Abstract
Rice grassy stunt virus (RGSV), a member of the genus Tenuivirus, causes serious rice disease in Southeast Asian countries. In this study, a green fluorescent protein (GFP)-based transient expression assay was conducted to show that p5, encoded on RNA5 in the viral sense, is a viral suppressor of RNA silencing (VSR). Protein-protein interactions (PPIs) between p5 and all RGSV proteins except pC1 and pC2 were investigated using Gal4-based yeast two-hybrid (Y2H) experiments. The results demonstrated that p5 interacts with itself and with p3 encoded on RNA3 in the viral sense. p5-p5 and p5-p3 interactions were detected by bimolecular fluorescence complementation (BiFC) assay, and the p5-p3 interaction was confirmed by subcellular co-localization and co-immunoprecipitation (Co-IP) assays. Using the Y2H system, we demonstrated that the p5-p3 interaction requires both the N-terminal (amino acid residues 1 to 99) and C-terminal (amino acid residues 94 to 191) domains of p5. In addition, either p5 or p3 could enhance the pathogenicity of potato virus X (PVX) in Nicotiana benthamiana plants. A much more significant enhancement of PVX pathogenicity and accumulation was observed when p5 and p3 were expressed together. Our data also showed that RGSV p3 does not function as a VSR, and it had no effect on the VSR activity of p5 or the subcellular localization pattern of p5 in plant cells from Nicotiana benthamiana.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice grassy stunt virus (RGSV) causes rice grassy stunt disease in rice in many Southeast Asian countries [11]. The RGSV was first found in the Philippines in 1963 [17, 20]. The virus is transmitted by the brown planthopper (BPH; Nilaparvata lugens Stal) in a circulative and propagative manner [25]. RGSV infects rice and induces symptoms including leaf yellowing, stunting, and excess tillering [20, 21]. The RGSV genome has six ambisense RNA segments containing 12 open reading frames (ORFs) [18, 23]. RNAs 1, 2, 5 and 6 of RGSV correspond to RNAs 1, 2, 3 and 4, respectively, of rice stripe virus (RSV), the type member of the genus Tenuivirus [3]. The functions of three RGSV-encoded proteins are known. The protein pC1 (339.1-kDa), encoded on RNA 1 in the viral complementary sense (vcRNA1), may function as an RNA-dependent RNA polymerase (RdRp) and is similar to the RdRp of RSV, with 37 ± 9 % amino acid sequence identity over 2140 amino acid residues [23, 24]. The protein pC5 (35.9 kDa), encoded by vcRNA5, is the nucleocapsid protein (N) and accumulates in both RGSV-infected rice and viruliferous brown planthoppers [3]. The protein pC6 (36.4 kDa), encoded on vcRNA6, can trans-complement the cell-to-cell spread of a movement-detective TMV, suggesting that it may act as a movement protein [12]. The functions of some other RGSV proteins can be deduced by comparison with their counterparts in RSV. However, functional verification is essential, given the low amino acid sequence identity between these proteins [3, 23, 24].
Protein-protein interactions (PPIs) play a crucial role in many cellular processes. Identification and characterization of PPIs can provide important insights into protein functions at the cell level. For viruses, mapping the interaction networks of viral proteins contributes considerably to understanding the mechanisms of viral pathogenicity, the virus infection cycle, and virus-host interactions. Intra-viral PPIs have been reported for southern rice black-streaked dwarf fijivirus (SRBSDV) and RSV [13, 14]. It is reasonable to assume that the intra-viral PPIs play fundamental roles in the replication cycle of SRBSDV and RSV, as in the case of many other viruses. Thus, it is of interest to test whether these PPIs are conserved among members of the genus Tenuivirus and to investigate whether PPIs exist between RGSV-specific proteins and proteins shared with RSV. RSV is a well-studied tenuivirus. It causes dramatic loss in rice production in China, Japan, and Korea [5, 11, 29]. It has been reported that the RSV infects not only rice plants but also maize, wheat, oat, foxtail millet, weeds and Arabidopsis [7, 15, 22]. Until now, several host factors have been identified that interact with RSV proteins or RNA, and those factors are thought to be recruited or manipulated by RSV and used to complete its life cycle [6, 16, 33]. Notably, the RSV nonstructural protein p2, a viral silencing suppressor, can bind to a rice host protein that is homologous to the suppressor of gene silencing in Arabidopsis (AtSGS3) [6]. Previously, is was shown that the nonstructural protein p3 of RSV, encoded by RNA3, also functions as a viral suppressor of RNA silencing [31]. Wu and colleagues hypothesized that the p3 protein has the dual functions of facilitating viral infection as a VSR and inhibiting pathogenic development as an inducer of the host defense system [27].
In this paper, we investigate whether the p5 protein of RGSV has silencing suppressor activity. Additionally, PPIs between the p5 protein and 10 other proteins encoded by the RGSV genome were examined to explore the functions of these proteins. We found that the p5 protein is a functional homologue of the p3 protein of RSV and interacts with the RGSV-specific protein p3. The RGSV p5 and p3 proteins could function additively to enhance the pathogenicity of potato virus X (PVX) in Nicotiana benthamiana.
Materials and methods
Plant materials, virus, and vectors
Rice plants infected with the RGSV were collected from Fuzhou, Fujian Province, China, in 2010. RGSV was maintained by serial passages, transmitted by brown planthoppers, in rice plants (Oryza sativa var. Taichung Native 1) in the greenhouse at the Institute of Plant Virology at the Fujian Agriculture and Forestry University. The seeds of N. benthamiana plants and line 16c plants were stored in our lab and cultured in an illumination incubator. All of the transient expression vectors used in our experiments, including pDONR221, pEarleyGate101, pEarleyGate102, pEarleyGate201, pYFPN, and pYFPC, were kindly provided by Professor Wang from the Southern Canada Agriculture Plant Protection and Food Research Center, and the vectors were not modified. The yeast expression vectors pGBKT7, pGADT7, and PVX vector pGR107 were stored in our lab and were not modified. The vectors driven by the CaMV 35S promoter had an HA-tag and a Myc-tag sequence inserted at the KpnI and SmaI sites at the N-terminus of the multiple cloning site (MCS).
Plasmid construction
To perform the GFP-based transient expression assay, full-length ORFs of p5 and p3 were amplified by using specific primers containing homologous recombination sequences at the 5’ termini of both the forward and reverse primers. PCR products were inserted into the entry vector pDONR221 using BP ClonaseTM II Enzyme (Invitrogen, USA). Positive recombinant plasmids were identified by colony PCR and were sequenced. Subsequently, extracted plasmids were digested with MluI (Takara, Japan), and the purified product containing p5 or p3 gene was inserted into the plant transient expression vector pEarleyGate201. The potyvirus VSR HC-Pro was used as a positive control, and the original vector was used as a negative control. Another control was constructed with an untranslatable mutant of p5 in which the second codon TCT was replaced with a TAG stop codon. The five constructs were named RGSV p5, RGSV p3, HC-Pro, Vector and RGSV Δp5.
To investigate PPIs using a Y2H assay, full-length ORFs of each RGSV gene, except pC1 and pC2, were amplified using primers with the appropriate restriction sites (Supplementary Table 1). PCR products were cloned into the vector pMD18T (Takara, Japan) and sequenced. Plasmids that were verified by sequencing were digested with the corresponding restriction enzymes. The purified enzyme-digested products were inserted into the yeast expression vectors, pGBKT7 and pGADT7, using T4 DNA ligase (Beijing TransGen Biotech Co., Ltd., China), creating pGBK-p5, pGAD-p1, pGAD-p2, pGAD-p3, pGAD-pC3, pGAD-p4, pGAD-pC4, pGAD-p5, pGAD-pC5, pGAD-p6, pGAD-pC6 and pGBK-p3. To identify the regions of p5 that are necessary for interaction with p3, p5 was separated into three overlapping domains: the N-terminus of p5 (p5N) containing amino acid residues 1-99; p5M, containing amino acid residues 49-140; and p5C, containing amino acid residues 94-191. Each domain of p5 was inserted into the pGBKT7 vector, and the constructs were named pGBK-p5N, pGBK-p5M and pGBK-p5C.
By using the same method described for the GFP-based transient expression assay, full-length ORFs of p5 and p3 were recombined into pYFPN, pYFPC, pEarleyGate101 and pEarleyGate102, respectively, for BiFC and cellular localization studies, creating pYFPN-p5, pYFPN-p3, pYFPC-p5, pYFPC-p3, YFP-p3 and CFP-p5. A construct expressing SV40NLS-mCherry was used in cotransfection experiments to label the nucleus [32]. In the Co-IP assay, the full-length ORFs of p5 and p3 were inserted into the Myc-tagged and HA-tagged expression vectors, respectively, driven by the CaMV 35S promoter.
To study the roles of p5 and p3 in viral infection in the context of plant-virus interaction, pGR107, a PVX vector, was utilized to express p5 and p3. Full-length ORFs of p5 and p3 were cloned into the PVX vector, creating PVX-p5 and PVX-p3. The primers used in plasmid construction are listed in Supplementary Table 1.
Y2H assay
A Gal4-based yeast two-hybrid system was used for the investigation of PPIs between p5 and 10 other proteins. The plasmids pGBKT7-p5 and pGADT7-X were used to co-transform 100 μL of freshly prepared AH109 competent cells simultaneously with the help of 5 μL of herring carrier DNA (Takara, Japan) using the lithium acetate method [2, 8, 9]. The transformants were uniformly plated onto the double dropout (DDO) medium SD/-Leu-Trp. Plates were incubated in a constant-temperature incubator at 30 °C. Yeast cells co-transformed with pGBK7-53/pGAD7-T and pGBKT7-Lam/pGADT7-T were used as positive and negative controls, respectively. Yeast co-transformants that grew on the DDO medium were selected to plate on triple dropout (TDO) medium SD/-Leu-Trp-His and quadruple dropout (QDO) medium SD/-Leu-Trp-His-Ade, respectively. Interactions were detected using an α-galactosidase activity assay on QDO medium containing the substrate α-galactosidase (X-α-gal).
Agrobacterium infiltration, GFP imaging and confocal microscopy
Agrobacterium infiltration was carried out as described previously [6, 26, 28, 31]. Briefly, each of the plant expression constructs was introduced individually by transformation into Agrobacterium strain EHA105 by freeze-thawing with liquid nitrogen. Cultures of EHA105 harboring each binary plasmid were grown in LB medium containing rifampicin (50 mg/ml) and kanamycin (50 mg/ml) at 28 °C for 48 h. Positive transformants were identified using colony PCR with the corresponding primers. Cultures of EHA105 strains containing each recombinant plasmid were resuspended and diluted to an optical density at 600 nm (OD600) of 0.8 with infiltration medium (5 mg of glucose per mL, 10 mM MES, 10 mM Na3PO4, 200 mM acetosyringone). For the GFP-based transient expression assay, five-week-old transgenic N. benthamiana plants of line 16c were used. Nicotiana benthamiana line 16c plants were kept in growth chambers at 22-24 °C. Agrobacterium containing the RGSV p5 protein, the RGSV p3 protein, HC-Pro, the original vector, or RGSV Δp5 was mixed with a strain containing the green fluorescent protein (GFP) transgene (35S-ssGFP) at a volume ratio of 1:1 and co-infiltrated into leaf epidermal cells of transgenic N. benthamiana line 16c. GFP fluorescence in the co-infiltrated leaves was visualized using a hand-held UV light (Black Ray model B 100A; UV Products) and photographed using a Nikon D70 digital camera at two and five days post-infiltration (dpi). To perform the BiFC assay, five- to eight-week-old N. benthamiana plants were used [10, 30]. Agrobacterium cells transformed with pYFPN-p5/pYFPC-p5, pYFPN-p3/pYFPC-p5 or pYFPN-p5/pYFPC-p3 were mixed at a volume ratio of 1:1 and inoculated into the leaf epidermal cells of N. benthamiana. For co-localization assays, Agrobacterium cells transformed with YFP-p3/CFP-p5 were mixed as described above and inoculated into the leaf epidermal cells of N. benthamiana. To examine the localization pattern of p5 or p3, Agrobacterium cells transformed with YFP-p3 or CFP-p5 were individually inoculated into the leaf epidermal cells of N. benthamiana. The infiltrated epidermal cells of N. benthamiana were examined for fluorescence under a Leica DS6000 confocal microscope at 48 h post-infiltration. For Co-IP assay, Agrobacterium cells transformed with RGSV HA-p3/RGSV p5-Myc were mixed as described above and inoculated into the leaf epidermal cells of N. benthamiana, and HA-GFP/RGSV p5-Myc were used as a negative control.
RNA extraction, RT-qPCR analysis and northern blot analysis
Total RNA from leaves of rice plants or N. benthamiana plants was isolated following the directions of the manufacturer of the Plant RNA Kit (OMEGA, USA). The first-strand complementary DNA (cDNA) was synthesized following the instructions of the manufacturer of the First Strand cDNA Synthesis Kit (TOYOBO CO., LTD., Japan). To analyze the transcript levels of the PVX CP gene by quantitative real-time reverse transcription PCR (RT-qPCR), ReverTra Ace® qPCR RT Master Mix with a gDNA Remover Kit (TOYOBO CO., LTD., Japan) was used for cDNA synthesis. qPCR was performed according to the manufacturer’s instructions using the THUNDERBIRD SYBR qPCR Mix (TOYOBO CO., LTD., Japan).
A northern blot analysis with total RNAs or sRNAs was performed as described previously [19, 29]. 32P-end-labeled oligonucleotide probes complementary to GFP mRNA or sRNAs were used for northern blots (Supplementary Table 1).
Protein extraction, immunoprecipitation and western blot analysis
Infiltrated leaf tissue (100 mg) from N. benthamiana was excised and homogenized in 200 μL of extraction buffer (50 mM Tris-HCl [pH 6.8], 9 M urea, 4.5 % SDS and 7.5 % β-mercaptoethanol). The crude extracts were centrifuged for 15 min at 12,000 g. The supernatant was precipitated with two volumes of acetone and centrifuged at 12,000 g for 5 min. The pellets were resuspended in double distilled water. Proteins were separated by 12 % SDS-PAGE and transferred to PVDF membranes. In the GFP-based transient expression assay to detect the accumulation of GFP, the PVDF membranes were probed with a commercially available mouse anti-GFP monoclonal antibody (Abmart Inc., Shanghai) and with anti-actin antibodies (Abmart Inc., Shanghai) to detect the expression of actin proteins, which were used as an internal reference. The secondary antibody was a goat anti-mouse IgG conjugated with alkaline phosphatase (Sigma, St. Louis, MO, USA) and used at a 1:8000 (v/v) dilution. Proteins were visualized by incubating the membranes in an NBT-BCIP solution (Promega, USA). For the Co-IP assay, an Immunoprecipitation Kit (Life, USA) was used according to the manufacturer’s instructions. Anti-HA antibodies (Abmart Inc., Shanghai) were first cross-linked with Dynabeads Protein A, and the target antigen complex was then immunoprecipitated. Anti-Myc antibodies (Abmart Inc., Shanghai) were used to perform Western blot analysis to verify the expression of p5 and the p5-p3 interaction. In the study on the pathogenicity of p5 and p3, protein expression was detected with anti-p5 and anti-p3 polyclonal antibodies, and the secondary antibody was a goat anti-rabbit IgG.
Results
RGSV p5, but not p3, functions as a viral suppressor of RNA silencing
A green fluorescent protein (GFP)-based transient expression assay was used to test whether p5 has silencing suppressor activities. Each of the five plasmids (RGSV p5, RGSV p3, HC-Pro, Vector and RGSV Δp5) was co-agrointroduced with a plasmid carrying the 35S-ssGFP segment into leaves of 16c transgenic N. benthamiana plants through an Agrobacterium-mediated infiltration procedure described previously. Notably, 16c plants co-infiltrated with an Agrobacterium harboring 35S-ssGFP and an Agrobacterium harboring either an original vector or a vector containing HC-Pro were used as negative and positive controls in this study. GFP fluorescence in the co-infiltrated leaves was observed at two and five days post-infiltration (dpi). At 2 dpi, the strong fluorescence of GFP was observed in all leaves of transgenic N. benthamiana line 16c. By 5 dpi, however, the fluorescence was significantly diminished in the leaves co-infiltrated with Agrobacterium carrying 35S-ssGFP and Agrobacterium carrying the RGSV p3, the original vector, or the RGSV Δp5, but strong GFP fluorescence was also seen in the leaves co-expressing 35S-ssGFP and the RGSV p5 or HC-Pro at 5 dpi (Fig. 1A).
The RGSV p5 protein, but not the p3 protein, inhibits local silencing triggered by ssGFP. (A) Leaves shown in panel A were co-infiltrated with 35S-ssGFP and each of the different plasmids indicated around the images. “Vector” is the original vector that was used as a negative control. HC-Pro, the first identified VSR of potato virus Y, was used as a positive control. RGSV Δp5 is an untranslatable mutant of p5 in which the second codon, TCT, was replaced with a TAG stop codon. GFP fluorescence in the co-infiltrated leaves was visualized with a hand-held UV light (Black Ray model B 100A; UV Products) and photographed using a Nikon D70 digital camera at five days post-infiltration (dpi). (B) Northern blot analysis of the levels of GFP mRNA extracted from differently infiltrated patches. The lanes (from left to right) contained samples from leaves co-infiltrated with 35S-ssGFP and vector, p3, Δp5, p5, p5 plus p3 and HC-Pro, respectively. rRNA and tRNA were used as loading controls for the detection of GFP mRNA and GFP siRNA, respectively. (C) The accumulation of GFP in the infiltrated zones was detected with anti-GFP antibody by Western blotting at 5 dpi. The results were in agreement with those of the northern blot analysis. Actin was used as an internal reference
Northern blot analysis showed negligible accumulation of GFP-specific siRNAs and high accumulation of GFP mRNA in leaves infiltrated with 35S-ssGFP plus either RGSV p5 or HC-Pro (Fig. 1B) at 5 dpi, further demonstrating that the RGSV p5 could indeed suppress local RNA silencing triggered by GFP ssRNA. Conversely, leaves infiltrated with 35S-ssGFP plus the RGSV p3, the original vector, or the RGSV Δp5 showed high accumulation of GFP siRNA, and the accumulation of GFP mRNA was not detected in these samples by 5 dpi (Fig. 1B), indicating that the RGSV p3 was not a VSR.
A Western blot was used to examine the expression levels of GFP. The results showed that the accumulation of GFP was similar at 2 dpi in all of the co-infilterated leaves. However, by 5 dpi, the GFP protein levels dramatically decreased in leaves co-infiltrated with the Agrobacterium carrying 35S-ssGFP/RGSV p3, 35S-ssGFP/vector or 35S-ssGFP/RGSV Δp5. In contrast, the GFP protein levels were markedly higher in leaves that were co-infiltrated with the Agrobacterium carrying either 35S-ssGFP/RGSV p5 or 35S-ssGFP/HC-Pro than in those carrying 35S-ssGFP/RGSV p3, 35S-ssGFP/vector, or 35S-ssGFP/RGSV Δp5. The actin protein was used as an internal reference, and its accumulation remained unchanged throughout the experiments (Fig. 1C). These results indicated that RGSV p5 could suppress local RNA silencing triggered by GFP ssRNA, suggesting that it functions as a VSR, whereas the RGSV p3 did not possess a congruent function with the RGSV p5.
p5 interacts with itself and p3 in yeast
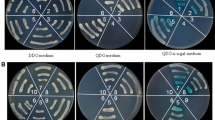
The Y2H system was used to study the interaction of p5 with all RGSV proteins except pC1 and pC2. Only the yeast cells co-transformed with pGBK-p5 and pGAD-p5 or pGBK-p5 and pGAD-p3 could grow on TDO and QDO media. To confirm the interaction of p5 and p3 in yeast cells, we constructed pGAD-p5 and pGBK-p3 and introduced them into yeast cells by transformation. The co-transformants could grow on TDO and QDO media as well. p5-p5- and p5-p3-interactions were also confirmed by detecting α-galactosidase activity. Blue yeast colonies were observed on QDO medium containing X-α-gal, the substrate of α-galactosidase, within four days, indicating that p5-p5 and p5-p3 interaction occurred in yeast cells (Fig. 2A). We observed that the p5-p3 interaction was weaker when p5 was expressed in pGADT7 and when p3 was expressed in pGBKT7. As shown in Fig. 2B, the same amount of yeast cells transformed with pGBK-p5/pGAD-p3 or pGAD-p5/pGBK-p3 from DDO medium was resuspended in 20 μL of sterile water and diluted in sterile water at ratios of 1:1, 1:10, 1:102, 1:103 and 1:104. Five μL of each dilution was placed in QDO-X-α-gal medium. Interestingly, the interaction between pGBK-p5 and pGAD-p3, but not the interaction between pGAD-p5 and pGBK-p3, could be detected at the 1:104 dilutions (Fig. 2B). Following the method used previously for the Y2H assay, the interaction between the three deletion mutants of p5 and p3 was analyzed. The results showed that p3 interacts with both p5N and p5C, but no interaction was detected between p3 and p5M (Fig. 2C). Taken together, these observations indicated that RGSV p5 not only interacts with itself but also interacts with protein p3 through its N-terminal and C-terminal regions in yeast.
The RGSV p5 protein interacts with itself and p3 in yeast. (A). Yeast strain AH109 was transformed with the bait plasmid pGBKT7 (pGBK) carrying p5 or p3 together with the prey plasmid pGADT7 (pGAD) carrying p3 or p5. Transformants were selected on TDO, QDO and QDO-X-α-gal minimal medium: 1) pGAD-p5/pGBK-p5, 2) pGBK-p5/pGAD-p3, 3) pGAD-p5/pGBK-p3, 4) pGBKT7-Lam/pGADT7-T, and 5) pGBKT7-53/pGADT7-T. Samples 4 and 5 were used, respectively, as negative and positive controls. (B) Concentration gradient tests examining the different interaction strength between pGBK-p5/pGAD-p3 (top) and AD-p5/BD-p3 interaction (bottom) were conducted as described in the text. (C) p5N and p5C interacted with p3 but not p5M. p5N, p5M and p5C are truncated mutants of p5 corresponding to the N-, middle, and C-terminal portions of the protein, respectively. p5N, amino acids residues 1-99; p5M, amino acids residues 49-140; p5C, amino acids residues 94-191
p5 interacts with itself and p3 in plant cells
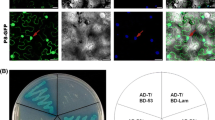
A bimolecular fluorescence complementation (BiFC) assay was conducted to confirm the p5-p5 and p5-p3 interactions. pYFPN-p5 and pYFPC-p5, pYFPN-p3 and pYFPC-p5 or pYFPN-p5 and pYFPC-p3 were used to co-agroinfiltrate N. benthamiana plants. By 48 hours post-infiltration (hpi), the strong fluorescence of yellow fluorescent protein (YFP) in cells co-infiltrated with pYFPN-p5/pYFPC-p5 or pYFPN-p3/pYFPC-p5 was observed under a confocal microscope. Similar results were obtained when p3 was fused with the C-terminal and p5 with the N-terminal fragment of YFP (Fig. 3). A construct expressing SV40NLS-mCherry was cotransfected to label the nucleus of N. benthamiana epidermal cells. The BiFC assay indicated that the self-interaction of p5 occurred in the nucleus and plasma membrane (PM) of N. benthamiana epidermal cells, whereas the interaction between p5 and p3 was not fully consistent with the interaction between p5 and itself. The p5-p3 interaction was mostly found in the cytoplasm in granular structures. Nevertheless, using the BiFC assay, we confirmed the self-interaction of p5 and the p5-p3 interaction.
Bimolecular fluorescence complementation analysis showing p5-p5- interaction, primarily in the nuclei of plant cells, and p5-p3- interaction, mainly in cytoplasmic granular structures. pYFPN-p5/pYFPC-p5, pYFPN-p3/pYFPC-p5, and pYFPN-p5/pYFPC-p3 were co-expressed in N. benthamiana through agroinfiltration. Fluorescence was detected by confocal microscopy at 48 hours post-infiltration (hpi). Cells were co-transfected with a construct expressing SV40NLS-mCherry was to label the nucleus. These experiments were repeated three times with similar results. The scale bars represent 12 μm. YFP, yellow fluorescent protein; BF, bright field
Subcellular co-localization of p5 and p3 protein
To further characterize the p5-p3 protein complex in plant cells, a subcellular co-localization assay was conducted. N. benthamiana plants were co-agroinfiltrated with YFP-p3, CFP-p5 or YFP-p3/CFP-p5. The cells were cotransfected a construct expressing SV40NLS-mCherry with to label the nucleus of N. benthamiana epidermal cells. By 48 hpi, cyan fluorescence was exclusively observed in the nucleus and the plasma membrane of N. benthamiana epidermal cells infiltrated with CFP-p5. This localization of RGSV p5 is consistent with the localization of RGSV p3, which was also mainly observed in the nucleus and the plasma membrane in N. benthamiana epidermal cells infiltrated with YFP-p3. Moreover, the co-localization of p5 and p3 was monitored in N. benthamiana leaf epidermal cells co-expressing YFP-p3/CFP-p5 (Fig. 4). The subcellular co-localization assay showed that RGSV p5 has a localization pattern similar to that of RGSV protein p3, which also indirectly suggested a p5-p3 interaction.
Subcellular localization assay exhibiting the localization of p5 and p3 in the cells of N. benthamiana plants. N. benthamiana plants were infiltrated with Agrobacterium harboring plasmids as indicated at the left of the images. Fluorescence was detected by confocal microscopy at 48 hours post-infiltration (hpi). Cells were co-transfected with a construct expressing SV40NLS-mCherry to label the nucleus. The yellow or cyan fluorescence indicates accumulation of p3: YFP fusion or p5: CFP fusion in the nucleus and the plasma membrane in N. benthamiana epidermal cells. These experiments were repeated three times with similar results. The scale bars represent 12 μm (color figure online)
Co-immunoprecipitation assay confirming the p5-p3-interaction
The p5-p3-interaction was confirmed by a co-immunoprecipitation assay in vivo. N. benthamiana plants were co-agroinfiltrated with RGSV HA-p3 and RGSV p5-Myc or HA-GFP and RGSV p5-Myc. As shown in Fig. 5, p5-Myc, with a size of almost 22 kDa, was significantly enriched in the HA-p3 precipitates (lane 1), but not the HA-GFP precipitate (lane 2), pulled down by using anti-HA-tag mouse monoclonal antibody (Fig. 5, top). Furthermore, using the Western blotting with the anti-HA-tag mouse monoclonal antibody, the expression of HA-p3 (lane 1) and HA-GFP (lane 2), of almost 23 kDa and 28 kDa in size, respectively, were detected from the anti-HA immunoprecipitate (Fig. 5, middle). Additionally, the expression of RGSV p5-Myc in leaves of N. benthamiana was verified by Western blotting with anti-Myc antibodies (Fig. 5, bottom). As expected, these results also suggested that RGSV p5 can associate with RGSV p3 in plant cells.
Co-immunoprecipitation assay confirming the p5-p3 interaction. Total proteins were extracted from N. benthamiana leaves co-expressing RGSV HA-p3 together with RGSV p5-Myc and leaves co-expressing HA-GFP and RGSV p5-Myc. HA-GFP/RGSV p5-Myc was used as a negative control. The immune complexes were pulled down by using anti-HA-tag mouse monoclonal antibody, and subsequently, the 22-kDa RGSV p5-Myc was detected by Western blotting with an anti-Myc-tag mouse monoclonal antibody from the co-precipitation of RGSV HA-p3 (lane 1, top). HA-tagged RGSV p3 (lane 1) and GFP (lane 2), approximately 23 kDa and 28 kDa in size, respectively, were detected in the anti-HA immunoprecipitate (middle). The expression of p5-Myc (approximately 22 kDa) in co-inoculated leaves of N. benthamiana was detected by Western blotting with an anti-Myc-tag mouse monoclonal antibody (bottom)
p5 and p3 are pathogenicity determinants in the PVX heterologous system
To explore the biological role of p5 and p3 in RGSV infection, the PVX system was applied because the biological roles of proteins encoded by tenuiviruses cannot be investigated directly due to the lack of infectious RSV cDNA clones for reverse genetics studies of gene functions [31]. N. benthamiana plants were agroinfiltrated with PVX empty vector, PVX-p5 or PVX-p3. By 9 dpi, mild symptoms such as chlorosis and necrotic mottling were observed on N. benthamiana plants inoculated with Agrobacterium containing PVX-p5 or PVX-p3. However, plants inoculated with the empty PVX showed no visible symptoms. As shown in Fig. 6, by 12 dpi, severe necrotic spots appeared on the leaves of N. benthamiana plants inoculated with PVX-p5 or PVX-p3, whereas symptoms on N. benthamiana plants inoculated with PVX were much milder than in those inoculated with PVX-p5 or PVX-p3. To analyze the biological role of the p5-p3 interaction in plants, the leaves of N. benthamiana plants were co-inoculated with PVX-p5/PVX-p3. When compared with the symptoms induced by PVX, PVX-p5 or PVX-p3, more-severe symptoms with necrotic spots and leaf deformity was observed in some cases in leaves co-inoculated with PVX-p5 together with PVX-p3 (Fig. 6A). To verify that the symptoms were indeed induced by the expression of RGSV p5 and p3, the accumulation of p5 and p3 was detected by Western blotting (Fig. 6B). RT-qPCR also conducted to examine differences in the expression levels of PVX CP mRNA in leaves infiltrated with PVX, PVX-p3, PVX-p5 and PVX-p3 plus PVX-p5. The results indicated that the PVX CP mRNA accumulation level was about five times higher in plants co-inoculated with PVX-p5/PVX-p3 than in plants inoculated with the empty PVX, and nearly 1.8 times as high as those in plants inoculated with PVX-p5 or PVX-p3 (Fig. 6C). There was a significant difference in the PVX CP mRNA accumulation level between PVX and either PVX-p3 or PVX-p5 (P ≤ 0.05). There was also a highly significant difference between PVX and PVX-p3/PVX-p5 (P ≤ 0.01). These results indicated that either p5 or p3 could enhance the pathogenicity of potato virus X (PVX) in N. benthamiana plants, and it appears that these two proteins could work synergistically by interacting together.
Analysis of the pathogenicity p5 and p3 in the heterologous PVX system. (A) Symptoms on N. benthamiana plants infiltrated with PVX, PVX-p5, PVX-p3 and PVX-p5/PVX-p3, respectively, were observed and photographed at 12 days post-infiltration (dpi). (B) The 22-kDa p5 (top) and 23-kDa p3 (middle) were detected in leaves infiltrated with PVX, PVX-p3, PVX-p5 and PVX-p3/PVX-p5 by Western blotting with anti-p5 and anti-p3 polyclonal antibodies, respectively. Actin protein was used as an internal reference (bottom). (C) RT-qPCR was conducted to detect the relative expression levels of the PVX CP gene at 12 dpi in the indicated plants. Data were analyzed by one-way ANOVA. The average (± standard deviation) values from three biological repeats of qRT-PCR are shown. P ≤ 0.05 indicates a significant difference between the two experimental treatments, and P ≤ 0.01 indicates a highly significant difference between the two experimental treatments
Discussion
It was suggested previously that RGSV should be classified as a member of a new genus because the proteins encoded by RNA5 and RNA6 are distantly related to the corresponding proteins of other members of the genus Tenuivirus [23]. Phylogenetic analysis of the nucleocapsid protein and the 94-kDa virion membrane-glycoprotein-like protein of RGSV, RSV, MStV and RHBV showed that RGSV is a monophyletic group that is set apart from the other members of the genus Tenuivirus [24]. Early studies demonstrated that the properties of the RGSV were probably different from those of other members of the genus Tenuivirus. We found that the RGSV p5 protein shares low sequence similarity with the RSV p3 protein (13.6 % amino acid sequence identity). However, we found that several general characteristics of were shared between RGSV and RSV, the type member of the genus Tenuivirus. First, the RGSV p5 functions as a silencing suppressor with its counterpart, p3 of RSV. Second, the two proteins have a similar subcellular localization pattern. A previous report demonstrated that the RSV p3 has a functional nuclear localization signal (NLS) that consists of four amino acids 173KKRH176 and targets the nucleus of plant cells [31]. We show that RGSV p5 is also localized in the nucleus, and the predicted NLS 100KNKMVNKASTKNCWICSLYKR120 may be required for nuclear localization. Additionally, we found that the two proteins are pathogenicity determinants in the heterologous PVX system, resulting in leaf chlorosis and necrotic mottling in N. benthamiana plants [31]. It has been shown that the two silencing suppressors, RGSV p5 and RSV p3, can interact with themselves [4, 14]. We confirmed that self-interaction of p5 by BiFC and subcellular co-localization assays. However, whether the VSR self-interaction is required for silencing suppressor activity remains unclear. Taken together, we hypothesize that RGSV and RSV have a common ancestor and that their proteins evolved to adapt to the changing environment in different ways, while their silencing suppressors possess a conserved mechanism to defend against host RNA silencing.
In a previous study conducted by Chomchan and colleagues, no interaction between p5 and p3 was reported [4]. The most plausible explanation to this discrepancy is that different Y2H systems were employed in their study and our study [1]. Another interesting observation is that the interaction between pGBK7-p5 and pGAD7-p3 was much stronger than the interaction between pGAD7-p5 and pGBK7-p3. This was also observed by Sen and colleagues in their study on the interactions between the nucleocapsid protein (NP) of RSV and its mutants [14]. This was perhaps due to the PPIs being affected by the position of the fusion, which may have an effect on protein folding or the exposure of binding sites. Also, there is a discrepancy between the subcellular localizations of p5-p3 interactions based on the BiFC assay and the co-localization assay. In the BiFC assay, it appeared that the p5-p3 interaction changed the cellular localization pattern of p5. In the subcellular co-localization assay, however, this alteration in the cellular localization of p5 in the presence of p3 was not observed. We also confirmed this result by expressing the HA-tagged p3 together with p5 in N. benthamiana plants, which showed that the localization of p5 alone is in agreement with that of the HA-tagged p3 together with p5. This suggests that p3 cannot change the subcellular localization pattern of p5 in the subcellular localization assay. These experiments were repeated three times with similar results. A possible explanation is that the expression vectors used in these experiments and the principle of the two assays are quite different. The results obtained with the BiFC assay were more reliable because the fluorescence can be monitored only if p3 and p5, fused with the two protein fragments of YFP, interacted in N. benthamiana. Another possibility is that p5 and p3 fused with full-length fluorescent proteins in the subcellular localization assay are expressed constitutively.
Although the RGSV p5 protein shares several similar characteristics with RSV p3, it is possible that p5 has some specific functions. In our study, p5 interacts with another viral protein, p3, through the N-terminus and C-terminus. It remains to be determined whether p3 has an effect on the silencing suppressor activity and localization pattern of p5 by interacting with it. The silencing suppressor activity of p5 was not affected by p3 (Fig. 1A), and neither was the subcellular localization pattern of p5 (Fig. 4). The biological function of the p5-p3 interaction is still unclear, but we found that p3 is also a pathogenicity determinant in the heterologous PVX system. The co-expression of p5 and p3 in N. benthamiana plants caused a more serious infection, which suggested that the pathogenicity effects of p5 and p3 are additive. Ongoing work in our laboratory has shown that p3 is also a pathogenic factor in the native host of RGSV. Transgenic rice plants overexpressing p3 show an apparent alteration in plant growth and morphology (data not published). Thus, it is important to investigate whether p5 and p3 work synergistically to generate the conditions for virus infection, replication, and induction of symptoms in rice.
References
Brückner A, Polge C, Lentze N, Auerbach D, Schlattner U (2009) Yeast two-hybrid, a powerful tool for systems biology. Int J Mol Sci 10:2763–2788
Causier B, Davies B (2002) Analysing protein-protein interactions with the yeast two-hybrid system. Plant Mol Biol 50:855–870
Chomchan P, Miranda G, Shirako Y (2002) Detection of rice grassy stunt tenuivirus nonstructural proteins p2, p5 and p6 from infected rice plants and from viruliferous brown planthoppers. Arch Virol 147:2291–2300
Chomchan P, Li S-F, Shirako Y (2003) Rice grassy stunt tenuivirus nonstructural protein p5 interacts with itself to form oligomeric complexes in vitro and in vivo. J Virol 77:769–775
Du P, Wu J, Zhang J, Zhao S, Zheng H, Gao G, Wei L, Li Y (2011) Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. PLoS Pathog 7:e1002176
Du Z, Xiao D, Wu J, Jia D, Yuan Z, Liu Y, Hu L, Han Z, Wei T, Lin Q, Wu Z, Xie L (2011) p2 of rice stripe virus (RSV) interacts with OsSGS3 and is a silencing suppressor. Mol Plant Pathol 12:808–814
Falk BW, Tsai JH (1998) Biology and molecular biology of viruses in the genus Tenuivirus. Annu Rev Phytopathol 36:139–163
Gietz RD, Woods RA (1998) Transformation of yeast by the lithium acetate/single-stranded carrier DNA/PEG method. Methods Microbiol 26:53–66
Gietz RD, Schiestl RH (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2:31–34
Goodin MM, Zaitlin D, Naidu RA, Lommel SA (2008) Nicotiana benthamiana: its history and future as a model for plant-pathogen interactions. Mol Plant Microbe Interact 21:1015–1026
Hibino H (1996) Biology and epidemiology of rice viruses. Annu Rev Phytopathol 34:249–274
Hiraguri A, Netsu O, Shimizu T, Uehara-Ichiki T, Omura T, Sasaki N, Nyunoya H, Sasaya T (2011) The nonstructural protein pC6 of rice grassy stunt virus trans-complements the cell-to-cell spread of a movement-defective tomato mosaic virus. Arch Virol 156:911–916
Li J, Xue J, Zhang H-M, Yang J, Lv M-F, Xie L, Meng Y, Li P-P, Chen J-P (2013) Interactions between the P6 and P5-1 proteins of southern rice black-streaked dwarf fijivirus in yeast and plant cells. Arch Virol 158:1649–1659
Lian S, Cho WK, Jo Y, Kim S-M, Kim K-H (2014) Interaction study of rice stripe virus proteins reveals a region of the nucleocapsid protein (NP) required for NP self-interaction and nuclear localization. Virus Res 183:6–14
Lian SJM, Cho WK, Choi HS, Je YH, Kim KH (2011) Generation of antibodies against Rice stripe virus proteins based on recombinant proteins and synthetic polypeptides. Plant Pathol J 27:37–43
Lu L, Du Z, Qin M, Wang P, Lan H, Niu X, Jia D, Xie L, Lin Q, Xie L, Wu Z (2009) Pc4, a putative movement protein of Rice stripe virus, interacts with a type I DnaJ protein and a small Hsp of rice. Virus Genes 38:320–327
Mayo M, De Miranda J, Falk B, Goldbach R, Haenni A, Toriyama S (2000) Genus Tenuivirus. Virus taxonomy. Academic Press, New York, pp 904–908
Miranda GJ, Azzam O, Shirako Y (2000) Comparison of nucleotide sequences between northern and southern Philippine isolates of rice grassy stunt virus indicates occurrence of natural genetic reassortment. Virology 266:26–32
Qi Y, Denli AM, Hannon GJ (2005) Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell 19:421–428
Rivera C, Ou S, Iida T (1966) Grassy stunt disease of rice and its transmission by the planthopper Nilaparvata lugens Stal. Plant Dis Rep 50:453–456
Satoh K, Yoneyama K, Kondoh H, Shimizu T, Sasaya T, Choi I-R, Yoneyama K, Omura T, Kikuchi S (2013) Relationship between gene responses and symptoms induced by Rice grassy stunt virus. Front Microbiol 4:313
Sun FYX, Zhou T, Fan Y, Zhou Y (2011) Arabidopsis is susceptible to rice stripe virus infections. J Phytopathol 159:767–772
Toriyama S, Kimishima T, Takahashi M (1997) The proteins encoded by rice grassy stunt virus RNA5 and RNA6 are only distantly related to the corresponding proteins of other members of the genus Tenuivirus. J Gen Virol 78:2355–2363
Toriyama S, Kimishima T, Takahashi M, Shimizu T, Minaka N, Akutsu K (1998) The complete nucleotide sequence of the rice grassy stunt virus genome and genomic comparisons with viruses of the genus Tenuivirus. J Gen Virol 79:2051–2058
Usugi HHT, Oraura T, Tsuchizaki T, Shohara K, Iwasaki M (1985) Rice grassy stunt virus: a planthopper-borne circular filament. Phytopathology 75:894–899
Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33:949–956
Wu G, Wang J, Yang Y, Dong B, Wang Y, Sun G, Yan C, Yan F, Chen J (2014) Transgenic rice expressing rice stripe virus NS3 protein, a suppressor of RNA silencing, shows resistance to rice blast disease. Virus Genes 48:566–569
Wu J, Wang C, Du Z, Cai L, Hu M, Wu Z, Li Y, Xie L (2011) Identification of Pns12 as the second silencing suppressor of Rice gall dwarf virus. Sci China Life Sci 54:201–208
Wu J, Yang Z, Wang Y, Zheng L, Ye R, Ji Y, Zhao S, Ji S, Liu RF, Xu L, Zheng H, Zhou Y, Zhang X, Cao X, Xie L, Wu Z, Qi Y, Li Y (2015) Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife 4. doi:10.7554/eLife.05733
Wydro M, Kozubek E, Lehmann P (2006) Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotiana benthamiana. Acta Biochim Pol 53:289
Xiong R, Wu J, Zhou Y, Zhou X (2009) Characterization and subcellular localization of an RNA silencing suppressor encoded by rice stripe tenuivirus. Virology 387:29–40
Ye R, Wang W, Iki T, Liu C, Wu Y, Ishikawa M, Zhou X, Qi Y (2012) Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Mol Cell 46:859–870
Zheng L, Du Z, Lin C, Mao Q, Wu K, Wu J, Wei T, Wu Z, Xie L (2014) Rice stripe tenuivirus p2 may recruit or manipulate nucleolar functions through an interaction with fibrillarin to promote virus systemic movement. Mol Plant Pathol
Acknowledgments
This work was supported by grants from the National Basic Research Program 973 (2014CB138402 and 2013CBA01403), Natural Science Foundation of China (31272018, 31201491 and 31171821) and Natural Science Foundation of Fujian Province of China (2013J01089). We thank Professor James L. Starr (Texas A&M University) and Doctor Zhenguo Du for helping in correcting the English of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
C. Zhang and X. Liu contributed equally to the report.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, C., Liu, Xj., Wu, Kc. et al. Rice grassy stunt virus nonstructural protein p5 serves as a viral suppressor of RNA silencing and interacts with nonstructural protein p3. Arch Virol 160, 2769–2779 (2015). https://doi.org/10.1007/s00705-015-2560-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-015-2560-6