Abstract
We report here the effect of adding different types of carbohydrate-binding modules (CBM) to a single-module GH7 family cellobiohydrolase Cel7A from a thermophilic fungus Talaromyces emersonii (TeCel7A). Both bacterial and fungal CBMs derived from families 1, 2 and 3, all reported to bind to crystalline cellulose, were used. Chimeric cellobiohydrolases with an additional S–S bridge in the catalytic module of TeCel7A were also made. All the fusion proteins were secreted in active form and in good yields by Saccharomyces cerevisiae. The purified chimeric enzymes bound to cellulose clearly better than the catalytic module alone and demonstrated high thermal stability, having unfolding temperatures (T m) ranging from 72 °C to 77 °C. The highest activity enhancement on microcrystalline cellulose could be gained by a fusion with a bacterial CBM3 derived from Clostridium thermocellum cellulosomal-scaffolding protein CipA. The two CBM3 fusion enzymes tested were more active than the reference enzyme Trichoderma reesei Cel7A both at moderate (45 °C and 55 °C) and at high temperatures (60 °C and 65 °C), the hydrolysis yields being two- to three-fold better at 60 °C, and six- to seven-fold better at 65 °C. The best enzyme variant was also tested on a lignocellulosic feedstock hydrolysis, which demonstrated its potency in biomass hydrolysis even at 70 °C.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cellulase research has recently focused particularly on the depolymerisation of cellulose-containing feedstocks to fermentable sugars for the second-generation bioethanol production. One of the challenges still remaining is the resistance of the lignocellulosic substrates towards enzymatic action. Particularly lignin and the crystalline nature of the substrate restrict access of the enzymes to the polysaccharides, and high amounts of (hemi)cellulases are needed for efficient deconstruction. Cost-effective production of ethanol would clearly benefit from improving the currently used enzyme components, having higher thermal stability and activity on cellulosic substrates.
Most cellulases are composed of several modules, including a catalytic module dedicated to hydrolysis, and one or more non-catalytic modules involved in substrate binding [carbohydrate-binding module (CBM)] (Davies and Henrissat 1995). The distinct modules are often connected via linker sequences of varying length. Both the catalytic modules and the CBMs can be classified into different sequence-based protein families (http://www.cazy.org/): To date, there are over 130 glycoside hydrolase (GH) and over 60 CBM families. Cellulase catalytic modules are found in 13 GH families, and CBMs having the ability to bind to cellulose have been reported in 18 CBM families.
The major cellulase component secreted by the industrial fungal production hosts is the cellobiohydrolase I (CBHI), which is essential for the crystalline cellulose deconstruction. The most studied cellobiohydrolase is that secreted from the mesophilic, filamentous ascomycetes Trichoderma reesei, TrCel7A. It is composed of a GH7 family catalytic module connected to the family-1 CBM via an O-glycosylated linker peptide. The 3D structures of both modules of TrCel7A have been determined, and extensive biochemical characterisation of both the wild-type and mutated versions of the intact enzyme, as well as of the individual modules, has been carried out (Viikari et al. 2012). The active site in the TrCel7A catalytic module is located in a cellulose-binding tunnel, and the main product from the hydrolysis is cellobiose, released from the reducing end of the cellulose chain. This processive cellulose hydrolysis by TrCel7A leads to thinning of the microcrystals (Imai et al. 1998; Igarashi et al. 2009).
Different roles for the cellulase CBMs derived from different families have been suggested, including the increase of local concentration of the intact enzyme on cellulose surface as well as targeting to a specific feature of the substrate, and disruption of crystalline cellulose architecture (Carrard et al. 2000; Fox et al. 2013; McLean et al. 2002; Guillén et al. 2010). The main role for cellulose-binding CBMs seems to be to direct the catalytic module onto the substrate surface, while not having more active role in cellulose deconstruction, as shown most extensively for the family-1 CBM of TrCel7A (Linder et al. 1995; Lehtiö et al. 2003; Igarashi et al. 2006, 2009). It has also been observed that CBM1 family members can enhance the stability of the Cel7 cellulases (Voutilainen et al. 2009; Hall et al. 2011). The recent high-speed AFM studies have for the first time demonstrated in real-time velocity measurements that although the catalytic module of TrCel7A has clearly lower affinity to the crystalline substrate, the sliding velocity is similar to that of the intact, 2-module enzyme, thus further confirming that the actual processive hydrolysis carried out by the TrCel7A catalytic module is not enhanced by the presence of CBM (Igarashi et al. 2009, 2011).
Yeast Saccharomyces cerevisiae has proven a potent host for heterologous expression of cellulases, as a tool for protein engineering but also in consolidated bioprocess (CBP) for the production of fuels and chemicals from lignocellulosic raw materials (van Zyl et al. 2007; Voutilainen et al. 2009, 2010). Heterologous expression of fungal cellobiohydrolases in yeast in fully active form has, however, remained challenging. This has been addressed to many disulphide bridges existing in GH7 cellobiohydrolases and the differences particularly in the N-glycosylation. Our recent study demonstrated that it is nevertheless possible to find CBHs that can be expressed in high yields in active form in S. cerevisiae (Ilmén et al. 2011). Among the 14 different fungal CBHIs tested, the most successful was the Talaromyces emersonii Cel7A (TeCel7A), which is a thermostable enzyme consisting only of the catalytic module. Subsequently, four different fungal CBM1+ linker regions were also tested either as C- or N-terminal fusions to the TeCel7A catalytic module in order to improve the activity on crystalline substrates. These fusions affected to the expression level and Avicel hydrolysis activity, measured directly from the yeast supernatant, to various extents. The best production and most efficient Avicel hydrolysis yield was achieved with a yeast strain expressing chimeric CBHI composed of a CBM1+ linker region from TrCel7A attached to the C-terminus of TeCel7A (Ilmén et al. 2011).
Encouraged from the results obtained with the heterologous expression of TeCel7A in yeast, in the current study, we wanted to explore further the possibility of fusing CBMs derived from different CBM families to the C-terminus of different TeCel7A catalytic modules to create thermostable chimeric CBHIs. We also wanted to carry out characterisation of the purified fusion proteins, particularly in terms of activity and thermostability. As the catalytic module in these chimeric CBHI proteins, we used either the TeCel7A wt or a S–S bridge mutant (SS TeCel7A) containing mutations N54C/P191C that we have shown earlier to increase the thermostability (Voutilainen et al. 2010). The three CBMs were chosen from families of which biochemical and structural data is available of their potency in crystalline cellulose degradation (Boraston et al. 2004). Additionally, earlier studies have suggested that the chosen three CBMs are relatively thermostable and have some differences in their binding affinities and specificities (Carrard et al. 2000; Fox et al. 2013). CBM1 family contains small CBMs found almost exclusively in fungal enzymes (cellulases and hemicellulases), while CBM2 and CBM3 family members are bigger in size and found in a large number of bacterial enzymes including cellulases, chitinases and xylanases. Each CBM was connected to the C-terminal end of the catalytic module using the linker region from TrCel7A, and the enzyme variants were expressed in yeast S. cerevisiae.
Materials and methods
DNA manipulations
Standard DNA techniques were used in the study (Sambrook and Russel 2001). Enzymes for the DNA modifications were purchased from New England Biolabs (Ipswich, MA, USA) and Finnzymes (Espoo, Finland). Sequencing reactions were performed using the Big Dye Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA) and analysed by an ABI Prism 3100 Genetic Analyzer automated DNA sequencer (Applied Biosystems). The Escherichia coli XL1-blue strain (Stratagene, La Jolla, CA, USA) was used as the bacterial cloning host.
Construction of the yeast expression plasmids
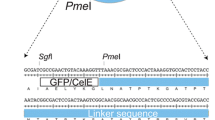
All the Tecel7A gene fusions, including the N-terminal sequence coding for signal peptide (amino acids 1–18), were codon-optimised for expression in S. cerevisiae and synthetised by GenScript (Piscataway, NJ, USA). The first three variants were constructed by combining the Tecel7A wt sequence (Grassick et al. 2004) with CBM1 from TrCel7A (Uniprot P62694), CBM2 from Cellulomonas fimi xylanase 10A, CfXyn10A (Uniprot P07986) and CBM3 from Clostridium thermocellum cellulosomal‐scaffolding protein, CtCipA (Uniprot Q06851). For the codon-optimised nucleotide sequences, see “GenBank accession numbers” section. Each CBM was connected to the C-terminal end of T. emersonii Cel7A using the linker peptide sequence from TrCel7A. The synthetic genes included about 40 bp regions on both 5′ and 3′ end overlapping with the vector to allow cloning of the gene into yeast expression vector by in vivo yeast homologous recombination (Orr-Weaver et al. 1981). The genes were cloned under ENO1 promoter in an expression vector containing 2 μ replicon for autonomous replication and URA3 selection marker (Ilmén et al. 2011) by transforming them together with the linearised plasmid pSVEmpty_ENO (digested with EcoRI and XhoI) into S. cerevisiae strain EGY48 (α, his3, trp1, ura3, 3xLexAoperator-LEU2) (Invitrogen, Foster City, CA, USA). The wt Tecel7A gene was cloned (without codon optimisation) in a similar manner by amplifying the gene from plasmid pSVTE4 (Voutilainen et al. 2010).
Yeast transformations were carried out with a modified LiAc method (Gietz and Woods 2002), and the transformation solution was plated on SC (synthetic complete)-Ura plates (Sherman 2002) containing 2 % (w/v) glucose. After 3 days of growth at 30 °C, transformants were picked and grown in 3 ml of SC-Ura media, buffered to pH 6 with 170 mM succinate and supplemented with 2 % glucose, for 3 days at 30 °C. The transformants were initially tested for cellobiohydrolase production by measuring the cellulase activity of the yeast supernatants on a soluble cellulase substrate, 4-methylumbelliferyl-β-d-lactoside (MULac, Sigma-Aldrich, St. Louis, MO, USA) as described earlier (Voutilainen et al. 2010). Plasmids from the cellobiohydrolase-positive transformants were isolated by first breaking the yeast cells with glass beads (Sigma-Aldrich) and then using Nucleospin (Macherey-Nagel, Düren, Germany) alkaline lysis method for plasmid isolation. Plasmid DNA was then transformed into E. coli XL1-blue strain (Stratagene), isolated and analysed by restriction enzyme digestions and DNA sequencing. The plasmid containing the TeCel7A fusion with the CBM1 from TrCel7A was named pSNR2, and the corresponding purified protein produced is called TeCel7A-CBM1. The TeCel7A fusion with the CBM2 from CfXyn10A is called TeCel7A-CBM2 (plasmid name pSNR4), and the TeCel7A fusion with CBM3 from CtCipA is called TeCel7A-CBM3 (plasmid pSNR5).
Site-specific mutagenesis was performed to create an additional disulphide bridge N54C/P191C to the catalytic module in the TeCel7A-CBM1 and TeCel7A-CBM3 proteins. Cysteine mutations were generated with QuickChange® II XL Site-Directed Mutagenesis Kit (Stratagene) according to manufacturer’s instructions. The primers were synthetised by Sigma-Aldrich and are listed in Table S1. The corresponding variants are called SS TeCel7A-CBM1 (plasmid pSNR8) and SS TeCel7A-CBM3 (plasmid pSNR6).
Protein production and purification
For production of TeCel7A wt or fusion proteins, 1 l (2 × 500 ml in 2-l bottles) of SCD-Ura (pH 6) media was inoculated with an overnight pre-culture and incubated in 30 °C for 3 or 4 days. The supernatants were harvested by removing the cells by centrifugation for 15 min at 4,000 × g. The supernatants were concentrated, and the buffer was exchanged to 50 mM NaAc, pH 5, using Vivaflow 200 (Vivascience, Sigma-Aldrich) ultrafiltration system. Each protein was purified with anion exchange chromatography using DEAE Sepharose FF material (GE Healthcare, Little Chalfont, UK) as described in Voutilainen et al. (2010). The purified TeCel7A wt and the fusion proteins were deglycosylated by endoglycosidase F1 (EndoF1, Sigma-Aldrich) treatment as described in Voutilainen et al. (2010). The production and purification of the TrCel7A was done as described earlier (Suurnäkki et al. 2000).
Characterisation of the purified proteins
Protein concentrations were calculated from the measured A280 value using the theoretical extinction coefficients, which were calculated on the ExPASy Server from the raw sequences (Gasteiger et al. 2003). Circular dichroism (CD) spectra were recorded from 240 to 190 nm using a 1-mm cell and a bandwidth of 1 nm with Chirascan CD spectrophotometer (Applied Photophysics, Leatherhead, UK). The measurements were performed in 10 mM NaAc, pH 5.0, using a protein concentration of 3 μM. The measurements were first done at 20 °C to record the spectra of the folded proteins. The CD spectra of the unfolded proteins were recorded at 80 °C, after which the refolding of the enzyme was studied by cooling the solution to 20 °C and recording the spectrum again. In addition, the unfolding curves were measured at 202 nm using the temperature ramping mode with a gradient of 2 °C/min until a temperature of 90 °C was reached.
Enzyme kinetics and inhibition studies at different temperatures
Kinetic constants (K m, k cat) and cellobiose inhibition constant (K i) for the TeCel7A wt and SS TeCel7A-CBM1 were determined using 2-chloro-4-nitrophenyl-β-d-lactoside (CNPLac) as substrate in different temperatures (22 °C, 45 °C and 60 °C) in 50-mM sodium phosphate buffer, pH 5.7. In each temperature, six different cellobiose concentrations (0–500 μM) were used. Ten different substrate concentrations (30–5,000 μM) were used in 22 °C and 45 °C, while the enzyme concentration was invariably 1.0 μM. The change of absorbance at 405 nm was measured continuously using Varioscan spectrofluorometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). The assays in the higher temperature (60 °C) were conducted in Eppendorf Thermomixer using eight different substrate concentrations (30–5,000 μM) and 0.5 μM enzyme. Linear initial reaction rates were determined at each substrate/inhibitor concentration over 5 min. Reactions were stopped at each time point (1, 2, 3, 4 and 5 min) by adding 0.5 M Na2CO3, and the absorbance was measured at 405 nm. Standard curves were prepared from 2-chloro-4-nitrophenyl (CNP) (0–200 μM). The K m and k cat constants were calculated by fitting the initial rate data to the Michaelis–Menten equation using the programme of Origin 6.0 (Microcal, GE Healthcare). Lineweaver–Burk plots and Hanes plots were used to determine the inhibition type. Lineweaver–Burk replot of their slopes against cellobiose concentration were used to estimate the inhibition constants (K i).
The specific activities of the TeCel7A wt and mutant proteins were measured in ambient temperature (22 °C) in 50-mM NaAc buffer (pH 5.0) basically as described in Voutilainen et al. (2007) using 2 mM MULac and 0.05 μM enzyme concentration. The results were calculated from a standard curve from 0 to 10 μM MU (4-methylumbelliferone, Sigma-Aldrich). The measurements were performed in duplicate.
Activity on insoluble substrates
The microcrystalline cellulose (Avicel PH105, FMC Biopolymer, Mechanicsburg, PA, USA) hydrolysis assays were performed at 45–65 °C by following the reactions up to 48 h as outlined earlier (Voutilainen et al. 2007). Reaction mixtures contained 10 mg/ml of Avicel and 1.4 μM of enzyme in 50-mM NaAc, pH 5.0, and the experiments were performed as duplicates. All the reactions were supplemented with 500 nkat/g Thermoascus aurantiacus β-glucosidase (received from Roal Oy). The formation of soluble reducing sugars was determined by the para-hydroxybenzoic acid hydrazide (PAHBAH) method (Lever 1972) using a glucose standard curve (50 to 800 μM cellobiose). Avicel hydrolysis was also conducted in high temperatures (60 °C, 70 °C and 75 °C) similarly as described above, except that no β-glucosidase was used, and the reducing sugars were measured with PAHBAH reagent against cellobiose standards. Hydrolysis of 1 % steam pre-treated Arundo donax, Giant cane (obtained from Chemtex, Italy), by the SS TeCel7A-CBM3 was followed at 70 °C for 48 h in pH 5 using the same enzyme concentration as for the Avicel hydrolysis tests. The soluble reducing sugars were detected with PAHBAH reagent using a cellobiose standard curve.
Adsorption on Avicel
Binding of TeCel7A variants to 1 % Avicel PH105 in 50 mM NaAc, pH 5, at 45 °C and 60 °C was measured under similar conditions as to those used for hydrolysis. The Avicel suspension containing 1.4 μM enzyme was shaken for 15, 30 and 90 min, and then filtered through Millex GV13 0.22-μm membranes to terminate the reaction. The protein amount was measured with spectrofluorometer (ex. 280 nm, em. 345 nm) before and after the binding. The percentage of the adsorbed enzyme was calculated from the initial protein concentration. The measurements were done as duplicates.
GenBank accession numbers
The nucleotide sequences for the TeCel7A wt and all codon-optimised variants used in this study can be found in the GenBank with the following accession numbers: AAL33603.2 for TeCel7A wt, KF170892 for TeCel7A-CBM1, KF170893 for TeCel7A-CBM2, KF170894 for SSTeCel7A-CBM1, KF170895 for TeCel7A-CBM3 and KF170896 for SSTeCel7A-CBM3.
Results
Design of the CBM fusions
The TeCel7A chimeric enzymes were designed to attach different CBMs to the C-terminal end of the catalytic module. The first fusion partner was a family-1 CBM (composed of 36 amino acids, see Fig. 1a) from TrCel7A. In addition, two different CBMs from bacterial origin were used: family-2 CBM from C. fimi xylanase 10A, CfXyn10A (110 amino acids, Fig. 1b) and family-3 CBM from C. thermocellum cellulosomal‐scaffolding protein, CtCipA (159 amino acids, Fig. 1c). In all cases, the linker peptide (27 amino acids) from TrCel7A was used to connect the CBM to the catalytic module of TeCel7A. In the constructed fusion proteins, the native TeCel7A (437 amino acids) ended at I430, and the linker region started from G427 in TrCel7A. Connecting the linker peptide sequence in this manner resulted to the elimination of N-glycosylation site N431 in the TeCel7A catalytic module. We also used one of the cysteine mutants (N54C/P191C) shown earlier to form a disulphide bond, and to improve both the thermostability and activity of TeCel7A wt (Voutilainen et al. 2010). These mutations create an additional S–S bridge between the adjacent loops1 and 3 which participate in forming the active site tunnel of TeCel7A. The variants are called as SS TeCel7A-CBM1 and SS TeCel7A-CBM3, respectively.
3D structures of the three CBMs used in this study. The aromatic and charged residues forming the cellulose-binding face are marked in red. The asparagine residues of the putative N-glycosylation sites are marked in orange. (a) CBM1 from T. reesei Cel7A; PDB code 2CBH; (b) CBM2 from C. fimi Xyn10A (1EXG); (c) CBM3 from C. thermocellum cellulosomal-scaffolding protein (1NBC), which contains a tightly bound Ca2+
Production and purification of the TeCel7A wt and variants
The TeCel7A wt and the five variants were all expressed in S. cerevisiae under a strong, constitutive ENO1 promoter, the culture supernatants were concentrated and the proteins were purified with an anion exchange column. The TeCel7A-containing fractions were pooled to two pools according to the amount of N-glycosylation detected on SDS-PAGE. Endo-β-N-acetylglucosaminidase F1 (EndoF1) treatment was used to verify that the higher molecular weight forms are due to N-glycosylation (data not shown). For all the characterisation work performed with the purified proteins, the less glycosylated pools were used (Fig. 2).
Enzyme kinetics on soluble substrates
The measured MULac activities of all five TeCel7A-CBM variants are similar, or possibly slightly improved when compared to the TeCel7A wt (Table 1), and we concluded that none of the mutations (fusions) affected to the overall fold of the catalytic module. The kinetic constants (K m, k cat) and cellobiose inhibition (K i) for the TeCel7A wt and SS TeCel7A-CBM1 were then determined on CNPLac at different temperatures (Table 2). As expected, both the K m and k cat constants increased when temperature was raised, while the product inhibition by cellobiose decreased. The cellobiose was a competitive inhibitor, as also reported earlier for TeCel7A wt (Tuohy et al. 2002). The level of TeCel7A product inhibition was moderate (K i = 95 μM at 22 °C) when compared to the inhibition constants published for other family GH7 cellobiohydrolases (Van Tilbeurgh and Claeyssens 1985; Voutilainen et al. 2008); e.g. TrCel7A, which was used as a reference enzyme in the Avicel hydrolysis studies (see below), has relatively high cellobiose inhibition (K i = 20 μM at 22 °C). Concerning the mutant SS TeCel7A-CBM1, the additional S–S bridge did not have any major effect to the cellobiose inhibition constant, whereas the activity (on CNPLac) seemed to be slightly improved.
The CD measurements
The overall fold and thermostability of the TeCel7A wt and the five TeCel7A-CBM fusion proteins were characterised by CD spectroscopy. All the different CBM fusions and the wt (i.e. the catalytic module) had identical spectra at 20 °C (Fig. 3a) indicating a typical shape of a β-sheet secondary structure. However, the TeCel7A-CBM3 fusion showed an unexpected shape of the thermally unfolded protein at 80 °C (Fig. 3b), which clearly differs from the other proteins measured here, or studied previously (Boer et al. 2000; Voutilainen et al. 2009). Our previous studies have additionally shown that GH7 cellobiohydrolases can typically refold when cooled after heating (Boer et al. 2000; Voutilainen et al. 2008, 2009), as was also detected here with the TeCel7A wt and the fusions with CBM1 and CBM2. However, neither one of the two CBM3 fusion proteins was refolding when cooled down after the heating step. To confirm this, the MULac activities of the heat-treated and cooled samples were compared to the activities before the heat treatment. The TeCel7A wt and the CBM1 and CBM2 fusion enzymes regain most (≥ 60 %) of their activity after cooling contrary to the two TeCel7A-CBM3 mutants, which show basically no residual activity.
CD spectra and the unfolding measurements of the TeCel7A wt and CBM fusion proteins measured in 10 mM NaAc, pH 5. The CD spectra of the folded protein in 20 °C (solid line), unfolded protein in 80 °C (dashed line), and refolded protein in the sample cooled quickly down to 20 °C after heating (dotted line) are shown for (a) TeCel7A; and (b) TeCel7A‐CBM3. (c) Temperature‐induced unfolding was measured at 202 nm for TeCel7A wt and five different variants using the temperature scan mode with a gradient of 2 °C/min until a temperature of 90 °C was reached. The unfolding temperatures (Tm) were calculated from the raw data
Secondly, the thermal stability of the TeCel7A wt and variants was studied by measuring their temperature induced unfolding at pH 5.0 by monitoring the CD at 202 nm (Fig. 3c). The unfolding temperatures (T m) were estimated from the unfolding curves by taking the inflection point of the curve. The T m value for the TeCel7A wt is 73 °C, which is about 8 °C higher than that for the mesophilic TrCel7A (Table 1). All the five TeCel7A mutants have also similar or even higher T m values. The only mutant that looks slightly worse than the wt is TeCel7A-CBM2 (Fig. 3c). The additional S–S bridge in the catalytic module seems to improve the unfolding temperature by 1.5–2 °C, irrespective of the CBM fusion partner. This is in accordance with our earlier CD studies (Voutilainen et al. 2010). The highest T m of 77 °C was measured for the SS TeCel7A-CBM3 variant.
Adsorption to Avicel
The ability of the TeCel7A wt and five mutant enzymes to bind onto the microcrystalline substrate was measured at two different temperatures (45 °C and 60 °C) after three different incubation times (15, 30 and 90 min), using similar conditions as in the Avicel hydrolysis (see below). The amount of the bound protein seemed in each case to remain constant throughout the measured time period (data not shown). The TeCel7A wt (i.e. without any CBM) adsorbed to Avicel more weakly than any of the CBM fusion proteins (Fig. 4), suggesting that all three CBMs had been correctly folded. The most efficient binding was exhibited by the CBM3 fusions and the worst by the CBM2 fusion. The results also show that the temperature change from 45 °C to 60 °C brings relatively small changes to the binding affinities, and the additional S–S bridge in the catalytic module does not seem to affect to the binding of the chimeric enzymes.
Adsorption of TeCel7A wt and the four CBM fusion enzymes on microcrystalline cellulose (1 % Avicel) was measured using 1.4 μM enzyme at two different temperatures [45 °C (light grey) and 60 °C (grey)]. The protein concentrations were measured with fluorometer (ex. 280 nm, em. 345 nm). In each case, duplicate samples were measured after 15, 30 and 90 min incubation; the results are averaged from those six measurements
Cellulose hydrolysis
Since the best binding was obtained with the CBM3 fusions, and the CBM1 and CBM2 fusions seemed to be similar to each other, we chose for the following hydrolysis studies three CBM1 and CBM3 variants (see also “Discussion” section). The Avicel hydrolysis efficiency of TeCel7A wt and the three fusion proteins was measured in the presence of a thermostable Ta β-glucosidase at four different temperatures (45 °C, 55 °C, 60 °C and 65 °C) using TrCel7A as a reference enzyme (Fig. 5 and Fig. S1 in the Supplementary Material). Hydrolytic efficiency of the single-module TeCel7A (i.e. wt) was clearly improved by the presence of CBM, CBM3 fusions being more efficient than the CBM1 fusion at all four temperatures (Fig. 5). The TeCel7A wt and all three variants showed the highest hydrolytic activity at 60 °C, and the activity at 65 °C was almost as high, whereas the reference enzyme TrCel7A (i.e. the two-module version having the CBM1) showed the highest activity at 55 °C, after which the activity declined rapidly (Fig. 5 and Fig. S1 in the Supplementary Material). Both CBM3 fusions were more active than the TrCel7A, the hydrolysis efficiency being two- to three-fold better at 60 °C and six- to seven-fold better at 65 °C (Fig. 5). At these higher temperatures, also the CBM1 fusion was clearly more effective than the TrCel7A. It should be noted that all the hydrolysis experiments were conducted in the presence of a thermostable β-glucosidase to avoid product inhibition by cellobiose. However, even though the Ta β-glucosidase has been reported to have temperature optima at 65 °C, and to retain most of its activity even at 70 °C in short-term (60 min) incubation (Viikari et al. 2007), the long-term temperature stability has not been studied. Thus, the somewhat reduced activity of the TeCel7A variants at 65 °C can be at least to some extent due to thermal inactivation of the β-glucosidase.
Hydrolysis of microcrystalline cellulose (Avicel) by TeCel7A wt and the three variants in the presence of β-glucosidase (500 nkat/g) at different temperatures (at 45 °C, 55 °C, 60 °C and 65 °C). TrCel7A was used as a reference enzyme. The hydrolysis of 1 % Avicel with 1.4 μM enzyme was followed in each case for 48 h, taking samples at three time points, and is shown here after 24 h incubation. Soluble reducing sugars released from Avicel were measured with the PAHBAH reagent using glucose as a standard and calculated as micromolar glucose released. Solubilisation (w/w) was calculated as the mass of measured soluble glucose divided by the initial mass of Avicel (calculated as glucose). Error bars are showing the standard deviation over duplicate samples
In the next hydrolysis experiment, the Avicel activity of the three TeCel7A variants was measured at elevated temperatures (60 °C, 70 °C and 75 °C) without the added β-glucosidase (Fig. 6a and Fig. S2 in the Supplementary Material). Under these conditions, the cellobiose concentration can reach up to 3–4 mM during the course of hydrolysis, while the measured cellobiose inhibition constant for TeCel7A is around 0.2 mM (or slightly higher; Table 2). Comparison of Fig. 6a and Fig. 5 (panels at 60 °C) for the effect of added β-glucosidase shows that the accumulation of cellobiose is clearly lowering the hydrolytic activity of all three TeCel7A variants. Similar inhibiting effect was not detected with the TeCel7A wt or TrCel7A, presumably due to the low overall activity (i.e. cellobiose production) of these enzymes. The most active enzyme at 60 °C was TeCel7A-CBM3, and at 70 °C, the SS TeCel7A-CBM3 (Fig. 6a and Fig. S2 in the Supplementary Material). At 75 °C, all three TeCel7A variants showed only minor activity and TrCel7A no activity. Overall, the Avicel hydrolysis results seem to correlate with the measured thermostability (Fig. 3c and Table 1) as well as binding data (Fig. 4). As a final step, hydrolysis of a potential lignocellulosic feedstock, pre-treated A. donax (giant cane), was performed with the best mutant, SS TeCel7A-CBM3, to demonstrate that this variant could hydrolyse the substrate even at 70 °C (Fig. 6b).
(a) Hydrolysis of microcrystalline cellulose (1 % Avicel) by TeCel7A wt and the three variants in pH 5, at 60 °C, 70 °C and 75 °C. TrCel7A was used as a control. Avicel hydrolysis was in each case followed for 48 h, taking samples at three time points and using 1.4 μM enzyme. Soluble reducing sugars after 24 h are shown in the figure. Solubilisation (w/w) was calculated as the mass of measured soluble cellobiose divided by the initial mass of Avicel (calculated as cellobiose). Error bars are showing the standard deviation over duplicate samples. (b) Degradation of Arundo donax (1 %) by the purified SSTeCel7A-CBM3 enzyme (1.4 μM) in pH 5.0 at 70 °C. Soluble reducing sugars were measured at time points 4, 6, 24 and 48 h with the PAHBAH reagent using cellobiose as a standard. Solubilisation (w/w) is calculated as percentage of the theoretical maximum monosaccharide yield from 10 mg/ml A. donax. Error bars are showing the standard deviation over three samples
Discussion
Various CBMs have been used as an affinity tag for protein purification as well as making chimeric enzymes (Levy and Shoseyov 2002; Boraston et al. 2004). The idea has been to use a particular CBM to target the enzyme towards specific substrates known to be the binding ligand for the CBM. Studies by us and others have shown that chimeric endoglucanases, expressed in E. coli and created by combining different CBMs to a catalytic module, can lead to improved activities (Carrard et al. 2000; Kim et al. 2010). In addition, chimeric fungal cellobiohydrolases having family-1 CBMs have been created (Voutilainen et al. 2008, 2009; Ilmén et al. 2011). However, very few, if any, articles exist on expressing fungal cellobiohydrolase fusions with bacterial CBMs. This is due to the difficulties encountered frequently in heterologous expression. On one hand, fungal GH7 cellobiohydrolases cannot be expressed in a bacterial host, and on the other hand, expression of bacterial genes in eukaryotic hosts sets some limitations due to differences in glycosylation, which can interfere particularly with the ability to bind to cellulose. Furthermore, heterologous expression of native fungal GH7 cellobiohydrolases even in another fungal host, such as yeast, can be challenging as shown for example in our previous studies (Ilmén et al. 2011; Boer et al. 2000; Voutilainen et al. 2007). Here, we could successfully express the TeCel7A wt and all the five CBM fusion enzymes in active form in S. cerevisiae. This led to chimeric CBHIs having some of the highest thermostabilities reported for GH7 cellobiohydrolases. Our previous study had shown that the TeCel7A wt is, for reasons that still remain partially unknown, particularly suitable for heterologous expression in S. cerevisiae (Ilmén et al. 2011). Our current work further suggests that the N-terminal catalytic module of TeCel7A can also drive the expression of chimeric enzymes having, besides fungal, also bacterial CBMs fused at the C-terminus.
Despite of the vastness of the gene sequences coding for different types of cellulose-binding CBMs, there is relatively little biochemical characterisation data published on them. In addition, the binding-site preference of a particular CBM cannot be easily identified due to methodological constraints and the heterogeneity of the cellulosic materials. Concerning the currently existing 18 CBM families where cellulose-binding affinity has been reported, some of these CBMs only recognise amorphous cellulose while others prefer crystalline cellulose. This property seems not to be dictated by the CBM family (i.e. protein fold) alone as e.g. different family-2 CBM members can have different types of binding specificities (Boraston et al. 2004). We chose as the CBM fusion partner three candidates from which both biochemical and structural data is available (Fig. 1) suggesting that they are thermostable and bind particularly to crystalline cellulose. All three CBMs have a typical type A binding site topography (Boraston et al. 2004) containing a flat hydrophobic binding face formed by aromatic amino acid residues, which have been shown to be important for the interaction with the more hydrophobic surfaces of the cellulose crystals (Tormo et al. 1996; McLean et al. 2000; Reinikainen et al. 1992; Linder et al. 1995; Igarashi et al. 2011). Despite of the similar planar binding site topography recognising crystalline cellulose, the chosen three CBMs have also been reported to have differences in their binding properties (Carrard et al. 2000; McLean et al. 2002; Fox et al. 2013). The binding affinity of the CBM2 of CfXyn10A has been estimated to be roughly two-fold higher to that of CBM1 from TrCel7A (Tomme et al. 1995). Additionally, this CBM2 has been shown to release small particles from cotton and bind besides cellulose, α-chitin, unlike the CBM1 from TrCel7A (Tomme et al. 1995). On the other hand, the CBM3 of CtCipA has been shown to enhance the activity of crystalline substrates more than the CBM1 of TrCel7A, or CBM2 of CfXyn10A, when linked to a bacterial endoglucanase (Carrard et al. 2000). This latter result apparently reflects the capacity of CBM3 (of CtCipA) to recognise also some less organised binding sites on crystalline cellulose preparates when compared to the CBM2 or CBM1 (McLean et al. 2002; Fox et al. 2013).
The fungal CBM1 (36 aa) from TrCel7A has a knottin-like tertiary structure (Cheek et al. 2006), which is stabilised by two S–S bridges and is assumed to be a stable fold. We and others have notices that the CBM1 family members may additionally stabilise the catalytic module (Voutilainen et al. 2009; Hall et al. 2011). It can be speculated that also here both the selected CBM1 and CBM3 modules stabilised the TeCel7A catalytic module leading to slightly higher T m values (Table 1). The bacterial CBM2 (110 aa) from CfXyn10A has a β-sandwich fold, stabilised by one S–S bridge that connects the cysteine residues near N-terminus and C-terminus of the module (Xu et al. 1995). The T m of the intact, two-module enzyme CfXyn10A has been determined to be 64 °C in pH 7 (Nikolova et al. 1997). In the present study, it seemed that the CBM2 could refold after heating to 80 °C, or at least the CBM did not interfere with the folding of the TeCel7A catalytic module as identical CD spectra could be recorded to the refolded and native TeCel7A-CBM2 proteins. The CBM3 (159 aa) from the CtCipA protein has a similar β-sandwich fold as CBM2 but having only one single cysteine residue. The structure contains additionally a tight binding Ca2+ ion at the side of the cellulose-binding surface (Tormo et al. 1996; Fig. 1c). The optimum temperature for C. thermocellum growth is 60 °C, and the different cellulases have been reported to have temperature optimum between 60 °C and 70 °C (Demain et al. 2005). In our CD studies, the two TeCel7A-CBM3 variants had the highest T m values of 75 °C and 77 °C (Table 1). The CBM3 fusion proteins could not be refolded after the heat treatment, unlike the TeCel7A catalytic module or the other CBM fusion proteins, and we conclude that this unexpected behaviour was due to the CBM3 module. We speculate that the lack of any disulphide bridges and possibly the presence of a single cysteine residue in the CBM3 caused the problems in refolding.
We chose to express the codon-optimised TeCel7A wt and all the five variants in the eukaryotic host S. cerevisiae, as this would allow fast generation and purification of the enzymes, and as we also had earlier experience of expressing TeCel7A wt successfully in S. cerevisiae (Voutilainen et al. 2010; Ilmén et al. 2011). Heterologous expression of the fusion proteins in S. cerevisiae resulted in each case in good yields of purified active protein (10–20 mg/l of yeast culture supernatant). The specific activities of the different yeast produced CBM fusions on soluble substrate were similar to the TeCel7A wt (containing only the catalytic module) and also similar to the previously published MULac activity values (Voutilainen et al. 2010). The correct folding of the catalytic module was also verified by CD spectroscopy. Binding studies (Fig. 4) further suggest that also the CBM in the chimeric enzymes was properly folded. S. cerevisiae is known to overglycosylate fungal cellulases through N-glycosylation sites, as was also detected here. The TeCel7A wt mature sequence contains two putative N-glycosylation sites (N267 and N431), both occupied by N-acetylglucosamine (GlcNAc) residues in the 3D structure (Grassick et al. 2004). All the CBM fusion proteins in this study were designed so that the second N-glycosylation site (N431) at the end of the catalytic module was eliminated by the linker junction sequence from T. reesei Cel7A to lower the possible overglycosylation products. Moreover, only fractions containing less overglycosylated versions of the proteins were pooled after the column chromatography and used in the characterisation work (Fig. 2).
Besides the catalytic module, also the bacterial CBM2 and CBM3 sequences used in this study contain putative N-glycosylation sites. The CBM2 contains five putative N-glycosylation sites, of which three have shown to be glycosylated by Pichia pastoris and hinder the binding to cellulose (Boraston et al. 2001). These N-glycosylation sites are also located in proximity of the carbohydrate-binding face (Fig. 1b). Furthermore, the CBM2 has been found to be O-glycosylated, although the O-glycans apparently did not affect the binding (Boraston et al. 2003). Here, the TeCel7A-CBM2 variant was detected to bind well on Avicel, reaching almost similar quantity as the CBM1 fusion. However, as the binding of the TeCel7A-CBM2 variant seemed somewhat lower than would have been expected based on previous studies with E. coli-expressed CBMs (Tomme et al. 1995; Carrard et al. 2000) and it looked very similar to that of the CBM1 fusion protein, the Avicel hydrolysis tests were carried out without the CBM2 fusion variant. The bacterial CBM3 used here contains two putative N-glycosylation sites, but these are located on the sides of the cellulose-binding face and not expected to affect the adsorption on the cellulose. Comparison of P. pastoris-expressed CBM3 to the E. coli-expressed CBM3 has also suggested that the N-glycans do not hinder the binding on cellulose (Wan et al. 2011). Our binding data (Fig. 4) showed the highest binding for the CBM3 fusions, thus further supporting this notion.
Our results demonstrate that the activity of the fungal TeCel7 cellobiohydrolase on crystalline cellulose can be improved by a fusion of either a fungal or a bacterial CBM having the planar binding site topography. Furthermore, all the TeCel7A-CBM fusions tested were more active at temperatures above 55 °C than the well-studied TrCel7A. It appears that the improved action of TeCel7A-CBM fusions was due to a clearly higher thermostability (up to 12 °C) and also due to higher binding to crystalline cellulose. The engineered S–S bridge in the TeCel7A catalytic module improved the activity at higher temperatures by stabilising the fold, and possibly also by improving the specific activity of the enzyme. The best overall activity enhancement could be gained by a fusion with a bacterial CBM3, having also the highest affinity towards the crystalline substrate. Even though the CBM3 seemed to interfere with the refolding of the TeCel7A catalytic module after heat treatment at 80 °C, it was the most efficient CBM fusion partner in high temperature hydrolysis at 60–70 °C. Overall, our studies show that modular shuffling of cellobiohydrolases with fungal or bacterial CBMs could be a useful approach to reach optimum enzymes for application purposes. The TeCel7A catalytic module seems to offer a particularly good N-terminal fusion partner for this type of expression and protein engineering work in yeast S. cerevisiae.
References
Boer H, Teeri TT, Koivula A (2000) Characterization of Trichoderma reesei cellobiohydrolase Cel7A secreted from Pichia pastoris using two different promoters. Biotechnol Bioeng 69:486–494
Boraston A, Warren RA, Kilburn DG (2001) Glycosylation by Pichia pastoris decreases the affinity of a family 2a carbohydrate-binding module from Cellulomonas fimi: a functional and mutational analysis. Biochem J 358:423–430
Boraston AB, Sandercock L, Warren RA, Kilburn DG (2003) O-glycosylation of a recombinant carbohydrate-binding module mutant secreted by Pichia pastoris. J Mol Microbiol Biotechnol 5:29–36
Boraston AB, Bolam DN, Gilbert HJ, Davies GJ (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 382:769–781
Carrard G, Koivula A, Söderlund H, Beguin P (2000) Cellulose-binding domains promote hydrolysis of different sites on crystalline cellulose. Proc Natl Acad Sci U S A 97:10342–10347
Cheek S, Krishna S, Grishin NV (2006) Structural classification of small, disulfide-rich protein domains. J Mol Biol 359:215–237
Davies G, Henrissat B (1995) Structures and mechanisms of glycosyl hydrolases. Structure 3:853–859
Demain AL, Newcomb M, Wu JH (2005) Cellulase, clostridia, and ethanol. Microbiol Mol Biol Rev 69:124–154
Fox JM, Jess P, Jambusaria RJ, Moo GM, Liphardt J, Clark DS, Blanch HW (2013) A single-molecule analysis reveals morphological targets for cellulase synergy. Nat Chem Biol 9:356–361
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788
Gietz RD, Woods RA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350:87–96
Grassick A, Murray PG, Thompson R, Collins CM, Byrnes L, Birrane G, Higgins TM, Tuohy MG (2004) Three-dimensional structure of a thermostable native cellobiohydrolase, CBH IB, and molecular characterization of the cel7 gene from the filamentous fungus, Talaromyces emersonii. Eur J Biochem 271:4495–4506
Guillén D, Sánchez S, Rodríguez-Sanoja R (2010) Carbohydrate-binding domains: multiplicity of biological roles. Appl Microbiol Biotechnol 85:1241–1249
Hall M, Rubin J, Behrens SH, Bommarius AS (2011) The cellulose-binding domain of cellobiohydrolase Cel7A from Trichoderma reesei is also a thermostabilizing domain. J Biotechnol 155:370–376
Igarashi K, Wada M, Hori R, Samejima M (2006) Surface density of cellobiohydrolase on crystalline celluloses. A critical parameter to evaluate enzymatic kinetics at a solid–liquid interface. FEBS J 273:2869–2878
Igarashi K, Koivula A, Wada M, Kimura S, Penttilä M, Samejima M (2009) High speed atomic force microscopy visualizes processive movement of Trichoderma reesei cellobiohydrolase I on crystalline cellulose. J Biol Chem 284:36186–36190
Igarashi K, Uchihashi T, Koivula A, Wada M, Kimura S, Okamoto T, Penttilä M, Ando T, Samejima M (2011) Traffic jams reduce hydrolytic efficiency of cellulase on cellulose surface. Science 333:1279–1282
Ilmén M, den Haan R, Brevnova E, McBride J, Wiswall E, Froehlich A, Koivula A, Voutilainen SP, Siika-aho M, la Grange DC, Thorngren N, Ahlgren S, Mellon M, Deleault K, Rajgarhia V, van Zyl WH, Penttilä M (2011) High level secretion of cellobiohydrolases in Saccharomyces cerevisiae. Biotechnol Biofuels 4:30–45
Imai T, Boisset C, Samejima M, Igarashi K, Sugiyama J (1998) Unidirectional processive action of cellobiohydrolase Cel7A on Valonia cellulose microcrystals. FEBS Lett 432:113–116
Kim T, Chokhawala HA, Nadler D, Blanch HW, Clark DS (2010) Binding modules alter the activity of chimeric cellulases: Effects of biomass pretreatment and enzyme source. Biotechnol Bioeng 107:601–611
Lehtiö J, Sugiyama J, Gustavsson M, Fransson F, Linder M, Teeri TT (2003) The binding specificity and affinity determinants of family 1 and family 3 cellulose binding modules. Proc Natl Acad Sci U S A 100:484–489
Lever M (1972) A new reaction for colorimetric determination of carbohydrates. Anal Biochem 47:273–279
Levy I, Shoseyov O (2002) Cellulose-binding domains: biotechnological applications. Biotechnol Adv 20:191–213
Linder M, Mattinen ML, Kontteli M, Lindeberg G, Ståhlberg J, Drakenberg T, Reinikainen T, Pettersson G, Annila A (1995) Identification of functionally important amino acids in the cellulose-binding domain of Trichoderma reesei cellobiohydrolase I. Protein Sci 4:1056–1064
McLean BW, Bray MR, Boraston AB, Gilkes NR, Haynes CA, Kilburn DG (2000) Analysis of binding of the family 2a carbohydrate-binding module from Cellulomonas fimi xylanase 10A to cellulose: specificity and identification of functionally important amino acid residues. Protein Eng 13(11):801–809
McLean BW, Boraston AB, Brouwer D, Sanaie N, Fyfe CA, Warren RAJ, Kilburn DG, Haynes CA (2002) Carbohydrate-binding modules recognize fine substructures of cellulose. J Biol Chem 277:50245–50254
Nikolova PV, Creagh AL, Duff SJ, Haynes CA (1997) Thermostability and irreversible activity loss of exoglucanase/xylanase Cex from Cellulomonas fimi. Biochemistry 36:1381–1388
Orr-Weaver TL, Szostak JW, Rothstein RJ (1981) Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A 78:6354–6358
Reinikainen T, Ruohonen L, Nevanen T, Laaksonen L, Kraulis P, Jones TA, Knowles JK, Teeri TT (1992) Investigation of the function of mutated cellulose-binding domains of Trichoderma reesei cellobiohydrolase I. Proteins 14:475–482
Sambrook J, Russel DW (2001) Molecular cloning, 3rd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Sherman F (2002) Getting started with yeast. Methods Enzymol 350:3–41
Suurnäkki A, Tenkanen M, Siika-aho M, Niku-Paavola ML, Viikari L, Buchert J (2000) Trichoderma reesei cellulases and their core domains in the hydrolysis and modification of chemical pulp. Cellulose 7:189
Tomme P, Driver DP, Amandoron EA, Miller RC Jr, Warren RAJ, Kilburn DG (1995) Comparison of a fungal (family I) and bacterial (family II) cellulose-binding domain. J Bacteriol 177(15):4356–4363
Tormo J, Lamed R, Chirino AJ, Morag E, Bayer EA, Shoham Y, Steitz TA (1996) Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J 15:5739–5751
Tuohy MG, Walsh DJ, Murray PG, Claeyssens M, Cuffe MM, Savage AV, Coughlan MP (2002) Kinetic parameters and mode of action of the cellobiohydrolases produced by Talaromyces emersonii. Biochim Biophys Acta 1596:366–380
van Tilbeurgh H, Claeyssens M (1985) Detection and differentiation of cellulase components using low molecular mass fluorogenic substrates. FEBS Lett 187:283–288
van Zyl WH, Lynd LR, Den Haan R, McBride JE (2007) Consolidated bioprocessing for bioethanol production using Saccharomyces cerevisiae. Biofuels 108:205–235
Viikari L, Alapuranen M, Puranen T, Vehmaanperä J, Siika-Aho M (2007) Thermostable enzymes in lignocellulose hydrolysis. Adv Biochem Eng Biotechnol 108:121–145
Viikari L, Vehmaanperä J, Koivula A (2012) Lignocellulosic ethanol: from science to industry. Biomass Bioenergy 46:13–24
Voutilainen SP, Boer H, Linder MB, Puranen T, Rouvinen J, Vehmaanperä J, Koivula A (2007) Heterologous expression of Melanocarpus albomyces cellobiohydrolase Cel7B, and random mutagenesis to improve its thermostability. Enzyme Microb Tech 41:234–243
Voutilainen SP, Puranen T, Siika-Aho M, Lappalainen A, Alapuranen M, Kallio J, Hooman S, Viikari L, Vehmaanperä J, Koivula A (2008) Cloning, expression, and characterization of novel thermostable family 7 cellobiohydrolases. Biotechnol Bioeng 101:515–528
Voutilainen SP, Boer H, Alapuranen M, Jänis J, Vehmaanperä J, Koivula A (2009) Improving the thermostability and activity of Melanocarpus albomyces cellobiohydrolase Cel7B. Appl Microbiol Biotechnol 83:261–272
Voutilainen SP, Murray PG, Tuohy MG, Koivula A (2010) Expression of Talaromyces emersonii cellobiohydrolase Cel7A in Saccharomyces cerevisiae and rational mutagenesis to improve its thermostability and activity. Protein Eng, Des Sel 23:69–79
Wan W, Wang D, Gao X, Hong J (2011) Expression of family 3 cellulose-binding module (CBM3) as an affinity tag for recombinant proteins in yeast. Appl Microbiol Biotechnol 91:789–798
Xu GY, Ong E, Gilkes NR, Kilburn DG, Muhandiram DR, Harris-Brandts M, Carver JP, Kay LE, Harvey TS (1995) Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy. Biochemistry 34:6993–7009
Acknowledgments
Arja Kiema and Eila Leino are thanked for the skilful technical assistance. We also wish to thank Roal Oy, Finland, for providing the T. aurantiacus β-glucosidase and Chemtext, Italy, for providing the pre-treated A. donax material. This study was performed in NEMO, a project under EU’s seventh framework program, grant number 222699.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 31 kb)
Rights and permissions
About this article
Cite this article
Voutilainen, S.P., Nurmi-Rantala, S., Penttilä, M. et al. Engineering chimeric thermostable GH7 cellobiohydrolases in Saccharomyces cerevisiae . Appl Microbiol Biotechnol 98, 2991–3001 (2014). https://doi.org/10.1007/s00253-013-5177-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-5177-2