Abstract
A coordinated study of water chemistry, sediment mineralogy, and sediment microbial community was conducted on four >73°C springs in the northwestern Great Basin. Despite generally similar chemistry and mineralogy, springs with short residence time (~5–20 min) were rich in reduced chemistry, whereas springs with long residence time (>1 day) accumulated oxygen and oxidized nitrogen species. The presence of oxygen suggested that aerobic metabolisms prevail in the water and surface sediment. However, Gibbs free energy calculations using empirical chemistry data suggested that several inorganic electron donors were similarly favorable. Analysis of 298 bacterial 16S rDNAs identified 36 species-level phylotypes, 14 of which failed to affiliate with cultivated phyla. Highly represented phylotypes included Thermus, Thermotoga, a member of candidate phylum OP1, and two deeply branching Chloroflexi. The 276 archaeal 16S rDNAs represented 28 phylotypes, most of which were Crenarchaeota unrelated to the Thermoprotei. The most abundant archaeal phylotype was closely related to “Candidatus Nitrosocaldus yellowstonii”, suggesting a role for ammonia oxidation in primary production; however, few other phylotypes could be linked with energy calculations because phylotypes were either related to chemoorganotrophs or were unrelated to known organisms.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The Unites States Great Basin is a >500,000 km2 endorheic region in the western United States with widely distributed geothermal activity. The hot springs of the Great Basin contrast with geothermal systems that exist in volcanically driven hot spring systems such as Yellowstone, Japan, Iceland, Kamchatka, and Italy in several ways. First, although acidic hot springs are common in many volcanically driven geothermal fields, acid springs (pH < 6.0) do not exist in the Great Basin (Zehner et al. 2006). Acid hot springs form when pressure/temperature conditions drive vapour/condensation-mediated concentration of H2S, which is abiotically or microbiologically oxidized to sulphuric acid (Fournier 2005). The absence of sulphuric acid-buffered springs in the Great Basin limits the diversity of microorganisms that could inhabit these springs, because many known thermophiles are obligate acidophiles, for example, archaea in the order Sulfolobales (Reysenbach 2001). Second, geothermal activity is driven by a different mechanism. In Yellowstone and other volcanically active regions, meteoric water that feeds hot spring systems is heated in the subsurface by magmatic heat. In contrast, geothermal activity throughout most of the Great Basin is attributed to tectonically driven dilation of range-front faults, which allows deep circulation and subsequent heating of meteoric water (Faulds et al. 2006). Third, in contrast to the high aerial density of hot springs in Yellowstone, surface expressions of geothermal systems in the Great Basin are geographically separated. This geographical separation raises interesting questions about the geochemical diversity of Great Basin springs as well as the biogeography of resident microorganisms.
Despite the different geological settings of Yellowstone and the Great Basin hot spring systems, these two systems have not been systematically compared as habitats for microorganisms. Thus far, only a small body of research on the microbiology of Great Basin hot springs exists (Breitbart et al. 2004; Connon et al. 2008; Huang et al. 2007; Lee et al. 2007; Pearson et al. 2004; Zhang et al. 2006a, 2007). Work on arsenite-oxidizing biofilms in Alvord Hot Spring system in southeastern Oregon has described a source pool (78.2°C) with abundant Thermocrinis, Sulfurihydrogenibium, and Thermus, and a more diverse outflow (~73°C) with the former genera and phylum OP10, uncultivated Bacteroidetes, and a novel uncultivated lineage (Connon et al. 2008). These groups are commonly described in Yellowstone hot springs, although environments in which Thermocrinis and Sulfurihydrogenibium cohabitate may be uncommon in Yellowstone (Hall et al. 2008).
Other studies have been less focused on a particular hot spring system. A number of studies have focused on Crenarchaeota, employing combinations of 16S rRNA gene censuses, lipid biomarker analyses, and habitat geochemistry characterization in northern Nevada and northeastern California (Huang et al. 2007; Pearson et al. 2004; Pearson et al. 2008; Zhang et al. 2006a). These papers established the presence of the crenarchaeol biomarker in continental hot springs and, furthermore, established a higher incidence and a higher relative concentration of crenarchaeol in Great Basin hot springs compared with those in Yellowstone, Kamchatka, and Tengchong, China. Crenarchaeol correlated positively with high pH. Insofar as crenarchaeol has only been discovered in ammonia-oxidizing Crenarchaeota (de la Torre et al. 2008; Schouten et al. 2008), the abundance of this biomarker in Great Basin springs could possibly be explained by the higher pH, and therefore higher ammonia activity, in Great Basin hot springs.
We have recently begun to focus on hot springs in the Great Boiling Springs and Mud Springs geothermal systems in the northwestern Great Basin, with the long term goal of understanding whether the geological setting of Great Basin springs influences energy and nutrient cycling relative to volcanically associated systems. These two spring systems have been classified as circumneutral Na–Cl springs with high levels of dissolved Na+, Cl−, HCO3 −, and SiO2 (Anderson 1978). These two systems are fed by a single subterranean reservoir; however, hydrologic controls and the variable influence of cold, shallow groundwater drives variation in the >70 springs in the region (Anderson 1978). To gain a broad view of the microbial diversity and to guide hypothesis generation on possible microbial metabolisms in these springs, a coordinated cultivation-independent census and geochemical/thermodynamic study was conducted on the sources of four different springs, each of which was minimally influenced by shallow groundwater. The springs were chosen to represent extremes of flow, with water residence times of >1.5 days or <30 min, allowing the data to be viewed with respect to the different flow regimes, which we predicted to affect the redox condition of the spring and, therefore, the microbial community composition. The current study expands the research on the microbiology of Great Basin hot springs and is the first study of Great Basin hot springs to tie molecular censuses of microbial inhabitants with a detailed study of the geochemistry and thermodynamic landscape of the habitat in which they live. To our knowledge, this is also the first study that considers the effect of water residence time on hot spring microbial community composition and potential activity in the Great Basin.
Materials and methods
Site description and chemical measurements
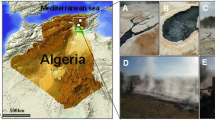
Samples for bulk water chemistry, sediment mineralogy, and sediment microbial community were coordinately sampled from the exact same locations in the shallows near hot spring sources (Fig. 1). Sampling for Great Boiling Spring (GBS)(also known as GBS17a) was done at GPS location N40° 39.689′ W119° 21.968′, Great Boiling Hot Springs 04b (G04b) at N40° 39.749′ W119° 21.986′, Sandy’s Spring West (SSW) at N40° 39.182′ W119° 22.496, and Sandy’s Spring East (SSE) at N40° 39.173′ W119° 22.471′ (datum: WGS84). Calculations of volume on GBS, SSW, and SSE were done assuming they were perfect cones, using V = 1/3πr 2 h and residence time was calculated by dividing the volume by the volumetric flow rate.
Field measurements made in situ using a multisystem probe (LaMotte, Chesterton, MD, USA) included temperature, pH, and conductivity. Concentrations of O2, NO2 −, NO3 −, total ammonia, alkalinity, and total sulphide were analyzed using portable kits and a spectrophotometer according to the manufacturer’s instructions (LaMotte, Chesterton, MD). Oxygen was analysed in samples that were not cooled. Sulphide was measured in samples that were diluted 1:3 with distilled water without cooling. All other measurements were made on samples that had cooled to ~25–35°C. Samples for major ions were filtered in the field through a pre-washed 0.2 μm Supor filter (hydrophilic polyether-sulfone, Pall Scientific) and collected in polypropylene conical tubes. Samples were stored on ice and brought back to the lab for analysis. Major cation and anion concentrations were measured using ion chromatography (anions: Dionex IonPac AS11 Analytical and IonPac AG11 Guard columns; cations: Dionex IonPac CS12A Analytical and IonPac SG11 Guard columns; conductivity detection). Samples for trace element analysis were collected in acid washed polypropylene vials, acidified to pH ~1 with nitric acid, filtered on site with a 0.2 μm Supor filter, and stored on ice. In the lab, samples were analyzed by high-resolution inductively coupled plasma mass spectrometry (ICP-MS) at ASU. Samples for dissolved gas measurements were collected using a gas stripping apparatus as described by (Spear et al. 2005). Gas samples were sent to Microseeps (Pittsburgh, PA) for quantification of gases.
Mineralogy
X-ray diffraction (XRD) analyses were made on clay fractions separated by centrifugation and sedimentation following rinsing with distilled water to achieve dispersion. Oriented pastes of K- and Mg-saturated clays (<2 μm) were prepared by smearing the clays onto glass slides (Theisen and Harward 1962). The K-saturated sample slides were examined by XRD at 25°C and after heating at 350 and 550°C for 2 h. The Mg-saturated samples were also analyzed at 25°C and after being placed in a desiccator containing a pool of ethylene glycol and heated at 65°C for 2 h. The desiccator vent was closed upon removal from the oven and the slides stored in the desiccator at least 12 h prior to XRD analysis. Samples were examined by XRD (CuKα radiation) using a PANalytical X’PERT Pro diffractometer, equipped with an X’Celerator detector. The minerals present were primarily identified based on their basal or d(001) spacings. The various Mg- and K-saturation, heat, and glycolation treatments were used to distinguish and identify the different clay minerals present.
Thermodynamic modelling
Chemical data gathered at each spring was used to determine chemical affinities by using the formula A = RT ln (K/Q), where R is the universal gas constant, T is the temperature in Kelvin, K represents the equilibrium constant, which can be calculated from the standard Gibbs free energy of the reaction, ΔG°r, at in situ temperature and pressure with the relation ΔG°r = −RT ln K, and Q is the activity product: the product of the activities of the products divided by the product of the activities of the reactants, each raised to its stoichiometric coefficient. Plausible metabolisms were based on those modelled by (Shock et al. 2005) and values for ΔG°r and Q were calculated using Supcrt92 (Johnson et al. 1992) and EQ3/6 (Wolery 1992), respectively. Chemical affinity was expressed in terms of kJ/mol e− transferred.
Molecular analyses
Sediment from the upper ~1 cm was collected from the shallows of springs using sterile spatulas, transferred into 1.5 ml Eppendorf tubes, stored on dry ice in the field, and transferred to a −80°C freezer in the lab. Samples were collected in May 2005 for GBS and May 2006 for G04b, SSW, and SSE. A sample from GBS was thawed and split into two vials for extraction using two different environmental DNA isolation kits: MoBio PowerMax Soil Kit (Carlsbad, CA) and the Qbiogene Fast DNA SPIN Kit for Soil (Irvine, CA). 16S rRNA gene terminal restriction fragment length polymorphism (T-RFLP) analysis using RsaI, MspI, and Taq1, separately, showed that the different polymerase chain reactions (PCR) with different template DNA yielded very similar 16S rRNA gene products (data not shown). Therefore, biases inherent in extracting DNA from microbial communities and amplifying SSU rRNA genes from a mixed template pool (Reysenbach et al. 1992; von Wintzingerode et al. 1997) were at least consistent. The Qbiogene kit was used for subsequent DNA isolations from 0.5 g sediment slurries. 16S rRNA genes were amplified by PCR using four different primer sets: for Bacteria: 9bF (Eder et al. 1999) and 1406uR or 1512uR (Eder et al. 2001); for Archaea: 8aF and 1406uR or 1512uR (Eder et al. 2001). PCR reaction mixtures contained 1 μL DNA extract, 1 × Taq reaction buffer, 6 nM of each primer, 800 μM each dNTP, and 0.65 U of GoTaq DNA polymerase (Promega). Cycling conditions were: denaturation at 96°C for 4 min followed by 35 cycles of denaturation (30 s at 94°C), primer annealing (30 s at 55°C), and elongation (1.5 min at 72°C), with a final elongation step (10 min at 72°C). Product from each primer set was ligated into a TA TOPO cloning vector (Invitrogen, Carlsbad, CA, USA) and sequenced using the appropriate forward PCR primer in a 96 well plate format at the Nevada Genomics Center using an Applied Biosystems (ABI) Prism 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA). Sequences were screened for chimeras and other anomalies using Bellerophon (Huber et al. 2004) and Mallard (Ashelford et al. 2006) and verified using Pintail (Ashelford et al. 2005). Sequences were aligned using clustalX (Thompson et al. 1997) and grouped into operational taxonomic units (OTUs) at the 97% identity level with the DOTUR nearest neighbor algorithm using only nucleotides with Phred scores > 20 (Schloss and Handelsman 2005). OTUs from each library were cross referenced to eliminate redundant OTUs and then representative phylotypes were sequenced with additional primers to obtain nearly complete 16S rDNA gene sequences. Contigs were assembled using EMBOSS (Rice et al. 2000) and checked again for anomalies. Alignments were made in ARB (Ludwig et al. 2004) and checked manually. Phylogenetic trees were constructed using ARB maximum likelihood (AxML), neighbor-joining (Kimura correction), and maximum parsimony with a heuristic search (Ludwig et al. 2004). Forward sequences from each hot spring were also compared using webLIBSHUFF (Singleton et al. 2001). Bacterial and archaeal trees were compared in Unifrac by using the Cluster Environments and Jackknife Environmental Clusters commands. All analyses were run with 1,000 permutations, including abundance weights, and with the “number of sequences to keep” set at 75% of the smallest library (Lozupone et al. 2006).
Nucleotide sequence accession numbers
Nearly complete 16S rDNA sequences from this study have been deposited in the Genbank database under accession numbers DQ490000–DQ490017 and EU635901–EU635954.
Results and discussion
Site description and mineralogy
Two springs were sampled in the Great Boiling Springs geothermal field: Great Boiling Springs (GBS; Fig. 1a) and Great Boiling Springs 04b (G04b; Fig. 1b). GBS is a large, roughly conical spring ~9.15 m deep and ~7.6 m wide. GBS has a flow rate of ~1.1 L/s and an estimated volume of ~1.39 × 105 L, yielding a water residence time of ~1.5 days. G04b is a deep elliptical feature ~7.6 by ~4 m. At most sampling times, this spring has no obvious surface outflow.
Two springs were sampled in the Mud Hot Springs geothermal field: Sandy’s Spring West (SSW; Fig. 1c) and Sandy’s Spring East (SSE). SSW is a conical vent ~1 m deep and ~2 m in diameter that discharges ~86°C water at ~3.3 L/s to a ~0.01 m deep, ~0.5 m wide outflow channel. SSW has a volume of ~1.05 × 103 L and the calculated residence time of the water is ~5 min. At ~42 m the outflow channel intersects a second vent, SSE, which is ~1.5 m deep and ~3 m in diameter with a temperature of ~77°C and a similar flow rate. SSW has a volume of ~3.53 × 103 L and the estimated residence time of the water is ~18 min. Together, SSW and SSE form a single ~0.5–1 m wide outflow channel.
Sediments from all four springs were uniformly compacted greenish grey clay with a ~1 cm cap of fluffy beige material. The mineralogical composition of the total clay (<2 μm) fraction was relatively uniform, consisting mainly of smectite, with lesser amounts of illite, the zeolite clinoptilolite, kaolinite, and K-feldspar. The amount of kaolinite was higher at SSW and SSE relative to GBS and G04b. Siliceous sinter occurs at the edges of springs in the northern parts of the GBS system, but the springs are not currently depositing sinter (Anderson 1978). Recent deposition of siderite (FeCO3) has been reported in most springs in the GBS system (Anderson 1978); however, it was not detected by XRD because it was confined to the edges of the springs instead of the sediments that were sampled. Conspicuous sediment-associated biomass was not evident at the time of sampling (e.g. streamers or well-developed microbial mats); however, microscopic analysis revealed a diversity of morphotypes in all four sediments that were sampled.
Aqueous chemistry
More than 60 analytes were measured in the source waters, including major (Table 1) and minor elements (Table S1) and dissolved gases (Table 1). The dominant solutes were Na+ and Cl−, with only minor amounts of HCO3 −, consistent with the model that the source water for these springs is deeply circulating ancient meteoric water; the high Na+ and Cl− content reflects long contact time with basin fill (Anderson 1978). The similar chemical composition of these springs with respect to conservative species is consistent with the model that springs from both geothermal fields are fed by a single geothermal reservoir at depth (Anderson 1978). Minor differences in redox-inactive cations (e.g. Na+/Ca2+ + Mg2+) are likely to be due to differences in reaction progress of alteration reactions, cation exchange during discharge, or composition of reacting rocks.
In contrast, major differences in redox-active species that are potential electron donors or acceptors for microbial respiration varied with water residence time. Springs form the Mud springs geothermal field exhibited a shorter water residence time than those from the Great Boiling Springs geothermal field. SSW, SSE, and GBS formed a series showing a progressive decrease in potential electron donors for chemolithotrophy (Table 1; e.g. CH4, 4.95–0.48 μM; total ammonia, 61–18.3 μM; total sulphide 2.9 to <0.5 μM; H2, 130–7.7 nM). From these data, it is likely that the source waters of these three springs were reduced, similar to SSW. The more oxidized chemistry in the long residence time pools reflects increased oxygen penetration and, most likely, complete nitrification of ammonia that was supplied by the source water. Particularly noteworthy is the roughly equimolar ratio of reduced to oxidized inorganic nitrogen compounds in GBS, which is near the upper temperature limit of growth for the recently identified ammonia oxidizer “Candidatus Nitrosocaldus yellowstonii” (de la Torre et al. 2008) but not as high as the hottest springs from which putative archaeal ammonia monooxygenase large subunit genes (amoA) have been amplified (94°C, Reigstad et al. 2008) or the highest temperature at which nitrification has been demonstrated by 15NO3 − pool dilution experiments (85°C, Reigstad et al. 2008). Whether the lower methane, hydrogen, and sulphide concentrations in SSE and GBS reflect biological oxidation, degassing, or in the case of sulphide, abiotic oxidation, is uncertain.
G04b had high concentrations of ammonia (124 μM) and hydrogen (6.5 μM) during this sampling trip, despite concentrations of volatile hydrocarbons and redox-inactive ions that were similar to GBS. Lower concentrations of ammonia (18–67 μM) and hydrogen (18 nM) have been measured at this spring during other sampling trips, and this spring can be extremely variable in temperature (18–80°C) (Hedlund and Romanek unpublished data; Anderson 1978).
Thermodynamic modelling of potential metabolic reactions
Following the general model used in geobiology (Amend and Shock 2001; Inskeep et al. 2005; Shock et al. 2005), empirical geochemical data (Table 1, Table S1) were used to calculate the overall Gibbs free energy of reaction for 122 known and plausible chemolithotrophic metabolisms in the bulk water of the spring. When the data were normalized to the number of moles of electrons transferred during electron transport, they sorted according to the electron acceptor, consistent with the standard reduction potential of the reduction half reactions (Fig. 2, Table S2). Thus, in all four springs, reactions involving O2 as the oxidant yielded the greatest energy followed by a large decrease in Gibbs free energy to NO3 −, and NO2 − reductions (20–35 kJ/mol e−). Elemental sulphur, if present, would also be a good electron acceptor. Reactions involving elemental sulphur as the electron acceptor could provide 38.7–46.8 kJ/mol e− for reduction to pyrite or 14.7–21.8 kJ/mol e− for reduction to sulphide. Although elemental sulphur was not detected by XRD, abiotic or biological sulphide oxidation could deposit sulphur. The pyrite-forming reactions that were calculated are not known to be used for microbial dissimilatory metabolism, although microbial pyrite formation from FeS and H2S has been observed (Schink 2002) and pyrite has been observed in magnetosomes (Bazylinski et al. 1994). In contrast, sulphur reduction to sulphide is widely distributed among cultivated thermophiles (Huber and Stetter 2006; Huber et al. 2000).
Potential metabolic reactions and calculated chemical affinities using chemical concentrations present in bulk spring water (Table 1). Chemical affinities are expressed in terms of kJ/mole of e− transferred and are normalized per mole of electrons participating in electron transport. Reactions are ordered from the most thermodynamically favorable (left) to the least thermodynamically favorable (right). Data points are coded by electron acceptor. Reactions and chemical affinity values are in Table S2
Reductions of sulphate, carbon dioxide, and the ferric iron-containing minerals magnetite, goethite, and hematite yielded less than 10 kJ/mol e−, which is below the energy yield generally accepted to maintain energy charge (20 kJ/mol e−) (Schink 1997); however, recent work has shown that syntrophs can operate well below that threshold (Jackson and McInerney 2002) and thermophiles are known to respire sulphate, CO2, and a variety of ferric iron-containing minerals (Huber et al. 2000). Furthermore, these reactions may be more favorable in the sediment pore water, which may have higher concentrations of certain electron donors than the bulk water. Thermophiles have also been shown to respire ferric iron in smectite (Kashefi et al. 2008), which is the dominant mineral in the clay fraction of these springs.
In contrast, in all four springs, the differences in energy yield from oxidation of different electron donors for chemolithotrophy were less well ordered. For example, the well known electron donors for aerobes CH4, S0, H2, Fe2+, and sulphide, where detectable, were among the most favorable reactions and were all within 12 kJ/mol e− of each other (91.9–103.5 kJ/mol e− for aerobic respirations). Thus, when normalized to electron flow, free energy yield did not suggest that a single electron-donating metabolism is dominant in the springs, as has been suggested for many Yellowstone springs (Spear et al. 2005). Sulphide was below the detection limit of our methods in GBS and G04b. Since all of these springs derive source water from a single subterranean reservoir (Anderson 1978), it is possible that sulphide was in the source water for GBS and G04b but was oxidized in the subsurface due to the longer residence time of the GBS springs. Other well known aerobic metabolisms such as ammonia oxidation (41.6–43.6 kJ/mol e−) and nitrite oxidation (32.9–34.9 kJ/mol e−) were significantly less exergonic than other aerobic oxidations when the data were normalized this way.
The broad thermodynamic landscape in these springs was similar between the four springs and to that described for Obsidian Pool, including the ordering of metabolisms according to electron acceptor and the two thermodynamic “steps” between aerobic and anaerobic metabolisms and between sulphur reductions and less favorable electron acceptors (Shock et al. 2005). It is noteworthy that between-system variability in chemical affinity was low despite differences in measured substrate and product concentrations as large as three orders of magnitude (Table 1).
Community diversity
Two different 16S rRNA gene libraries, each derived from a PCR using a different reverse primer, were constructed from surface sediment of each spring and sequenced to assess bacterial diversity (~48 16S rDNAs from each library, 298 total non-chimeric sequences). The different primer pairs yielded libraries with significantly different homologous and heterologous sequence coverage curves from G04b (Table S3), justifying the use of multiple primer pairs. Libraries from the other three springs were not significantly different. Well-documented limitations of PCR-based microbial censuses notwithstanding (Reysenbach et al. 1992; von Wintzingerode et al. 1997), bacterial diversity statistics were calculated on the combined datasets from each spring. Individual springs were moderately rich, with 14–16 observed and 16–25 predicted species-level OTUs per spring (Table 2) [97% 16S rRNA gene percent identity (PID)]. At this PID, coverage varied from 63 to 88%, indicating that additional species-level phylotypes would be discovered with additional sampling effort. The springs were almost as rich at the phylum level (<80% 16S rRNA gene PID), with 10–15 observed and 11–20 predicted groups per spring (Table 2). Phylum level coverage varied from 75 to 98%, suggesting that additional sampling would uncover new groups, consistent with high phylum level bacterial diversity observed in Yellowstone hot springs and other geochemically complex environments (Hugenholtz et al. 1998; Ley et al. 2006).
At the species level, bacterial evenness was low, 0.50–0.59, relative to comparable studies in soils and water columns, which range from 0.70 to 0.98 (Table 2) (Dunbar et al. 1999; Liao et al. 2007; Tarlera et al. 1997; Wu et al. 2008; Zhang et al. 2006b). Although we are not aware of evenness calculations reported for Yellowstone springs, the Great Basin hot spring sediments were probably much more even than comparable sediment or streamer communities from Yellowstone National Park, which are typically dominated by the phylum Aquificae (D’Imperio et al. 2008; Dojka et al. 1998; Reysenbach et al. 1994; Spear et al. 2005).
Archaeal 16S rRNA gene libraries were made and analyzed in parallel. Two different 16S rRNA gene libraries were sequenced for GBS and G04b, yet only a single library was analyzed for each of SSW and SSE (~48 16S rDNAs from each library, 276 total non-chimeric sequences). For GBS and G04b, libraries prepared using different reverse primers were not significantly different (Table S3). GBS and G04b had extremely low richness even at the species level, with six observed and predicted OTUs in GBS and five observed and predicted OTUs in G04b (Table 2). In contrast, in SSW and SSE, archaea were as rich as bacteria, with 16–19 observed and 28 predicted species-level groups and 7–12 observed and 7.5–13.5 predicted order- to class-level groups (<80% PID). Even though the number of OTUs was identical to the Chao1 richness estimator in GBS and G04b, the well-documented increase in Chao1 diversity estimate that accompanies increased sampling (Sloan et al. 2007), suggests additional sampling would be needed to saturate the archaeal census. Coverage in SSW and SSE was 68 and 57%, respectively. The archaeal communities were similarly even at the species level as compared with the bacterial communities, 0.46–0.64.
To compare the composition of the four sediment communities, independent phylogenetic trees of all bacteria and archaea were used to make a Unifrac distance matrix, which was used to make a UPGMA tree (Lozupone et al. 2006). In both cases, GBS and G04b grouped to the exclusion of SSW and SSE with >99.9% jackknife support, showing that both the bacterial and archaeal compositions of springs in the GBS geothermal field (GBS and G04b) were significantly different from those in the Mud Hot Springs geothermal field (SSW and SSE) (data not shown). This was evident at the phylum level (Fig. 3). Deinococcus-Thermus and Planctomycetes were only recovered in libraries from GBS and G04b, whereas Thermotogae, Thermodesulfobacteria, Dictyoglomi, Firmicutes, Euryarchaeota, and “Korarchaeota” were only represented in libraries from SSW and SSE.
Bacterial community composition
In total, bacteria represented 36 species-level phylotypes. Twenty-two phylotypes belonged to nine formally named phyla: Aquificae, Thermus-Deinococcus, Thermotogales, Chloroflexi, Firmicutes, Planctomycetes, Dictyoglomi, Firmicutes, and Bacteroidetes. The other 14 phylotypes represented 3 candidate phyla, OP1, OP9, and OP10, or did not affiliate with known phyla (GBS_L1_A05, SSW_L1_C03, GBS_L4_E12, GBS, L1_B05, and SSW_L1_A03) (Figs. 3, 4). Together, sequences representing novel phyla comprised 18–40% of bacterial libraries (Fig. 3). Most were rare in libraries (<10%); however, phylotype SSW_L2_A03, belonging to OP1, was an exception, representing 25% of 16S rDNAs in libraries from SSW. 16S rDNAs representing candidate phylum OP10 were only distantly related to the newly described isolates from thermal soils (Stott et al. 2008).
Maximum likelihood phylogenetic tree of bacteria from this study and close relatives. The tree was constructed using E. coli nucleotide positions 107–1346 (Brosius et al. 1978) in ARB (Ludwig et al. 2004). Node support using neighbor joining and maximum parsimony is shown with closed squares (node supported using all three methods) or open squares (node supported using two of three methods). Sequences present in different springs are shown using letters: dark G for GBS, light G for G04b, dark S for SSW, light S for SSE. Relative abundance of sequences in each library are shown as percentages with pie charts and detailed in Table S4. All OTUs are shown at the 97% identity level as determined using DOTUR (Schloss and Handelsman 2005)
Chloroflexi was the most abundant bacterial phylum represented in libraries from all four springs, yet five of the six identified Chloroflexi 16S rDNAs were phylogenetically novel. Phylotype GBS_L1_A03 was dominant in GBS libraries (38%) yet phylotype SSE_L1_E01, was dominant in libraries from SSE (26%) and SSW (37%). Other deeply branching Chloroflexi 16S rDNAs were present in G04b; however, Thermus was the dominant phylotype in libraries from that spring (22%). 16S rDNAs representing Planctomycetes and Bacteroidetes were also novel.
16S rDNAs from GBS and G04b that did affiliate with known genera were close relatives of Thermus thermophilus, Thermomicrobium roseum, Rhodothermus marinus, and Thermocrinis ruber (Fig. 4). Thermus thermophilus is an aerobic organotroph, although denitrifiers (Ramirez-Arcos et al. 1998), mixotrophic sulphur oxidizers (Skirnisdottir et al. 2001), and chemoorganotrophic iron reducers (Balkwill et al. 2004) are also known within the genus. Thermomicrobium roseum is also a facultatively anaerobic heterotroph (Perry 2006). Rhodothermus marinus is an obligately aerobic chemoorganotroph capable of growth on diverse polysaccharides (Bjornsdottir et al. 2006). Aquificae 16S rDNAs were related to Thermocrinis ruber, which is capable of chemolithotrophic growth on hydrogen, thiosulfate, or elemental sulphur with oxygen as an electron acceptor, or growth on formate (Huber et al. 1998). Aquificae in the genera Thermocrinis, Sulfurihydrogenibium, or Hydrogenobaculum typically dominate sediment and streamer communities from high temperature Yellowstone hot springs (D’Imperio et al. 2008; Dojka et al. 1998; Reysenbach et al. 1994; Spear et al. 2005) as well as sediments and mats from Alvord Hot Spring in the Great Basin (Connon et al. 2008), so their low abundance in libraries described here was surprising, especially in light of the thermodynamic favorability of hydrogen and sulphur oxidation in these springs. It is possible that Thermocrinis and other members of the Aquificae are more prevalent in other parts of these hot spring systems. For example, it is possible that Aquificae may be more prevalent in the subsurface of these springs, where the redox environment might be more conducive to their growth. Hydrogen oxidation by certain Sulfurihydrogenibium strains is inhibited by high oxygen concentrations (Takai et al. 2003) and Aquificales are typically grown under low oxygen tension (<1% O2 by volume)(Eder and Huber 2002).
In SSW and SSE, 16S rDNAs related to known genera affiliated with Thermomicrobium roseum, Rhodothermus marinus, and Thermocrinis ruber, as in GBS and G04b, and also Caloramator fervidus, Thermotoga petrophila, Thermotoga hypogea, Dictyoglomus thermophilum, Geothermobacterium ferrireducens, and Thermodesulfobacterium commune (Fig. 4). Caloramator, Thermotoga, and Dictyoglomus are fermenters capable of hydrolyzing complex polymers (Morris et al. 1998; Tarlera et al. 1997; Vanfossen et al. 2008). Geothermobacterium and Thermodesulfobacterium are facultatively chemolithoautotrophic hydrogen oxidizing iron and sulphate reducers, respectively (Jeanthon et al. 2002; Kashefi et al. 2002).
Archaeal community composition
All archaeal libraries were dominated by Crenarchaeota (Figs. 3, 5). Libraries from long residence time springs, GBS and G04b, were very similar and composed exclusively of Crenarchaeota. In GBS and G04b the dominant phylotype, 58 and 33%, respectively, was represented by phylotype SSE_L4_B03, almost identical to the newly discovered ammonia oxidizer “Candidatus Nitrosocaldus yellowstonii” (de la Torre et al. 2008). The predominance of the putative ammonia oxidizer in GBS and G04b was consistent with the higher NO3 − and NO2 − concentrations in those springs, and suggests that nitrification may provide important electron acceptors to anaerobes in underlying sediment. Phylotype GBS_L3_B06, 9 and 4%, in GBS and G04b, respectively, grouped within the Desulfurococcales (Figs. 3, 5). Cultivated members of the Desulfurococcales oxidize H2 using S0, S2O3 2−, NO2 −, NO3 −, or O2, as electron acceptors when growing autotrophically or ferment or respire S0 chemoorganotrophically (Huber and Stetter 2006; Huber et al. 2000). Other sequences were related to 16S rDNAs from other hot springs, but were distant from cultivated organisms or 16S rDNAs from other habitats.
Maximum likelihood phylogenetic tree of archaea from this study and close relatives. The tree was constructed using E. coli nucleotide positions 100–1,378 (Brosius et al. 1978) in ARB (Ludwig et al. 2004). Node support using neighbor joining and maximum parsimony is shown with closed squares (node supported using all three methods) or open squares (node supported using two of three methods). Sequences present in different springs are shown using letters: dark G for GBS, light G for G04b, dark S for SSW, light S for SSE. Relative abundance of sequences in each library are shown as percentages with pie charts and detailed in Table S4. All OTUs are shown at the 97% identity level as determined using DOTUR (Schloss and Handelsman 2005)
Libraries from the short resident time springs, SSW and SSE, were much richer, and yet were also dominated by Crenarchaeota that branched outside the Thermoprotei (Table 2; Figs. 3, 5). 16S rDNAs related to “Candidatus Nitrosocaldus yellowstonii” were present in SSE but not detected in SSW, which is consistent with the extremely low concentrations of nitrate and nitrite in SSW. It is possible that nitrate respiration capacity is limited by nitrification in SSW, consistent with the increased proportion of plausible fermeters (Thermotogae, Firmicutes, Dictyoglomus, and “Korarchaeota”) and sulphate or iron reducers (Archaeoglobales, Thermodesulfobacteria), as well as the absence of Thermus, many of which can respire nitrate, in libraries from those springs. One OTU was related to Thermofilum, a strict anaerobe that uses elemental sulphur as a terminal electron acceptor (Huber et al. 2006). Three different OTUs represented the Desulfurococcales were most closely related to the polysaccharide and peptide fermenters Thermosphaera (Huber et al. 1995) and Ignisphaera (Niederberger et al. 2006), or were deeply branching within the order.
The SSW archaeal library contained one phylotype that grouped in the “Korarchaeota”. SSW also had one phylotype that branched deeply within the DHVE2 group, which is represented by only one thermoacidophilic representative, Aciduloprofundum boonei, and several environmental sequences obtained from acidic deep-sea hydrothermal vents (Reysenbach et al. 2006; Takai and Sako 1999). SSE_L4_E01 (5% in SSE and 2.5% in SSW) was closely related to Archaeoglobus fulgidus, which couples H2 or organic substrates with the reduction of SO4 2− to S2− (Stetter 1988).
Conclusions
Here we describe the geochemistry, thermodynamics, and microbial communities in four >73°C Great Basin hot springs with a single subterranean source but with different hydrology. The thermodynamic landscape of the four springs were broadly similar to that described for Obsidian Pool (Shock et al. 2005) in that metabolisms were well-sorted according to electron acceptor but less resolved by electron donor. From this, we predict that aerobic metabolisms predominate in the bulk water and surface sediment, although the electron donor(s) cannot be easily predicted based on geochemical data and thermodynamic modelling alone. In sediments below the oxic/anoxic interface (~1 cm) microorganisms are likely to respire electron acceptors in the order defined by the electronegativity of the oxidized/reduced form of the electron donor (Fig. 2; Table S2) although nitrate respiration in short residence time springs may be limited by nitrification. In contrast, the long residence time springs GBS and G04b had highly abundant “Nitrosocaldales” in libraries and higher concentrations of nitrate and nitrite.
Libraries from the four hot spring sediments were unusual in the high proportion of OTUs representing uncultivated phyla, the abundance of novel phyla in the libraries, and the low number of Aquificae 16S rDNAs. Similarly, archaeal libraries contained high proportions of Crenarchaeota unrelated to cultivated organisms. The reason that these springs host many yet-uncultivated microorganisms is unknown, as are the roles of these novel taxa in microbial processes in the springs. Therefore, it was particularly difficult to link 16S rRNA gene phylotypes with plausible metabolisms in these systems. This begs the question of why these springs are so microbiologically unusual and justifies further study of these geothermal systems with the goal of linking these unusual phylotypes with their activities in nature.
An exception to this problem was the recovery of high numbers of 16S rRNA gene sequences that were closely related to “Candidatus Nitrosocaldus yellowstonii” in three of the springs. This implies an important role for nitrification in primary production and is consistent with the abundance of crenarchaeol (Pearson et al. 2004, 2008; Zhang et al. 2006a) and novel archaeal 16S rRNA gene phylotypes in cooler Great Basin springs (Huang et al. 2007).
References
Amend JP, Shock EL (2001) Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiol Rev 25:175–243
Anderson JP (1978) A geochemical study of the southwest part of the Black Rock Desert and its geothermal areas; Washoe, Pershing, and Humboldt Counties, Nevada. Colo Sch Mines Q 73:15–22
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2005) At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol 71:7724–7736
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2006) New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol 72:5734–5741
Balkwill DL, Kieft TL, Tsukuda T, Kostandarithes HM, Onstott TC, Macnaughton S, Bownas J, Fredrickson JK (2004) Identification of iron-reducing Thermus strains as Thermus scotoductus. Extremophiles 8:37–44
Bazylinski DA, Garratt-Reed AJ, Frankel RB (1994) Electron microscopic studies of magnetosomes in magnetotactic bacteria. Microsc Res Tech 27:389–401
Bjornsdottir SH, Blondal T, Hreggvidsson GO, Eggertsson G, Petursdottir S, Hjorleifsdottir S, Thorbjarnardottir SH, Kristjansson JK (2006) Rhodothermus marinus: physiology and molecular biology. Extremophiles 10:1–16
Breitbart M, Wegley L, Leeds S, Schoenfeld T, Rohwer F (2004) Phage community dynamics in hot springs. Appl Environ Microbiol 70:1633–1640
Brosius J, Palmer ML, Kennedy PJ, Noller HF (1978) Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA 75:4801–4805
Connon SA, Koski AK, Neal AL, Wood SA, Magnuson TS (2008) Ecophysiology and geochemistry of microbial arsenic oxidation within a high arsenic, circumneutral hot spring system of the Alvord desert. FEMS Microbiol Ecol 64:117–128
D’Imperio S, Lehr CR, Oduro H, Druschel G, Kuhl M, McDermott TR (2008) The relative importance of H2 and H2S as energy sources for primary production in geothermal springs. Appl Environ Microbiol 74:5802–5808
de la Torre JR, Walker CB, Ingallis AE, Konneke M, Stahl DA (2008) Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol 10:810–818
Dojka MA, Hugenholtz P, Haack SK, Pace NR (1998) Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol 64:3869–3877
Dunbar J, Takala S, Barns SM, Davis JA, Kuske CR (1999) Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl Environ Microbiol 65:1662–1669
Eder W, Huber R (2002) New isolates and physiological properties of the Aquificales and description of Thermocrinis albus sp. nov. Extremophiles 6:309–318
Eder W, Ludwig W, Huber R (1999) Novel 16S rRNA gene sequences retrieved from highly saline brine sediments of Kebrit Deep, Red Sea. Arch Microbiol 172:213–218
Eder W, Jahnke LL, Schmidt M, Huber R (2001) Microbial diversity of the brine-seawater interface of the Kebrit Deep, Red Sea, studied via 16S rRNA gene sequences and cultivation methods. Appl Environ Microbiol 67:3077–3085
Faulds JE, Coolbaugh MF, Vice GS, Edwards ML (2006) Characterizing structural controls on geothermal fields in the northwestern Great Basin: a progress report. GRC Trans 30:69–76
Fournier RO (2005) Geochemistry and dynamics of the Yellowstone National Park Hydrothermal System. In: Inskeep WP, McDemott TR (eds) Geothermal biology and geochemistry in Yellowstone National Park. Montana State University Publications, Bozeman
Hall JR, Mitchell KR, Jackson-Weaver O, Kooser AS, Cron BR, Crossey LJ, Takacs-Vesbach CD (2008) Molecular characterization of the diversity and distribution of a thermal spring microbial community using rRNA and metabolic genes. Appl Environ Microbiol 74:4910–4922
Huang Z, Hedlund BP, Wiegel J, Zhou J, Zhang CL (2007) Molecular phylogeny of uncultivated Crenarchaeota in Great Basin hot springs of moderately elevated temperature. Geomicrobiol J 24:535–542
Huber H, Stetter KO (2006) Desulfurococcales. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds) The prokaryotes. Springer, New York
Huber R, Burggraf S, Mayer T, Barns SM, Rossnagel P, Stetter KO (1995) Isolation of a hyperthermophilic archaeum predicted by in situ RNA analysis. Nature 376:57–58
Huber R, Eder W, Heldwein S, Wanner G, Huber H, Rachel R, Stetter KO (1998) Thermocrinis ruber gen. nov., sp. nov., a pink-filament-forming hyperthermophililc bacterium isolated from Yellowstone National Park. Appl Environ Microbiol 64:3576–3583
Huber R, Huber H, Stetter KO (2000) Towards the ecology of hyperthermophiles: biotopes, new isolation strategies and novel metabolic properties. FEMS Microbiol Rev 24:615–623
Huber T, Faulkner G, Hugenholtz P (2004) Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319
Huber H, Huber R, Stetter KO (2006) Thermoproteales. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds) The prokaryotes. Springer, New York, pp 10–22
Hugenholtz P, Pitulle C, Hershberger KL, Pace NR (1998) Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol 180:366–376
Inskeep WP, Ackerman GG, Taylor WP, Kozubal M, Korf S, Macur RE (2005) On the energetics of chemolithotrophy in nonequilibrium systems: case studies of geothermal springs in Yellowstone National Park. Geobiology 3:297–313
Jackson BE, McInerney MJ (2002) Anaerobic microbial metabolism can proceed close to thermodynamic limits. Nature 415:454–456
Jeanthon C, L’Haridon S, Cueff V, Banta A, Reysenbach AL, Prieur D (2002) Thermodesulfobacterium hydrogeniphilum sp. nov., a thermophilic, chemolithoautotrophic, sulfate-reducing bacterium isolated from a deep-sea hydrothermal vent at Guaymas Basin, and emendation of the genus Thermodesulfobacterium. Int J Syst Evol Microbiol 52:765–772
Johnson JW, Oelkers EH, Helgeson HC (1992) SUPCRT92: a software package for calculating the standard molal thermodynamic properties of minerals, gases, aqueous species, and reactions from 1 to 5,000 bar and 0 to 1,000° C. Comput Geosci 18:899–947
Kashefi K, Holmes DE, Reysenbach AL, Lovley DR (2002) Use of Fe(III) as an electron acceptor to recover previously uncultured hyperthermophiles: isolation and characterization of Geothermobacterium ferrireducens gen. nov., sp. nov. Appl Environ Microbiol 68:1735–1742
Kashefi K, Shelobolina ES, Elliott WC, Lovley DR (2008) Growth of thermophilic and hyperthermophilic Fe(III)-reducing microorganisms on a ferruginous smectite as the sole electron acceptor. Appl Environ Microbiol 74:251–258
Lee MH, Keams JL, Helzer DW, Leiser OP, Ochoa MA, Connon SA, Magnuson TS, Watwood ME (2007) Evaluation of viral and prokaryotic community dynamics in Alvord Desert hot springs, Oregon, USA. Aquatic Microb Ecol 48:19–26
Ley RE, Harris JK, Wilcox J, Spear JR, Miller SR, Bebout BM, Maresca JA, Bryant DA, Sogin ML, Pace NR (2006) Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl Environ Microbiol 72:3685–3695
Liao PC, Huang BH, Huang S (2007) Microbial community composition of the Danshui River estuary of Northern Taiwan and the practicality of the phylogenetic method in microbial barcoding. Microb Ecol 54:497–507
Lozupone C, Hamady M, Knight R (2006) UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371
Morris DD, Gibbs MD, Chin CW, Koh MH, Wong KK, Allison RW, Nelson PJ, Bergquist PL (1998) Cloning of the xynB gene from Dictyoglomus thermophilum Rt46B.1 and action of the gene product on kraft pulp. Appl Environ Microbiol 64:1759–1765
Niederberger TD, Gotz DK, McDonald IR, Ronimus RS, Morgan HW (2006) Ignisphaera aggregans gen. nov., sp. nov., a novel hyperthermophilic crenarchaeote isolated from hot springs in Rotorua and Tokaanu, New Zealand. Int J Syst Evol Microbiol 56:965–971
Pearson A, Huang Z, Ingalls AE, Romanek CS, Wiegel J, Freeman KH, Smittenberg RH, Zhang CL (2004) Nonmarine crenarchaeol in Nevada hot springs. Appl Environ Microbiol 70:5229–5237
Pearson A, Pi Y, Zhao W, Li W, Li Y, Inskeep W, Perevalova A, Romanek C, Li S, Zhang CL (2008) Factors controlling the distribution of archaeal tetraethers in terrestrial hot springs. Appl Environ Microbiol 74:3523–3532
Perry JJ (2006) The genus Thermomicrobium. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (eds) The prokaryotes. Springer, New York, pp 849–853
Ramirez-Arcos S, Fernandez-Herrero LA, Berenguer J (1998) A thermophilic nitrate reductase is responsible for the strain specific anaerobic growth of Thermus thermophilus HB8. Biochim Biophys Acta 1396:215–227
Reigstad LJ, Richter A, Daims H, Urich T, Schwark L, Schleper C (2008) Nitrification in terrestrial hot springs of Iceland and Kamchatka. FEMS Microbiol Ecol 64:167–174
Reysenbach A-L (2001) Class I. Thermoprotei class. nov. In: Boone DR, Castenholz RW (eds) Bergey’s manual of systematic bacteriology. Springer, New York
Reysenbach AL, Giver LJ, Wickham GS, Pace NR (1992) Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol 58:3417–3418
Reysenbach AL, Wickham GS, Pace NR (1994) Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol 60:2113–2119
Reysenbach AL, Liu Y, Banta AB, Beveridge TJ, Kirshtein JD, Schouten S, Tivey MK, Von Damm KL, Voytek MA (2006) A ubiquitous thermoacidophilic archaeon from deep-sea hydrothermal vents. Nature 442:444–447
Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277
Schink B (1997) Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev 61:262–280
Schink B (2002) Synergistic interactions in the microbial world. Antonie Van Leeuwenhoek 81:257–261
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71:1501–1506
Schouten S, Hopmans EC, Baas M, Boumann H, Standfest S, Konneke M, Stahl DA, Sinninghe Damste JS (2008) Intact membrane lipids of “Candidatus Nitrosopumilus maritimus”, a cultivated representative of the cosmopolitan mesophilic group I Crenarchaeota. Appl Environ Microbiol 74:2433–2440
Shock E, Holland M, Meyer-Dombard DR, Amend JP (2005) Geochemical sources of energy for microbial metabolism in hydrothermal ecosystems: Obsidian Pool, Yellowstone National Park. In: Inskeep WP, McDermott TR (eds) Geothermal biology and geochemistry in Yellowstone National Park. Montana State University Publications, Bozeman, pp 95–109
Singleton DR, Furlong MA, Rathburn SL, Whitman WB (2001) Quantitative comparisons of 16S rDNA sequence libraries from environmental samples. Appl Environ Microbiol 67:4373–4376
Skirnisdottir S, Hreggvidsson GO, Holst O, Kristjansson JK (2001) Isolation and characterization of a mixotrophic sulfur-oxidizing Thermus scotoductus. Extremophiles 5:45–51
Sloan WT, Woodcock S, Lunn M, Head IM, Curtis TP (2007) Modeling taxa-abundance distributions in microbial communities using environmental sequence data. Microb Ecol 53:443–455
Spear JR, Walker JJ, McCollom TM, Pace NR (2005) Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc Natl Acad Sci USA 102:2555–2560
Stetter KO (1988) Archaeoglobus fulgidus gen. nov., sp. nov. a new taxon of extremely thermophilic Archaebacteria. Syst Appl Microbiol 10:172–173
Stott MB, Crowe MA, Mountain BW, Smirnova AV, Hou S, Alam M, Dunfield PF (2008) Isolation of novel bacteria, including a candidate division, from geothermal soils in New Zealand. Environ Microbiol 10:2030–2041
Takai K, Sako Y (1999) A molecular view of archaeal diversity in marine and terrestrial hot water environments. FEMS Microbiol Ecol 28:177–188
Takai K, Kobayashi H, Nealson KH, Horikoshi K (2003) Sulfurihydrogenibium subterraneum gen. nov., sp. nov., from a subsurface hot aquifer. Int J Syst Evol Microbiol 53:823–827
Tarlera S, Muxi L, Soubes M, Stams AJ (1997) Caloramator proteoclasticus sp. nov., a new moderately thermophilic anaerobic proteolytic bacterium. Int J Syst Bacteriol 47:651–656
Theisen AA, Harward ME (1962) A paste method for preparation of slides for clay mineral identification by X-ray diffraction. Soil Sci Soc Am Proc 26:90–91
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Vanfossen AL, Lewis DL, Nichols JD, Kelly RM (2008) Polysaccharide degradation and synthesis by extremely thermophilic anaerobes. Ann NY Acad Sci 1125:322–337
von Wintzingerode F, Gobel UB, Stackebrandt E (1997) Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev 21:213–229
Wolery TJ (1992) EQ3/6: software package for geochemical modeling of aqueous systems: package overview and installation guide (version 7.0). Lawrence Livermore National Laboratory Report UCRL-MA-110662 PT I, Livermore, CA
Wu T, Chellemi DO, Graham JH, Martin KJ, Rosskopf EN (2008) Comparison of soil bacterial communities under diverse agricultural land management and crop production practices. Microb Ecol 55:293–310
Zehner RE, Coolbaugh MF, Shevenell L (2006) Regional groundwater geochemical trends in the Great Basin: implications for geothermal exploration. Geotherm Resour Counc Trans 30:117–124
Zhang CL, Pearson A, Li YL, Mills G, Wiegel J (2006a) Thermophilic temperature optimum for crenarchaeol synthesis and its implication for archaeal evolution. Appl Environ Microbiol 72:4419–4422
Zhang R, Jiang J, Gu JD, Li S (2006b) Long term effect of methylparathion contamination on soil microbial community diversity estimated by 16S rRNA gene cloning. Ecotoxicology 15:523–530
Zhang CL, Huang Z, Li Y-L, Romanek CS, Mills G, Wiegel J, Culp R, Noakes J, White DC (2007) Lipid biomarkers and carbon-isotope signatures of bacteria in Nevada hot springs. Geomicrobiol J 24:519–534
Acknowledgments
We are grateful to David and Sandy Jamieson for access to the field site and Joy Hallmark and Nicole Fester for assistance in sample collection. We also thank Jeremy A. Dodsworth for assistance in the editorial process and Natasha Zolotova and Panjai Prapaipong at ASU for help with IC and ICP-MS. Harriet Brady and students from Pyramid Lake Junior and Senior High School were important participants in this research. This work was supported by NSF Grant Number MCB-054865 and start up funds from UNLV to BPH. KCC received fellowship support through NSF Grants 0447416 and 0724226. JBN received fellowship support through NIH grant P20 RR16464. CLZ was supported by NSF Grant Number MCB-0348180. Support to the Nevada Genomics Center also made this work possible through Grant Number P20 RR01646 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by T. Matsunaga.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Costa, K.C., Navarro, J.B., Shock, E.L. et al. Microbiology and geochemistry of great boiling and mud hot springs in the United States Great Basin. Extremophiles 13, 447–459 (2009). https://doi.org/10.1007/s00792-009-0230-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-009-0230-x