Abstract
Amyotrophic lateral sclerosis (ALS), the most common motor neuron disease, appears to result from the combination of genetic and environmental factors. Whether the rs2275294 polymorphism in the ZNF512B gene influences ALS risk is controversial. We meta-analysed the association between rs2275294 and ALS risk based on evidence published in the PubMed database. Five case–control studies involving 2559 patients with sporadic ALS and 5740 controls were analysed. Based on random-effects meta-analysis, the polymorphism rs2275294 was associated with increased risk of ALS disease in an allele model (C vs. T: OR 1.222, 95%CI 1.057 to 1.414, p = 0.007). The available evidence suggests that the ZNF512B polymorphism rs2275294 is associated with ALS risk. These results should be validated in large, well-designed studies, especially in non-Asian populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Amyotrophic lateral sclerosis (ALS), a typical neurological disease, is an adult-onset neurodegenerative disease in which motor neurons specifically deteriorate. It is associated with mean survival of 3–4 years [1], and it involves progressive wasting and weakness of limb, bulbar, and respiratory muscles [2]. Familial ALS accounts for 10% of global ALS cases, with the remainder classified as sporadic ALS [3], which may result from the combination of genetic and environmental factors [3]. Some genes have been implicated in familial ALS, including SOD1, OPN, C9orf72, TARDBP, FUS, and VCP [4]. Sporadic ALS has also been associated with several genetic risk factors identified in candidate gene studies and genome-wide association studies [5,6,7]. However, some of these genetic risk factors have not shown consistent relationships with ALS onset across different studies.
For example, the rs2275294 polymorphism in the ZNF512B gene has been associated with increased risk of sporadic ALS in a Japanese population [8] and in one Han Chinese population [9], but not in a second Chinese population [2]. This potential association is important to clarify, because several lines of indirect evidence implicate the ZNF512B gene in ALS. ZNF512B, originally identified as KIAA1196, encodes an 893-residue transcription factor that is expressed in the brain and spinal cord and that may regulate the TGF-β signaling pathway via SMAD2/3 [10]. This signaling pathway helps to protect neurons and prolong their survival in several neurodegenerative diseases, including ALS [11], ischemic stroke [12], Alzheimer’s disease, and Parkinson’s disease [13]. The rs2275294 polymorphism lies in an enhancer region, and the susceptibility allele (C) is associated with decreased ZNF512B transcription [14,15,16]. Thus, the rs2275294 polymorphism may increase risk of ALS by decreasing TGF-β signaling.
We meta-analysed the available literature in the PubMed database in order to gain a comprehensive understanding of whether the rs2275294 polymorphism is likely to be associated with ALS risk.
Materials and methods
Literature search strategy
We searched the PubMed, Web of Science, Embase, Chinese National Knowledge Infrastructure, Wanfang, and SinoMed databases for studies of the potential association between the single-nucleotide polymorphism (SNP) rs2275294 in ZNF512B and risk of ALS. We used the following search terms: “amyotrophic lateral sclerosis” or “ALS” and “zinc finger protein 512B” or “ZNF512B.” No language restriction was imposed. The final search was conducted on May 21, 2017.
Selection and exclusion criteria
To be included in our review, eligible studies had to (a) evaluate the potential association between the ZNF512B polymorphism rs2275294 and ALS risk among human, (b) use a case–control or genome-wide association design, and (c) report genotype frequency and other data necessary for estimating odds radios (ORs) with 95% confidence intervals (CIs). If populations in two studies overlapped, only the larger study was included. Studies were excluded if they did not report original research or if they were published only as abstracts, reviews, case reports, discussions, editorials, or letters to the editor.
Data extraction
Two authors independently assessed articles for inclusion/exclusion, and discrepancies were resolved by discussion with a third reviewer. The following data were extracted from each study: name of the first author, publication year, ethnicity (country), sample size, and genotype or allele distribution in cases and controls.
Statistical analysis
The potential association was assessed between ZNF512B rs2275294 and ALS risk using Stata 12.0, and the strength of the association was estimated in terms of OR and 95%CI. The OR was assessed using the allele model (C vs. T). A p value equal to or less than 0.05 was considered the threshold for statistical significance in all analyses.
Prior to meta-analysis, genotype distributions in each study were checked using the Hardy–Weinberg equilibrium test. Heterogeneity among studies was evaluated using the Q test and was quantified using I2. An I2 value below 25% was considered to indicate homogeneity; values of 25% to just under 50%, low heterogeneity; values of 50% to just under 75%, moderate heterogeneity; and values of at least 75%, substantial heterogeneity. We planned to use a fixed-effects model to meta-analyse pooled data classified as homogeneous or of low heterogeneity, and a random-effects model to meta-analyse data classified as showing moderate or substantial heterogeneity. Publication bias was assessed using Egger’s and/or Begg’s tests.
Results
Literature search and included studies
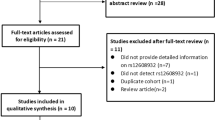
A total of seven potentially eligible articles were identified after searching the six databases above and removing duplicates. Then, three articles were excluded because the object of the study is not human (n = 1), the study design is cohort study (n = 1), and the type of the article is review (n = 1). After eliminating the three articles mentioned above based on the title and abstract, the remaining four were read in full and one is excluded because the authors did not report enough data to calculate minor allele frequency [17]. Ultimately, the three remaining publications were included [2, 8, 9], one of which [8] comprised a case–control study to analyse the association between ALS and rs2275294, followed by two independent case–control studies to validate the association. Therefore, the final meta-analysis included five case–control studies from three publications (Fig. 1).
Characteristics of included studies
Tables 1 and 2 summarise key characteristics of the studies included in the meta-analysis. Of the five studies, three were from Japan and involved 1305 cases and 4244 controls; the other two were from mainland China and involved 1254 cases and 1496 controls. Four studies [8, 9] involving 1606 ALS patients reported that individuals carrying the C allele of the rs2275294 polymorphism were at higher ALS risk than those carrying the T allele. The remaining study found no association between the polymorphism and disease risk [2] (Tables 1 and 2).
The studies did not report information about gender or type of onset (bulbar or spinal) in sufficient detail to allow subgroup analyses.
Heterogeneity
The heterogeneity test revealed obvious heterogeneity among studies in the allele model (C vs. T, I2 = 76.7%). Therefore, we meta-analysed the data using a random-effects model.
Meta-analysis
The meta-analysis suggests that rs2275294 in ZNF512B is associated with ALS risk in an allele model (C vs. T, OR 1.222, 95%CI 1.057–1.414, p = 0.007) (Fig. 2).
Assessment of publication bias and sensitivity analysis
Significant risk of publication bias was not observed in Egger’s test (p = 1.000) or Begg’s test (p = 0.365) (Fig. 3). Sensitivity analysis showed that none of the studies on its own significantly influenced these results (Fig. 4).
Discussion
This is, to our knowledge, the first meta-analysis assessing the available evidence on whether the rs2275294 polymorphism in ZNF512B is associated with ALS risk. Our results suggest that the polymorphism may be associated with increased risk of ALS, at least among Asians.
One of the articles in our review involved an initial case–control study suggesting an association between rs2275194 and increased risk of ALS in Japanese. This association was then validated in a different set of 249 ALS cases and 1030 controls from the Japanese Biobank (set 1; allele model, p = 1.8 × 10−3), as well as in a different Japanese group of 602 ALS cases and 2256 controls (set 2; allele model, p = 5.6 × 10−5). Our group confirmed this association in a southwest Chinese population of 301 ALS cases and 457 controls, but another team did not replicate it in a northern Chinese population of 953 ALS cases and 1039 controls. This discrepancy about genetic factors linked to ALS should not be surprising, given that ALS is a complex disease associated with genetic and environmental risk factors. The fact that different results were obtained with populations from different parts of China highlights the potential importance of geographic location.
The rs2275294 SNP decreases expression of ZNF512B, thereby reducing TGF-β signaling. Such a reduction in signaling can compromise the brain’s ability to stimulate the birth of new neurons to replace damaged tissue, and it can compromise the ability of TGF-β signaling to shield neurons from glutamate-mediated excitotoxicity, which may underlie various neurodegenerative disorders such as ALS. Interruption of TGF-β signaling is linked to motor neuron harm induced by polyglutamine in spinal and bulbar muscular atrophy [18]. TGF-β is upregulated in the astrocytes of ALS patients and mice, and it is a negative regulator of the neuroprotective inflammatory response [19, 20]. Activation of TGF-β signaling protects against aggregation of TAR DNA-binding protein that has mislocalised to the cytoplasm [21], making it a potential therapeutic approach to delay the progression of ALS. These various studies linking rs2275294 to attenuation of TGF-β signaling are consistent with our meta-analysis associating the SNP with elevated ALS risk.
An important question is whether the risk allele (C) at rs2275294 is associated with poorer prognosis. One study reported that survival time of C allele carriers was shorter by 72 months than the survival time of ALS patients without this allele [17]. Unfortunately, the studies in our review did not report survival time, so this question should be addressed in future work. Similarly, the lack of detailed data reporting prevented us from performing subgroup analyses to assess whether the risk allele at rs2275294 influences factors known to affect the prognosis of ALS patients, such as age at disease onset, site of symptom onset, and delay between first symptom and diagnosis [22,23,24,25].
While our meta-analysis offers the first comprehensive assessment of rs2275294 polymorphism and ALS risk, the results should be interpreted with caution in light of several limitations. One is that our meta-analysis did not include genome-wide association studies due to the lack of sufficient data on the genotype frequency of rs2275294 for cases and controls. Another limitation is that we could not perform subgroup analysis based on gender due to limited data reporting, or based on ethnicity because all the included studies involved Asians. In addition, one study [8] performed on Japanese subjects by Iida et al. provides three out of the five included populations, which may influence the present analysis. However, sensitivity analysis showed that none of the studies on its own significantly influenced these results (Fig. 4). We are sure that our meta-analysis is stable on the whole. Future work, especially with Caucasians and other non-Asian groups, is needed to validate and extend these findings about the association between rs2275294 and ALS risk.
References
van Es MA, Hardiman O, Chio A, Al-Chalabi A, Pasterkamp RJ, Veldink JH et al (2017) Amyotrophic lateral sclerosis. Lancet 390(10107):2084–2098
Ju XD, Liu T, Chen J, Li XG, Liu XX, Liu WC, Wang K, Deng M (2015) Single-nucleotide polymorphism rs2275294 in ZNF512B is not associated with susceptibility to amyotrophic lateral sclerosis in a large Chinese cohort. Chin Med J 128(24):3305–3309
Al-Chalabi A, Hardiman O (2013) The epidemiology of ALS: a conspiracy of genes, environment and time. Nat Rev Neurol 9(11):617–628
Ghasemi M, Brown RH Jr (2017) Genetics of amyotrophic lateral sclerosis. Cold Spring Harb Perspect Med. https://doi.org/10.1101/cshperspect.a024125
Saeed M, Siddique N, Hung WY, Usacheva E, Liu E, Sufit RL, Heller SL, Haines JL, Pericak-Vance M, Siddique T (2006) Paraoxonase cluster polymorphisms are associated with sporadic ALS. Neurology 67(5):771–776
Wills AM, Cronin S, Slowik A, Kasperaviciute D, Van Es MA, Morahan JM et al (2009) A large-scale international meta-analysis of paraoxonase gene polymorphisms in sporadic ALS. Neurology 73(1):16–24
Deng M, Wei L, Zuo X, Tian Y, Xie F, Hu P et al (2013) Genome-wide association analyses in Han Chinese identify two new susceptibility loci for amyotrophic lateral sclerosis. Nat Genet 45(6):697–700
Iida A, Takahashi A, Kubo M, Saito S, Hosono N, Ohnishi Y, Kiyotani K, Mushiroda T, Nakajima M, Ozaki K, Tanaka T, Tsunoda T, Oshima S, Sano M, Kamei T, Tokuda T, Aoki M, Hasegawa K, Mizoguchi K, Morita M, Takahashi Y, Katsuno M, Atsuta N, Watanabe H, Tanaka F, Kaji R, Nakano I, Kamatani N, Tsuji S, Sobue G, Nakamura Y, Ikegawa S (2011) A functional variant in ZNF512B is associated with susceptibility to amyotrophic lateral sclerosis in Japanese. Hum Mol Genet 20(18):3684–3692
Yang X, Zhao Q, An R, Zheng J, Tian S, Xu Y (2015) Association of the functional SNP rs2275294 in ZNF512B with risk of amyotrophic lateral sclerosis and Parkinson's disease in Han Chinese. Amyotroph Lateral Scler Frontotemporal Degener 17(1–2):142–147
Colland F, Jacq X, Trouplin V, Mougin C, Groizeleau C, Hamburger A, Meil A, Wojcik J, Legrain P, Gauthier JM (2004) Functional proteomics mapping of a human signaling pathway. Genome Res 14(7):1324–1332
Ilzecka J, Stelmasiak Z, Dobosz B (2002) Transforming growth factor-Beta 1 (tgf-Beta 1) in patients with amyotrophic lateral sclerosis. Cytokine 20(5):239–243
Krupinski J, Kumar P, Kumar S, Kaluza J (1996) Increased expression of TGF-beta 1 in brain tissue after ischemic stroke in humans. Stroke 27(5):852–857
van der Wal EA, Gomez-Pinilla F, Cotman CW (1993) Transforming growth factor-beta 1 is in plaques in Alzheimer and Down pathologies. Neuroreport 4(1):69–72
Henrich-Noack P, Prehn JH, Krieglstein J (1994) Neuroprotective effects of TGF-beta 1. J Neural Transm Suppl 43:33–45
Iwasaki Y, Shiojima T, Tagaya N, Kobayashi T, Kinoshita M (1997) Effect of transforming growth factor beta 1 on spinal motor neurons after axotomy. J Neurol Sci 147(1):9–12
Krieglstein K, Strelau J, Schober A, Sullivan A, Unsicker K (2002) TGF-beta and the regulation of neuron survival and death. J Physiol Paris 96(1–2):25–30
Tetsuka S, Morita M, Iida A, Uehara R, Ikegawa S, Nakano I (2013) ZNF512B gene is a prognostic factor in patients with amyotrophic lateral sclerosis. J Neurol Sci 324(1–2):163–166
Katsuno M, Adachi H, Banno H, Suzuki K, Tanaka F, Sobue G (2011) Transforming growth factor-beta signaling in motor neuron diseases. Curr Mol Med 11(1):48–56
Tetsuka S (2017) Difficulty in determining the association of a single nucleotide polymorphism in the ZNF512B gene with the risk and prognosis of amyotrophic lateral sclerosis. Rinsho Shinkeigaku 57(8):417–424
Endo F, Komine O, Fujimori-Tonou N, Katsuno M, Jin S, Watanabe S, Sobue G, Dezawa M, Wyss-Coray T, Yamanaka K (2015) Astrocyte-derived TGF-beta1 accelerates disease progression in ALS mice by interfering with the neuroprotective functions of microglia and T cells. Cell Rep 11(4):592–604
Nakamura M, Kaneko S, Ito H, Jiang S, Fujita K, Wate R, Nakano S, Fujisawa JI, Kusaka H (2013) Activation of transforming growth factor-beta/Smad signaling reduces aggregate formation of mislocalized TAR DNA-binding protein-43. Neurodegener Dis 11(4):182–193
Czaplinski A, Yen AA, Appel SH (2006) Amyotrophic lateral sclerosis: early predictors of prolonged survival. J Neurol 253(11):1428–1436
Talbot K (2009) Motor neuron disease: the bare essentials. Pract Neurol 9(5):303–309
Chio A, Mutani R, Mora G (2003) Prognosis in amyotrophic lateral sclerosis: a population-based study. Neurology 61(12):1826–1827 author reply 7
Atsuta N, Watanabe H, Ito M, Tanaka F, Tamakoshi A, Nakano I, Aoki M, Tsuji S, Yuasa T, Takano H, Hayashi H, Kuzuhara S, Sobue G, Research Committee on the Neurodegenerative Diseases of Japan (2009) Age at onset influences on wide-ranged clinical features of sporadic amyotrophic lateral sclerosis. J Neurol Sci 276(1–2):163–169
Funding
This work was funded by the Major Clinical Disease Research Program from the Health and Family Planning Commission of Sichuan Province (17ZD011) and the Sichuan Key Project of Science and Technology (2010SZ0086).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Rights and permissions
About this article
Cite this article
Ning, P., Yang, X., Yang, B. et al. Meta-analysis of the association between ZNF512B polymorphism rs2275294 and risk of amyotrophic lateral sclerosis. Neurol Sci 39, 1261–1266 (2018). https://doi.org/10.1007/s10072-018-3411-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-018-3411-5