, M. (2001). MEGA2: molecular evolutionary genetics analysis software. Bioinformatics, 17, 1244–1245.
Johnson, D. L., & Pilson, M. E. Q. (1972). Spectrophotometric determination of arsenite, arsenate and phosphate in natural waters. Analytica Chimica Acta, 58, 289–299.
Pirt, S. J. (1975). Principles of microbe and cell cultivation. Oxford: Blackwell.
Lowry, O. H., Rosenberg, N. J., Farr, A. L., & Randall, R. J. (1951). Estimation of protein by Lowry’s method. Journal of Biological Chemistry, 193, 265.
Anderson, C. R., & Cook, G. M. (2004). Isolation and characterization of arsenate-reducing bacteria from arsenic-contaminated sites of New Zealand. Current Microbiology, 48, 341–347.
Jackson, C. R., Harrison, K. G., & Dugas, S. L. (2005). Enumeration and characterization of culturable arsenate resistant bacteria in a large estuary. Systematic and Applied Microbiology, 28, 727–734.
Turner, A. W. (1954). Bacterial oxidation of arsenite. Description of bacteria isolated from arsenical cattle-dipping fluids. Australian Journal of Biological Sciences, 7, 452–478.
Abdrashitova, S. A., Mynbaeva, B. N., & Ilyaletdinov, A. N. (1981). Oxidation of arsenic by the heterotrophic bacteria Pseudomonas putida and Alcaligenes eutrophus. Mikrobiologiya, 50, 41–45.
Macur, R. E., Jackson, C. R., Botero, L. M., McDermott, T. R., & Inskeep, W. P. (2004). Bacteria populations associated with the oxidation and reduction of arsenic in an unsaturated soil. Environmental Science and Technology, 38, 104–111.
Ilyaletdinov, A. N., & Abdrashitova, S. A. (1981). Autotrophic oxidation of arsenic by a culture of Pseudomonas arsenitoxidans. Mikrobiologiya, 50, 197–204.
Joshi, D. N., Patel, J. S., Flora, S. J. S., & Kalia, K. (2008). Arsenic accumulation by Pseudomonas stutzeri and its response to some thiol chelators. Environmental Health and Preventive Medicine, 13(5), 257–263.
Rathinasabapathi, B., Raman, S. B., Kerthlis, G., & Ma, L. (2006). Arsenic resistant proteobacterium from the phyllosphere of arsenic-hyperaccumulating fern (Pteris vita L.) reduces arsenate to arsenite. Canadian Journal of Microbiology, 52(7), 695–700.
Salam, M. A., Hossain, M. S., Ali, M. E., Asad, M. A., & Ali, M. H. (2009). Isolation and characterization of arsenic resistant bacteria from different environment in South-West region of Bangladesh. Research Journal of Environment Science, 3(1), 110–115.
Drewniak, L., Styczek, A., Majder-Lopatka, M., & Sklodowska, A. (2008). Bacteria, hypertolerant to arsenic in the rocks of an ancient gold mine, and their potential role in dissemination of arsenic pollution. Environmental Pollution, 156, 1069–1074.
Drewniak, L., Styczek, A., & Sklodowska, A. (2007). Arsenic hypertolerant bacteria isolated from gold mine rocks biofilms. Advanced Materials Research, 576, 20–21.
Matlakowska, R., Hallberg, K. B., & Sklodowska, A. (2007). Isolation and characterization of microorganisms from copper bearing black shale of Lubin copper mine (Poland). Advanced Materials Research, 580, 20–21.
Matlakowska, R., Drewniak, L., & Sklodowska, A. (2008). Arsenic-hypertolerant pseudomonads isolated from ancient gold and copper-bearing black shale deposits. Geomicrobiology Journal, 25, 357–362.
Yamamura, S., Ike, M., & Fujita, M. (2003). Dissimilatory arsenate reduction by a facultative anaerobe, Bacillus sp. Strain SF-1. Journal of Bioscience and Bioengineering, 96(5), 454–460.
Fujita, M., Ike, M., Nishimoto, S., Takahashi, K., & Kashiwa, M. (1997). Isolation and characterization of a novel selenate-reducing bacterium, Bacillus sp. SF-1. Journal of Fermentation and Bioengineering, 83, 517–522.
Switzer Blum, J., Burns Bindi, A., Buzzelli, J., Stolz, J. F., & Oremland, R. S. (1998). Bacillus arsenicoselenatis, sp. nov., and Bacillus selenitireducens, sp. nov.: two haloalkaliphiles from Mono Lake, California that respire oxyanions of selenium and arsenic. Archives of Microbiology, 171, 19–30.
Stolz, J. F., Ellis, D. J., Switzer Blum, J., Ahmann, D., Lovley, D. R., & Oremland, R. S. (1999). Sulfurospirillum barnesii sp. nov. and Sulfurospirillum arsenophilum sp. nov., new members of the Sulfurospirillum clade of the epsilon Proteobacteria. International Journal of Systematic Bacteriology, 49, 1177–1180.
Switzer Blum, J., Stolz, J. F., Oren, A., & Oremland, R. S. (2001). Selenihalanaerobacter shriftii gen. nov., sp. nov., a halophilic anaerobe from deep sea sediments that respires selenate. Archives of Microbiology, 175, 208–219.
von Wintzingerode, F., Gobel, U. B., Siddiqui, R. A., Rosick, U., Schumann, P., Fruhling, A., et al. (2001). Salana multivorans gen. nov., sp. nov., a novel actinobacterium isolated from an anaerobic bioreactor and capable of selenate reduction. International Journal of Systematic and Evolutionary Microbiology, 51, 1653–1661.
Zhang, Y., Siddique, T., Wang, J., & Frankenberger, W. T., Jr. (2004). Selenate reduction in river water by Citrobacter freundii isolated from a selenium-contaminated sediment. Journal of Agricultural and Food Chemistry, 52, 1594–1600.
Prithiviraj Singh, S., Mishra, S. K., & Mahadevan, A. (2001). Functional analysis of chromosomal arsenic resistance operon in Pseudomonas fluorescens strain MSP3. Molecular Biology Reports, 28(2), 63–72.
Sizova, O. I., Kochetkov, V. V., & Boronin, A. M. (2006). The arsenic-phytoremediation potential of genetically modified Pseudomonas sp. In J. L. Morel, G. Echevarria & N. Goncharova (Eds.), Phytoremediation of metal contaminated soil (Vol. 68, pp. 327–334). Heidelberg: Springer.
Nemeti, B., & Gregus, Z. (2002). Reduction of arsenate to arsenite in hepatic cytosol. Toxicological Sciences, 70, 4–12.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Srivastava, D., Madamwar, D. & Subramanian, R.B. Pentavalent Arsenate Reductase Activity in Cytosolic Fractions of Pseudomonas sp., Isolated from Arsenic-Contaminated Sites of Tezpur, Assam. Appl Biochem Biotechnol 162, 766–779 (2010). https://doi.org/10.1007/s12010-009-8852-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-009-8852-0