Abstract
Objective
The 9_2 carbohydrate-binding module (C2) locates natively at the C-terminus of the GH10 thermophilic xylanase from Thermotoga marimita. When fused to the C-terminus, C2 improved thermostability of a GH11 xylanase (Xyn) from Aspergillus niger. However, a question is whether the C-terminal C2 would have a thermostabilizing effect when fused to the N-terminus of a catalytic module.
Results
A chimeric enzyme, C2-Xyn, was created by step-extension PCR, cloned in pET21a(+), and expressed in E. coli BL21(DE3). The C2-Xyn exhibited a 2 °C higher optimal temperature, a 2.8-fold longer thermostability, and a 4.5-fold higher catalytic efficiency on beechwood xylan than the Xyn. The C2-Xyn exhibited a similar affinity for binding to beechwood xylan and a higher affinity for oat-spelt xylan than Xyn.
Conclusion
C2 is a thermostabilizing carbohydrate-binding module and provides a model of fusion at an enzymatic terminus inconsistent with the modular natural terminal location.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hydrolases play a pivotal role in carbohydrate recyclization and renewable energy production. A hydrolase usually contains a carbohydrate-binding module (CBM) and 69 families of CBMs have been isolated (Carvalho et al. 2015). Xylanase (EC 3.2.1.8) is a hydrolase that breaks down the β-1,4-xylan backbone of a polysaccharide. It is widely used in food, feed, paper and pulp industry (van Gool et al. 2013; Zhao et al. 2013). An Aspergillus niger xylanase (Xyn) is a typical of family GH11 xylanase (GenBank: EU375728) (Chen et al. 2006), which usually contains no CBM. To fulfil biotechnological demands, the thermostability of Xyn needs to be improved because its thermal inactivation half-life (t1/2) is only 21 min at 50 °C. To fuse a thermophilic CBM is a straight-forward rational engineering strategy.
A thermophilic family 9 CBM (CBM9) locates natively at the C-terminus of a multimodular family GH10 xylanase 10A from Thermotoga marimita. The CBM9 was first recognized as a cellulose-binding domain for binding to insoluble microcrystalline cellulose and was later identified as a xylan-binding module (Meissner et al. 2000; Notenboom et al. 2001; Winterhalter et al. 1995). When fused to the C-terminus of Xyn, the CBM9 caused an improvement of 2 °C in enzyme optimal temperature (Topt) (Liu et al. 2011). The CBM9 contains two smaller modules, CBM9_1 (C1) and CBM9_2 (C2). When fused to the C-terminus, both the modules C1 and C2 improved the Xyn catalytic activity, and C2 additionally improved the enzyme’s thermostability (Liu et al. 2012b).
Which terminus to fuse is the first consideration in module fusion. Presently, the terminus is intuitively selected consisting with a CBM natural terminal location (Furtado et al. 2015; Li and Shao 2006; Liu et al. 2011, 2012b; Mai-Gisondi et al. 2015; Ye et al. 2011). A puzzle is whether the C-terminal C2 would have a thermostabilizing effect when fused to the N-terminus of an enzyme. Only when the C2 also improves enzyme thermostability at the N-terminus, can it be regarded as thermostabilizing. A thermostabilizing CBM is more attractive than those only binding substrate (Furtado et al. 2015; Khan et al. 2013; Kittur et al. 2003; Mai-Gisondi et al. 2015; Mangala et al. 2003). To elucidate this puzzle, the C2 is fused to the Xyn N-terminus through a natural 29 amino acid linker peptide selected from the T. marimita xylanase 10A because it had been successfully used in module fusion (Liu et al. 2011, 2012a, b). Besides engineering the Xyn properties and elucidating the C2 functions, the study also provides a model of fusing a CBM at another enzyme terminus inconsistent with its natural terminal location.
Materials and methods
Materials and regents
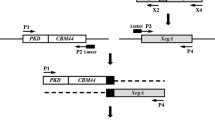
The C2-Xyn was created using the following primers synthesized by Genewiz Inc (Beijing, China): P1 (GAAGGAGATATACATATGATGGTAGCGACAGCAAAAT)/P2 (AGGACCTCAGGCTTGATGAGCCTGAGGTTAC)/P3 (GTAACCTCAGGCTCATCAAGCCTGAGGTCCT)/P4 (GTG GTGGTGCTCGAGAGAGGAGATC). The primers P2 and P3 had a homologous sequence to allow the related PCR products to combine with each other in an overlap extension process. For cloning into pET21a(+) (Novagen), the primers P1 and P4 respectively had restriction sites (shown in italics) digested by NdeI and XhoI (Takara). ExTaq DNA polymerase (Takara) was used to ensure DNA sequences correctly amplified.
Construction of the C2-Xyn
The C2 DNA fragment was amplified in 50 µl containing 1 µM primers P1/P2, 54 ng pET20b-Xyn-C2 template (Liu et al. 2012b), 1 U ExTaq DNA polymerase, 4 µmol of dNTPs, and polymerase buffer. The PCR procedure was: pre-denaturation at 94 °C for 5 min, 25 cycles of denaturation at 94 °C for 1 min, annealing at 64.9 °C for 1 min, and extension at 72 °C for 1 min. Containing the linker peptide sequence, the Xyn DNA fragment was amplified in 50 µl containing 1 µM P3/P4 primers, 49 ng pET20b-Glu-Xyn template (Liu et al. 2012a), 1 U ExTaq DNA polymerase, 4 µmol of dNTPs, and polymerase buffer. The PCR procedure was: pre-denaturation at 94 °C for 5 min, 25 cycles of denaturation at 94 °C for 1 min, annealing at 61.9 °C for 1 min, and extension at 72 °C for 1 min. The fragments C2 and Xyn were purified with a DNA extraction and clean kit (Qiagen).
The C2-Xyn DNA was created in an overlap extension PCR. A 50 µl mixture contained 1 µM primers P1/P4, 49.8 ng C2 and 68.4 ng Xyn fragments, 1 U ExTaq DNA polymerase, 4 µmol dNTPs, and polymerase buffer. The PCR procedure was: five cycles of self-extension by annealing at 68.3 °C for 1 min, and then 22 cycles of annealing at 63.8 °C for 1 min and extension at 72 °C for 1.5 min. The products were purified with a DNA extraction and clean kit. After being digested with NdeI and XhoI, the DNA fragments and pET21a(+) were ligated with T4 DNA ligase at 16 °C overnight. The ligation product was transformed to 200 µl E. coli BL21(DE3) competent cells using a standard procedure. Recombinant plasmid was extracted from positive transformants, and gene accuracy was confirmed by DNA sequencing with an ABI 3730 automated sequencer (Genewiz Inc). Transformant containing pET21a(+)-C2-Xyn was incubated until the cell OD600 reached 0.6. Enzyme expression was induced by 1 µM IPTG at 25 °C for 6 h. Cells were collected and lysed ultrasonically. Enzyme was collected and purified using Co2+-binding resin (Amersham Bioscience), because a 6-His tag was added at the xylanase C-terminus. Active fractions were collected and further purified using Sephadex G-25.
Enzyme properties
Enzyme activities were assayed in parallel with the wild-type Xyn using the DNS method (Liu et al. 2011). Each value was an average of three independent assays. Activities were assayed from pH 2.6 to 4.4 and from 38 to 54 °C on beechwood xylan (Sigma) in phosphate buffer. Enzyme thermostability was determined by assaying residual activities after incubation at 50 °C for 10 min; the t1/2 was calculated using the Arrhenius function (y = A × exp−x/t) (OriginPro 8). Kinetics was assayed under optimal conditions at substrate concentration ranging from 0.625 to 12.5 mg/ml, and the data were fitted to the Hill function: y = Vmax × [S]/(Km + [S]) (OriginPro 8). Kinetics were also assayed on oat spelt xylan to analyze enzyme affinity for insoluble substrate. Protein concentration was assayed by the Bradford method. One unit of enzyme activity was defined as the amount of enzyme that produced 1 µmol xylose after hydrolysis for 1 min under optimal conditions.
Results
Construction of the C2-Xyn
The chimeric C2-Xyn DNA created a band at ~1.2 kb on a 1.4 % agarose gel (Fig. 1), approximately the combined molecular masses of contributing gene fragments C2 (~750 bp) and Xyn (~700 bp). The C2-Xyn DNA was cloned in pET21a(+) and transformed BL21(DE3) E.coli competent cell. Recombinant plasmid pET21a(+)-C2-Xyn was extracted from a positive transformant, and gene accuracy was confirmed by DNA sequencing analysis. Having 403 amino acid residues, the chimeric enzyme C2-Xyn created a band at ~55 kDa (Fig. 1), about two times larger than the 185 residue Xyn (~29 kDa).
Enzyme properties
The C2-Xyn had maximal activity at pH 3.6 and 50 °C (Fig. 2; Table 1). The N-terminal C2 had caused a 0.2 unit shift in the optimal pH (pHopt) towards more acidic conditions. The N-terminal C2 caused an improvement of 2 °C in the optimal temperature (Topt) of the enzyme, similar to the fusion of CBM9 at the C-terminus (Liu et al. 2011). It seemed that the N-terminal C2 covered partly, and therefore protected, the xylanase at a 2 °C higher reaction temperature.
The C2-Xyn Topt and thermostability. Enzyme activities were assayed at temperatures from 38 to 54 °C (left) in phosphate buffer. The C2-Xyn had a 2 °C higher Topt than the Xyn. Residual activities were assayed after incubation at 50 °C for a 10 min interval (right). The data were fitted to the equation: y = A × exp(−x/t) (OriginPro8). Thermal in-activation half-life (t1/2) values were calculated to be 58.7 and 21 min for the C2-Xyn and Xyn, respectively
At 50 °C, the C2-Xyn t1/2 was 58.7 min, 2.8 times longer than that of the Xyn (Fig. 2). The N-terminal C2 improved both Topt and thermostability of the Xyn. In contrast, the C-terminal C2 only improved enzyme thermostability (Liu et al. 2012b). Topt and t1/2 are related but different parameters with the former indicating enzyme adaptivity to a higher temperature and the later, enzyme longevity at a certain temperature.
Enzyme kinetics
Using beechwood xylan as substrate, C2-Xyn exhibited a 155 % maximal catalytic velocity (Vmax) and a similar Km value with the Xyn (Fig. 3; Table 1). The similar Km value indicated that the N-terminal C2 contributed a little to enzyme affinity for the soluble substrate. The ~4.5 fold higher Kcat value showed that the N-terminal C2 greatly improved catalytic efficiency of the xylanase. This effect can be attributed to an enlarged catalytic range and cooperation of the modules C2 and Xyn. The C2-Xyn had a two fold larger size, therefore, a two fold larger catalytic range than the Xyn. In addition, C2 adsorbs with and transfers substrate to the Xyn module. Consistent with the result, all the fusions of CBM9, C2, and C1 at the C-terminus improved activity of the GH11 xylanase (Liu et al. 2011, 2012b). Deletion of the CBM32 indicated that it directly participated in substrate recognition and catalysis (Kimiya et al. 2014). Other CBM fusions also improved catalytic activities of the related enzymes (Furtado et al. 2015; Khan et al. 2013; Kittur et al. 2003; Li et al. 2015; Mai-Gisondi et al. 2015; Mangala et al. 2003; Ye et al. 2011).
Kinetics of the C2-Xyn. Enzyme activities were assayed on beechwood (left) and oat spelt xylan (right) at substrate concentration from 0.625 to 12.5 mg/ml. The data were fitted to the Hill’s function: y = Vmax × x/(km + x) (OriginPro 8). The Vmax and Km values on beechwood xylan were 9.33 and 6.02 µmol/min, 2.89 and 2.74 mg/ml for the enzymes C2-Xyn and Xyn, respectively. The Vmax and Km values on oat spelt xylan were 1.55 and 2.03 µmol/min, and 6.98 and 7.76 mg/ml for the enzyme C2-Xyn, respectively
Using oat spelt xylan as substrate, the C2-Xyn displayed a decreased Km value and a decreased Kcat value (Table 1, Fig. 3). The former and latter value, respectively, indicated that the C2-Xyn had an increased binding affinity and a slightly reduced catalytic efficiency. Perhaps, the increased affinity interfered with enzyme being released from oat spelt xylan. Consistent with the result, a sequential deletion mutants showed that Vmax of the T. maritima xylanase XynA increased in the order of XynA∆C < XynA∆A1C < XynA∆NC (Kleine and Liebl 2006). The N-terminal C2 thus improved enzyme affinity for insoluble substrate.
Intending to infer structural changes of the catalytic module of C2-Xyn, its structure was made using a homologous modeling procedure. The modeled structure displays a typical β-jelly roll of a family GH11 xylanase (Supplementary Fig 1), and is consistent with the assayed properties of C2-Xyn. Alignment analysis shows that the active sites are Glu297 (proton donor) and Glu388 (nucleophile). The thumb region is composed of ten amino acid residues (Asn335Glu336Pro337Ser338Ile339Thr340Gly341Thr342Ser343Thr344). The N-terminal C2 locates near to the substrate-binding residues (Asp255, Trp290, Tyr293, Tyr299, and Gln347), therefore, improves enzyme binding affinity for oat spelt xylan.
Discussion
Both the N- and C-terminal fusions of C2 improved the Xyn thermostability, indicating that the C2 is a thermostabilizing CBM distinct from those binding only to the substrate (Furtado et al. 2015; Khan et al. 2013; Kittur et al. 2003; Mai-Gisondi et al. 2015; Mangala et al. 2003). In addition, the N-terminal C2 improved enzyme Topt by 2 °C. Therefore, the N-terminal C2 is a little better for thermal adaptivity of the enzyme than the C-terminal C2. A slight difference between the N- and C-terminal fusions can be attributed to β jelly-roll structure of the GH11 xylanase and terminal effect of the C2. Probably, a jelly-roll structure folds not as easily as a (β/α)8 structure of GH10 xylanase.
It is no surprise that the C2-Xyn exhibited different affinities for beechwood and oat-spelt xylan. The C2 bound specifically to the reducing ends of amorphous cellulose, crystalline cellulose, and unmodified insoluble fraction of oat-spelt xylan, whilst binding weakly to soluble glucans, xyloglucan, and barley α-glucan (Boraston et al. 2001). Within a solvent-exposed slot sufficient enough to accommodate a disaccharide, a CBM interacts with a carbohydrate ligand through an intricate hydrogen-bonding network mainly involving charged residues (Fisher et al. 2015; Nishijima et al. 2015), as well as stacking interactions of Trp175 and Trp71 (Notenboom et al. 2001).
Modular function can be elucidated by truncation and fusion analysis. Truncation mutants have often been made (Kleine and Liebl 2006; Paloheimo et al. 2007; Wang et al. 2014). Removal of some CBMs affects the thermostabilities of the associated xylanases and, thereby, some CBM22 s were originally described as thermostabilizing domains (Charnock et al. 2000). These modules were later recognized as CBMs (Dias et al. 2004; Sunna et al. 2000), and the thermostabilizing effects of CBM deletions were regarded as lacking discrete linker peptides separating from the catalytic domains (Dias et al. 2004; Sunna et al. 2000). Recently, fusion mutants were more often made (Furtado et al. 2015; Khan et al. 2013; Kittur et al. 2003; Liu et al. 2011, 2012a, b; Mai-Gisondi et al. 2015; Mangala et al. 2003; Tang et al. 2014; Voutilainen et al. 2014; Ye et al. 2011), and fusion seems better for elucidating modular functions. Each module should be separated by a proper linker to provide sufficient space for a module to fold into an intact conformation. Lack of proper linkers might be a reason why transposition of the catalytic modules got unexpected results (An et al. 2005; Hong et al. 2006; Liu et al. 2012a). The natural 22 amino acid linker used in the present study is rich in prolines, glutamines, and valines.
Conclusion
Fusion of the C2 at the N-terminus improved the Topt by 2 °C: 2.8-times in thermostability, and 4.5-times in catalytic efficiency of the GH11 xylanase. Thus, the C2 was confirmed to be a thermostabilizing CBM acting at either terminus of the GH11 xylanase. The bifunctional C2 is useful for engineering enzyme thermostability and activity. A modular function might be elucidated better by fusion rather than deletion analysis.
References
An JM, Kim YK, Lim WJ, Hong SY, An CL, Shin EC, Cho KM, Choi BR, Kang JM, Lee SM, Kim HY (2005) Evaluation of a novel bifunctional xylanase-cellulase constructed by gene fusion. Enzyme Microb Technol 36:989–995
Boraston AB, Creagh AL, Alam MM, Kormos JM, Tomme P, Haynes CA, Warren RA, Kilburn DG (2001) Binding specificity and thermodynamics of a family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A. Biochemistry 40:6240–6247
Carvalho CC, Phan NN, Chen Y, Reilly PJ (2015) Carbohydrate-binding module tribes. Biopolymers 103:203–214
Charnock SJ, Bolam DN, Turkenburg JP, Gilbert HJ, Ferreira LM, Davies GJ, Fontes CM (2000) The X6 “thermostabilizing” domains of xylanases are carbohydrate-binding modules: structure and biochemistry of the Clostridium thermocellum X6b domain. Biochemistry 39:5013–5021
Chen H, Yan X, Liu X, Wang M, Huang H, Jia X, Wang J (2006) Purification and characterization of a novel bifunctional xylanase, XynIII, isolated from Aspergillus niger A-25. J Microb Biotechnol 16:1132–1138
Clarke JH, Davidson K, Gilbert HJ, Fontes C, Hazlewood GP (1996) A modular xylanase from mesophilic Cellulomonas fimi contains the same cellulose-binding and thermostabilizing domains as xylanases from thermophilic bacteria. FEMS Microbiol Lett 139:27–35
Dias FM, Goyal A, Gilbert HJ, Prates J, Ferreira LM, Fontes CM (2004) The N-terminal family 22 carbohydrate-binding module of xylanase 10B of Clostridium themocellum is not a thermostabilizing domain. FEMS Microbiol Lett 238:71–78
Fisher SZ, Schantz LV, Hakansson M, Logan DT, Ohlin M (2015) Neutron crystallographic studies reveal hydrogen bond and water mediated interactions between a carbohydrate-binding module and its bound carbohydrate ligand. Biochemistry 54:6435–6438
Furtado GP, Santos CR, Cordeiro RL, Ribeiro LF, de Moraes L, Damasio A, Polizeli TM, Lourenzoni MR, Murakami MT (2015) Enhanced xyloglucan-specific endo-1,4-glucanase efficiency in an engineered CBM44-XegA chimera. Appl Microbiol Biotechnol 99:1–13
Hong SY, Lee JS, Cho KM, Math RK, Kim YH, Hong SJ, Cho YU, Kim H, Yun HD (2006) Assembling a novel bifunctional cellulase-xylanase from Thermotoga maritima by end-to-end fusion. Biotechnol Lett 28:1857–1862
Khan MI, Sajjad M, Sadaf S, Zafar R, Niazi UH, Akhtar MW (2013) The nature of the carbohydrate binding module determines the catalytic efficiency of xylanase Z of Clostridium thermocellum. J Biotechnol 168:403–408
Kimiya M, Makiko S, Tetsuya K, Kazuo S (2014) Essential role of a family-32 carbohydrate-binding module in substrate recognition by Clostridium thermocellum mannanase CtMan5A. FEBS Lett 588:1726–1730
Kittur FS, Mangala SL, Rus’d AA, Kitaoka M, Tsujibo H, Hayashi K (2003) Fusion of family 2b carbohydrate-binding module increases the catalytic activity of a xylanase from Thermotoga maritima to soluble xylan. FEBS Lett 549:147–151
Kleine J, Liebl W (2006) Comparative characterization of deletion derivatives of the modular xylanase XynA of Thermotoga maritima. Extremophiles 10:373–381
Li X, Shao W (2006) The construction of Thermotoga maritima endoglucanase Cel12B fused with CBD and the characterization of chimeric enzyme. Acta Microbiol Sinica 46:726–729
Li S, Yang X, Bao M, Yu W, Yu W, Han F (2015) Family 13 carbohydrate-binding module of alginate lyase from Agarivorans sp. L11 enhances its catalytic efficiency and thermostability, alters its substrate preference and product distribution. FEMS Microbiol Lett 362:1–10
Liu L, Cheng J, Chen H, Li X, Wang S, Song A, Wang M, Wang B, Shen J (2011) Directed evolution of a mesophilic fungal xylanase by fusion of a thermophilic bacterial carbohydrate-binding module. Process Biochem 46:395–398
Liu L, Wang L, Zhang Z, Guo X, Li X, Chen H (2012a) Domain-swapping of mesophilic xylanase with hyper-thermophilic glucanase. BMC Biotechnol 12:28
Liu L, Zeng L, Wang S, Li X, Song A, Wu K, Chen H (2012b) Activity and thermostability increase of xylanase following transplantation with modules sub-divided from hyper-thermophilic CBM9_1-2. Process Biochem 47:853–857
Mai-Gisondi G, Turunen O, Pastinen O, Pahimanolis N, Master ER (2015) Enhancement of acetyl xylan esterase activity on cellulose acetate through fusion to a family 3 cellulose binding module. Enzyme Microb Technol 79–80:27–33
Mangala SL, Kittur FS, Nishimoto M, Sakka K, Ohmiya K, Kitaoka M, Hayashi K (2003) Fusion of family VI cellulose binding domains to Bacillus halodurans xylanase increases its catalytic activity and substrate-binding capacity to insoluble xylan. J Mol Catal B 21:221–230
Meissner K, Wassenberg D, Liebl W (2000) The thermostabilizing domain of the modular xylanase XynA of Thermotoga maritima represents a novel type of binding domain with affinity for soluble xylan and mixed-linkage β-1,3/β-1, 4-glucan. Mol Microbiol 36:898–912
Nishijima H, Nozaki K, Mizuno M (2015) Extra tyrosine in the carbohydrate-binding module of Irpex lacteus Xyn10B enhances its cellulose-binding ability. Biosci Biotechnol Biochem 79:738–746
Notenboom V, Boraston AB, Kilburn DG, Rose DR (2001) Crystal structures of the family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A in native and ligand-bound forms. Biochemistry 40:6248–6256
Paloheimo M, Mantyla A, Kallio J, Puranen T, Suominen P (2007) Increased production of xylanase by expression of a truncated version of the xyn11a gene from Nonomuraea flexuosa in Trichoderma reesei. Appl Environ Microbiol 73:3215–3224
Sunna A, Gibbs MD, Bergquist PL (2000) The thermostabilizing domain, XynA, of Caldibacillus cellulovorans xylanase is a xylan binding domain. Biochem J 346:583–586
Tang Z, Chen H, Chen L, Liu S, Han X, Wu Q (2014) Improving endoglucanase activity by adding the carbohydrate-binding module from Corticium rolfsii. J Microbiol Biotechnol 24:440–446
van Gool MP, Hinz S, Schols HA, Sinitsyn AP, Gruppen H (2013) Two novel GH11 endo-xylanases from Myceliophthora thermophila C1 act differently toward soluble and insoluble xylans. Enzyme Microb Technol 53:25–32
Voutilainen SP, Nurmi-Rantala S, Penttila M, Koivula A (2014) Engineering chimeric thermostable GH7 cellobiohydrolases in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 98:2991–3001
Wang J, Zeng D, Liu G, Wang S, Yu S (2014) Truncation of a mannanase from Trichoderma harzianum improves its enzymatic properties and expression efficiency in Trichoderma reesei. J Ind Microbiol Biotechnol 41:1–9
Winterhalter C, Heinrich P, Candussio A, Wich G, Liebl W (1995) Identification of a novel cellulose-binding domain within the multidomain 120 kDa xylanase XynA of the hyperthermophilic bacterium Thermotoga maritima. Mol Microbiol 15:431–444
Ye X, Zhu Z, Zhang C, Zhang Y-H (2011) Fusion of a family 9 cellulose-binding module improves catalytic potential of Clostridium thermocellum cellodextrin phosphorylase on insoluble cellulose. Appl Microbiol Biotechnol 92:551–560
Zhao L, Meng K, Bai YG, Shi PJ, Huang HQ, Luo HY, Wang YR, Yang PL, Song W, Yao B (2013) Two family 11 xylanases from Achaetomium sp Xz-8 with high catalytic efficiency and application potentials in the brewing industry. J Agric Food Chem 61:6880–6889
Acknowledgments
This work was financially supported by the Natural Science Foundation of China (30972123 and 31371831). The authors are grateful to Ian Riley for his sincere advice on the paper writing.
Supporting information
Supplementary Fig. 1—Modeled structure of the catalytic domain of C2-Xyn.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, W., Liu, Y., Ye, Y. et al. C-Terminal carbohydrate-binding module 9_2 fused to the N-terminus of GH11 xylanase from Aspergillus niger . Biotechnol Lett 38, 1739–1745 (2016). https://doi.org/10.1007/s10529-016-2149-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-016-2149-5