Abstract
Southern rice black-streaked dwarf virus (SRBSDV) is a recently described member of the genus Fijivirus, family Reoviridae. The roles of the proteins encoded by the SRBSDV genome have rarely been studied. In a yeast two-hybrid (YTH) assay in which SRBSDV P6, a putatively multifunctional protein, was used as bait and an SRBSDV cDNA library was used as prey, there was a strong interaction between the P6 and P5-1 proteins. The interaction was confirmed by bimolecular fluorescence complement (BiFC) assay in plant cells. YTH analysis using truncated mutants showed that the N-terminal region (amino acids 9-231) of P5-1 is necessary for binding P5-1 to P6 and that the N-terminal fragment (amino acids 1-93) of P6 is necessary for its interaction with P5-1. SRBSDV P5-1 formed granules positioned at the cell periphery in Nicotiana benthamiana leaves; P6 was present in both the cytoplasm and the nucleus and formed punctate bodies associated with the cell periphery. Immunogold labeling showed that both P6 and P5-1 localized within viroplasms in infected cells of rice plants. These results suggest that the interaction between P5-1 and P6 of SRBSDV may be involved in the formation of viroplasms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Southern rice black-streaked dwarf virus (SRBSDV) (or rice black-streaked dwarf virus 2), is a recently described member of the genus Fijivirus [1, 2], a genus of plant-infecting viruses in the family Reoviridae [3]. Like all known members of the genus, it can be propagatively transmitted to its hosts in a persistent manner by planthoppers and contains ten linear genomic segments of double-stranded RNA (dsRNA), ranging in size from approximately 1.4 to 4.5 kb and named S1-S10 according to their migration in PAGE. SRBSDV is a candidate member of Fijivirus group 2, which currently has four recognized species, Rice black-streaked dwarf virus(RBSDV), Maize rough dwarf virus (MRDV), Mal de Rio Cuarto virus (MRCV), and Pangola stunt virus (PaSV) [4]. PaSV has not been studied recently, but the remaining viruses of group 2 are very closely related. They have similar particle morphology, genomic profiles, and serological relationships [1, 5–8] and cause similar symptoms on maize plants [9–11]. The best-studied member of the group is RBSDV: the complete genomic sequences of two isolates have been determined [6, 7, 12], and the functions of some viral genes have been identified [13–18]. SRBSDV was first reported in 2001 in Yangjiang City, Guangdong Province, China, and was initially considered an isolate of RBSDV because of its particle morphology and serological similarity to RBSDV [19], but it is now considered distinct on the basis of sequence analysis and differences in vector specificity. The complete genomes of two SRBSDV isolates from Guangdong and Hainan Provinces, China, have been sequenced [20]. Most genome segments contain one open reading frame (ORF), but S5, S7 and S9 each contain two ORFs. Not much is known about the functions of the proteins encoded by the genome segments of SRBSDV. Similar to their homologues in RSBDV, the P7-1 and P9-1 proteins of SRBSDV are involved in the formation of tubular structures and viroplasms, respectively, and P6 is a silencing suppressor [13, 21–23, 38]. S6 is the least conserved of the fijivirus genome segments and is thought to encode a nonstructural protein that may be involved in several processes, such as viroplasm nucleation, virus morphogenesis, and virus pathogenicity [7, 17, 24]. S5 contains one major ORF (P5-1) and a partially overlapping ORF (P5-2), but the functions of these proteins remain unknown [20]. Genomic comparisons have shown that S1-S4, S8, and S10 of SRBSDV encode putative structural proteins: the RNA-dependent RNA polymerase, the major core structural protein, capping enzyme, the outer shell B-spike protein, the minor core protein, and the major outer capsid protein, respectively [1, 2, 20]. Viral protein-protein interactions play important roles in the life cycle of plant viruses, including genomic replication and packaging, virion assembly, cell-to-cell movement, and long-distance transport [25–27]. Here, we used SRBSDV P6 as bait and an SRBSDV cDNA library as prey and found a strong interaction between the P6 and P5-1 proteins. We then confirmed and investigated the interaction using bimolecular fluorescence complementation (BiFC) in plants.
Materials and methods
Construction of recombinant plasmids
The cDNA library of SRBSDV Vietnam isolate was constructed as described previously [1, 20]. The yeast vectors pGBKT7 and pGADT7 and the control plasmids pGADT7-T, pGBKT7-53 and pGBKT7-Lam were used for yeast two-hybrid (YTH) assays (Clontech, Palo Alto, CA). The binary expression vector pCV-GFP-N1, which harbours the eGFP-encoding gene, and the BiFC vectors pCV-nYFP-C and pCV-cYFP-C (for split YFP N-terminal/C-terminal fragment expression) were kindly provided by Dr. Fei Yan [28]. The primers used for construction of recombinant plasmids are listed in Table 1.
To construct plasmids for YTH analysis, the coding sequences of the intact SRBSDV P6 protein and its truncated mutants P6M1 (amino acids 1-661), P6M2 (amino acids 204-794), P6M3 (amino acids 1-359), P6M4 (amino acids 1-286), P6M5 (amino acids 153-359), P6M6 (amino acids 1-150), P6M7 (amino acids 1-93), P6M8 (amino acids 1-70) and P6M9 (amino acids 20-93) were amplified separately using primer pairs P6F/P6R, P6F/P6M1R, P6M2F/P6R, P6F/P6M3R, P6F/P6M4R, P6M5F/P6M3R, P6F/P6M6R, P6F/P6M7R, P6F/P6M8R and P6M9F/P6M7R, respectively. The products were then inserted into the NdeI/BamHI sites of pGADT7, creating the recombinant plasmids pGADT7-P6, pGADT7-P6M1, pGADT7-P6M2, pGADT7-P6M3, pGADT7-P6M4, pGADT7-P6M5, pGADT7-P6M6, pGADT7-P6M7, pGADT7-P6M8 and pGADT7-P6M9, respectively.
The coding sequences of the full-length SRBSDV P5-1 protein and its truncated mutants P5-1-1 (amino acids 1-540), P5-1-2 (amino acids 479-940), P5-1-1M1 (amino acids 1-392), P5-1-1M2 (amino acids 203-540), P5-1-1M3 (amino acids 1-257), P5-1-1M4 (amino acids 1-231), P5-1-1M5 (amino acids 1-202), P5-1-1M6 (amino acids 9-231) and P5-1-1M7 (amino acids 22-231) were amplified separately using primer pairs P5-1F/P5-1R, P5-1-1F/P5-1-1R, P5-1-2F/P5-1R, P5-1-1F/P5-1-1M1R, P5-1-1M2F/P5-1-1R, P5-1-1F/P5-1-1M3R, P5-1-1F/P5-1-1M4R, P5-1-1F/P5-1-1M5R, P5-1-1M6F/P5-1-1M4R and P5-1-1M7F/P5-1-1M4R, respectively. The PCR fragments were digested with BamHI/PstI or NdeI/BamHI and cloned into the pGBKT7 vector to generate the recombinant plasmids pGBKT7-P5-1, pGBKT7-P5-1-1, pGBKT7-P5-1-2, pGBKT7-P5-1-1M1, pGBKT7-P5-1-1M2, pGBKT7-P5-1-1M3, pGBKT7-P5-1-1M4, pGBKT7-P5-1-1M5, pGBKT7-P5-1-1M6 and pGBKT7-P5-1-1M7, respectively.
For BiFC analysis, the full-length and truncated mutant (amino acids 9-231) of SRBSDV P5-1 were amplified by PCR, using primer pairs Y5-1F/Y5-1R and Y5-1-1M6F/Y5-1-1M6R, respectively; the full-length and truncated mutant (amino acids 1-93) of SRBSDV P6 were amplified by PCR, using primer pairs Y6F/Y6R and Y6F/Y6M7R, respectively. PCR products were cloned into pCV-nYFP-C as a fusion with the N-terminal fragment of YFP or pCV-cYFP-C as a fusion with the C-terminal fragment of YFP via the BamHI/SacI sites, forming pCV-nYFP-P5-1, pCV-nYFP-P6, pCV-nYFP-P5-1-1M6, pCV-nYFP-P6M7, pCV-cYFP-P5-1, pCV-cYFP-P6, pCV-cYFP-P5-1-1M6 and pCV-cYFP-P6M7, respectively.
For subcellular localization, the full-length coding sequences of SRBSDV P5-1 and P6 were amplified by PCR using the primer pairs Y5-1F/G5-1R and Y6F/G6R, respectively. The products were subsequently digested with BamHI/KpnI and ligated into the corresponding sites of pCV-GFP-N1, generating recombinant plasmids pCV-P5-1-GFP and pCV-P6-GFP, respectively.
These PCR amplifications were conducted using LA Taq polymerase (TaKaRa Bio, Dalian, China), and the PCR amplification program was as follows: preheating for one cycle of 3 min at 94 °C; 30 cycles of 1 min at 94 °C, 50 s at 58 °C, 1-3 min at 72 °C; and a final extension at 72 °C for 10 min.
Yeast two-hybrid assays
YTH assays were performed using the Matchmaker™ Gold Yeast Two-Hybrid System (Clontech, Palo Alto, CA), according to the manufacturer’s protocols. Saccharomyces cerevisiae strain Y2HGold was co-transformed with bait and prey plasmids using the small-scale lithium acetate method [37]. The two plasmids supplied with the kits, pGBKT7-53 and pGADT7-T, were used as positive controls for co-transformation, whereas pGBKT7-Lam and pGADT7-T, pGBKT7 and pGADT7-P6, and pGBKT7-P5-1 and pGADT7 were used as negative controls for co-transformation. Co-transformants were first plated on synthetically defined (SD) medium lacking adenine, histidine, leucine, and tryptophan (SD/-Ade/-His/-Leu/-Trp), and positive yeast colonies that grew on the auxotrophic medium were then tested for α-galactosidase activity on SD/-Ade/-His/-Leu/-Trp medium supplemented with X-α-Gal and aureobasidin A (SD/-Ade/-His/-Leu/-Trp/X-α-Gal/AbA). All experiments were done at least three times.
Subcellular localization and BiFC assay
Agrobacterium tumefaciens strain C58C1 was transformed with different binary plasmids by electroporation. Agroinfiltration was done as described [29] with a few modifications. Briefly, cultures of C58C1 harbouring the relevant binary plasmid were grown in YEP medium containing rifampicin (50 μg/ml) and kanamycin (100 μg/ml) at 28 °C for 16 h. For subcellular localization, C58C1 strains containing recombinant plasmids pCV-P5-1-GFP and pCV-P6-GFP were resuspended and diluted to an OD600 of 0.6 with infiltration medium (10 mM MES, pH 5.6, 10 mM MgCl2, 200 mM acetosyringone). For the BiFC assay, the plasmid combinations pCV-nYFP/pCV-cYFP-P5-1, pCV-nYFP-P6/pCV-cYFP, pCV-nYFP/pCV-cYFP-P6, and pCV-nYFP-P5-1/pCV-cYFP were used as negative controls, and Agrobacterium cultures containing the BiFC plasmids were resuspended at a final OD600 of 0.8:0.8. The cells were incubated at room temperature for 2 to 4 h and then used to infiltrate 5- to 6-week-old Nicotiana benthamiana leaves. Underside epidermal cells of infiltrated leaves were assayed for fluorescence by confocal microscopy 48-72 h after infiltration [30].
Confocal laser scanning microscopy
Fluorescence analysis was performed using a Leica TCS SP5 confocal laser scanning microscope (Leica Microsystems, Heidelberg, Germany) with an argon laser. GFP was excited at 488 nm, and the emitted light was captured between 500-550 nm. YFP was excited at 514 nm, and the emitted light was captured between 530-600 nm.
Immuno-electron microscopy
For immunogold labeling, tissue samples from SRBSDV-infected or healthy rice plants were cut into small pieces and then fixed with a mixture of 4 % paraformaldehyde and 0.1 % glutaraldehyde for 1 h at 4 °C after a brief vacuum infiltration. The fixed samples were rinsed three times in phosphate buffer (PB) and dehydrated through a graded ethanol series (50 %, 70 %, 90 % and 100 %) at −20 °C. After a graded infiltration of resin (30 %, 70 % in ethanol) and pure Lowicryl K4M resin (Electron Microscopy Sciences, Fort Washington, PA), samples were polymerized under UV light (360 nm) for 72 h at −20 °C and then for 48 h at room temperature. Ultrathin sections were cut using a glass knife with a UC6 microtome (Leica, Vienna, Austria) and collected on nickel grids. For immunogold labeling, sections were first blocked with blocking buffer for 30 min at room temperature. The blocking buffer was 50 mM PB, pH 6.8, containing 1 % (wt/vol) bovine serum albumin (BSA) and 0.02 % polyethylene glycol 2000. Grids were then incubated for 2 h at 37 °C with polyclonal antibodies raised against P5-1 or P6 proteins, followed by 10-nm IgG-gold conjugate antibody (Sigma St. Louis, MO) for 2 h at 37 °C. After each antibody treatment grids were washed three times with water to remove nonspecific binding. Sections incubated with blocking buffer followed by IgG-gold were used as labeling controls. Finally, sections were stained with uranyl acetate for 15 min and then with lead citrate for 15 min before being examined under a H-7650 transmission electron microscope (TEM, Hitachi, Akagi, Japan) at 80 kV of accelerating voltage. Photographs were taken with a Gatan 830 CCD camera (Gatan, USA).
Results
Interaction between P5-1 and P6 in yeast cells
Using P6 as bait and an SRBSDV cDNA library as prey in a yeast two-hybrid (Y2H) assay, 267 independent clones were recovered following growth on selective media. Sequencing showed that 223 of these clones corresponded to the first open reading frame (P5-1) of genomic segment S5. The full-length coding regions of genome segments S5 and S6 were sequenced and deposited in the GenBank/EMBL/DDBJ databases with the accession numbers HQ731496 and HQ731497, respectively. To test the interaction of SRBSDV P5-1 with P6 in yeast, S. cerevisiae Y2HGold cells were co-transformed with pGBKT7-P5-1 and pGADT7-P6 plasmids containing the full-length coding region of SRBSDV P5-1 and P6 after eliminating the autoactivation and toxicity. The resultant transformants were selected on SD/-Ade/-His/-Leu/-Trp/X-α-Gal/AbA medium. Only transformants of pGBKT7-P5-1/pGADT7-P6 and the positive control grew well and turned blue. In contrast, no growth was observed in the negative controls (Fig. 1). The results confirmed a strong interaction between SRBSDV P5-1 and P6 in yeast cells.
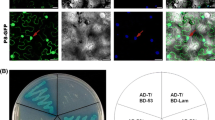
Interaction of SRBSDV P5-1 with P6 in transformed S. cerevisiae Y2HGold cells grown on SD/-Ade/-His/-Leu/-Trp/X-α-Gal/AbA. Segment 1, pGBKT7-53/pGADT7-T (positive control); segment 2, pGBKT7-Lam/pGADT7-T (negative control); segment 3, pGBKT7-P5-1/pGADT7-P6; segment 4, pGBKT7-P5-1/pGADT7 (negative control); and segment 5, pGBKT7/pGADT7-P6 (negative control)
Interaction between P5-1 and P6 in plant cells
Bimolecular fluorescence complementation (BiFC) assays were performed to confirm the interaction between P6 and P5-1 in living plant cells. The full-length coding sequences of P5-1 and P6 were cloned into BiFC transformation vectors pCV-nYFP-C and pCV-cYFP-C, and two pairs of combinations, pCV-nYFP-P6/pCV-cYFP-P5-1 and pCV-nYFP-P5-1/pCV-cYFP-P6, were generated. These plasmid combinations were used to agroinfiltrate N. benthamiana leaves. Three days after agroinfiltration, the infiltrated N. benthamiana leaves were examined for YFP fluorescence using laser confocal scanning microscopy. As shown in Fig. 2 (panels A and B), strong reconstitution of YFP fluorescence was detected in leaf epidermal cells of N. benthamiana co-infiltrated with pCV-nYFP-P6/pCV-cYFP-P5-1 or pCV-nYFP-P5-1/pCV-cYFP-P6. No fluorescence was observed in leaves co-infiltrated with pCV-nYFP/pCV-cYFP-P5-1, pCV-nYFP-P6/pCV-cYFP, pCV-nYFP/pCV-cYFP-P6, or pCV-nYFP-P5-1/pCV-cYFP (Fig. 2C, D, E, F). These results confirmed that there were strong interactions between P5-1 and P6 in plant cells.
Interaction of SRBSDV P5-1 with P6 in living plant cells. N. benthamiana leaves were co-infiltrated with pCV-nYFP-P6/pCV-cYFP-P5-1 (A), pCV-nYFP-P5-1/pCV-cYFP-P6 (B), pCV-nYFP/pCV-cYFP-P5-1 (C), pCV-nYFP-P6/pCV-cYFP (D), pCV-nYFP/pCV-cYFP-P6 (E) or pCV-nYFP-P5-1/pCV-cYFP (F). The white arrow points to a granule. The results were observed 72 h after infiltration. Scale bar, 25 μm
The reconstituted granular YFP fluorescence observed was randomly distributed in the cytoplasm and ranged from 0.5 to 10 μm in diameter (Fig. 2A, B). No fluorescence was seen in the nuclei. The fluorescence appeared similar to the viroplasm-like structures (VLS) that were formed when MRCV P9-1 was expressed in Sf9 cells [31] or when RBSDV P6 was expressed in onion epidermal cells [17].
The region of SRBSDV P5-1 necessary for its interaction with P6
To analyze the domains of SRBSDV P5-1 that mediate its interaction with P6, native P5-1 protein was divided into two fragments, P5-1-1 (amino acids 1-540) and P5-1-2 (amino acids 479-940), as shown in Fig. 3A. Both fragments were subcloned into pGBKT7 and used together with pGADT7-P6 to transform S. cerevisiae Y2HGold cells. As shown in Fig. 3B, yeast cells co-transformed with the plasmid pair pGBKT7-P5-1-1/pGADT7-P6 grew well and turned blue on SD/-Ade/-His/-Leu/-Trp/X-α-Gal/AbA, similar to those co-transformed with pGBKT7-P5-1/pGADT7-P6, while no visible growth was observed for cells co-transformed with pGBKT7-P5-1-2/pGADT7-P6, indicating that only the N-terminal part of P5-1 contains the domains that are indispensable for interacting with P6.
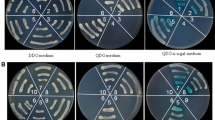
Mapping of the P5-1 region involved in the P5-1-P6 interaction. (A) Schematic representation of P5-1 mutants: P5-1, full-length P5-1 (amino acids 1-940); P5-1-1, amino acids 1-540; P5-1-2, amino acids 479-940; P5-1-1M1, amino acids 1-392; P5-1-1M2, amino acids 203-540; P5-1-1M3, amino acids 1-257; P5-1-1M4, amino acids 1-231; P5-1-1M5, amino acids 1-202; P5-1-1M6, amino acids 9-231; P5-1-1M7, amino acids 22-231. The ability of P5-1 mutants to interact with intact P6 in YTH assays is shown on the right (+, positive; −, negative). (B) Interaction of SRBSDV P6 and mutants of P5-1 in transformed S. cerevisiae Y2HGold cells grown on SD/-Ade/-His/-Leu/-Trp/X-α-Gal/AbA. Segment 1, pGBKT7-P5-1/pGADT7-P6; segment 2, pGBKT7-P5-1-1/pGADT7-P6; segment 3, pGBKT7-P5-1-2/pGADT7-P6; segment 4, pGBKT7-P5-1-1M1/pGADT7-P6; segment 5, pGBKT7-P5-1-1M2/pGADT7-P6; segment 6, pGBKT7-P5-1-1M3/pGADT7-P6; segment 7, pGBKT7-P5-1-1M4/pGADT7-P6; segment 8, pGBKT7-P5-1-1M5/pGADT7-P6; segment 9, pGBKT7-P5-1-1M6/pGADT7-P6; segment 10, pGBKT7-P5-1-1M7/pGADT7-P6; segment 11, pGBKT7-53/pGADT7-T (positive control); segment 12, pGBKT7/pGADT7-P6 (negative control)
Similar experiments with a series of truncated mutants showed that interactions were obtained with P5-1-1 fragments comprising amino acids 1-392 (P5-1-1M1), 1-257 (P5-1-1M3) or 1-231 (P5-1-1M4) but not with the fragment comprising amino acids 203-540 (P5-1-1M2) (Fig. 3). This was further refined by showing an interaction with P5-1-1M6 (amino acids 9-231) but not with P5-1-1M5 (amino acids 1-202) or P5-1-1M7 (amino acids 22-231) (Fig. 3). These results show that amino acids 9-231 of P5-1 are crucial for its interaction with P6.
The region of SRBSDV P6 necessary for its interaction with P5-1
To determine the domain of P6 responsible for its interaction with P5-1, nine truncated mutants (P6M1-P6M9; Fig. 4A) were subcloned in pGADT7 and then used together with pGBKT7-P5-1 to transform S. cerevisiae Y2HGold cells. In the YTH assay (Fig. 4B), the interaction with P5-1 was abolished when the 203 amino acid residues in the N-terminal part of P6 (i.e. P6M2 mutant) were removed, but fragments comprising amino acids 1-661 (P6M1), 1-359 (P6M3) or 1-286 (P6M4) were able to interact. This was further refined by showing an interaction with P6M6 (amino acids 1-150) and P6M7 (amino acids 1-93) but not with P6M5 (amino acids 153-359), P6M8 (amino acids 1-70) or P6M9 (amino acids 20-93) (Fig. 4B). Therefore, the region of P6 involved in the interaction with P5-1 was located within the N-terminal fragment (P6M7, amino acids 1-93).
Mapping of the P6 region involved in the P5-1-P6 interaction. (A) Schematic representation of P6 mutants: P6, full-length P6 (amino acids 1-794); P6M1, amino acids 1-661; P6M2, amino acids 204-794; P6M3, amino acids 1-359; P6M4, amino acids 1-286; P6M5, amino acids 153-359; P6M6, amino acids 1-150; P6M7, amino acids 1-93; P6M8, amino acids 1-70; P6M9, amino acids 20-93. The ability of P6 mutants to interact with intact P5-1 in YTH assays is shown on the right (+, positive; −, negative). (B) Interaction of SRBSDV P5-1 and mutants of P6 in transformed S. cerevisiae Y2HGold cells grown on SD/-Ade/-His/-Leu/-Trp/X-α-Gal/AbA. Segment 1, pGBKT7-P5-1/pGADT7-P6; segment 2, pGBKT7-P5-1/pGADT7-P6M1; segment 3, pGBKT7-P5-1/pGADT7-P6M2; segment 4, pGBKT7-P5-1/pGADT7-P6M3; segment 5, pGBKT7-P5-1/pGADT7-P6M4; segment 6, pGBKT7-P5-1/pGADT7-P6M5; segment 7, pGBKT7-P5-1/pGADT7-P6M6; segment 8, pGBKT7-P5-1/pGADT7-P6M7; segment 9, pGBKT7-P5-1/pGADT7-P6M8; segment 10, pGBKT7-P5-1/pGADT7-P6M9; segment 11, pGBKT7-53/pGADT7-T (positive control); segment 12, pGBKT7-P5-1/pGADT7 (negative control)
Interaction between the reactive minimal mutants of SRBSDV P5-1 and P6 in yeast and plant cells
Interaction between the minimal reactive mutants of both proteins (P5-1-1M6 and P6M7) was then confirmed both by YTH assay and BiFC. In YTH (Fig. 5A), transformants of pGBKT7-P5-1-1M6/pGADT7-P6M7 and the positive control pGBKT7-53/pGADT7-T grew well and turned blue on SD/-Ade/-His/-Leu/-Trp/X-α-Gal/AbA medium, while there was no significant cell growth for the negative controls (pGBKT7/pGADT7-P6M7, pGBKT7-P5-1-1M6/pGADT7 and pGBKT7-Lam/pGADT7-T). To confirm the interaction in plant cells, we cloned P5-1-1M6 and P6M7 into BiFC transformation vectors pCV-nYFP-C and pCV-cYFP-C to generate pCV-nYFP-P5-1-1M6, pCV-nYFP-P6M7, pCV-cYFP-P5-1-1M6 and pCV-cYFP-P6M7, respectively. All BiFC constitutive vectors were used individually to transform A. tumefaciens strains C58C1, and then used for co-infiltration of leaves of N. benthamiana. YFP fluorescence was observed at 3 days post-infiltration (dpi) in the cells co-expressing pCV-nYFP-P6M7/pCV-cYFP-P5-1-1M6 and pCV-nYFP-P5-1-1M6/pCV-cYFP-P6M7 (Fig. 5B, C), but not in the control cells co-expressing pCV-nYFP/pCV-cYFP-P5-1-1M6, pCV-nYFP-P6M7/pCV-cYFP, pCV-nYFP/pCV-cYFP-P6M7 or pCV-nYFP-P5-1-1M6/pCV-cYFP (data not shown). These results indicate that the minimal reactive mutant of P5-1 (P5-1-1M6) interacts with the minimal reactive mutant of P6 (P6M7) in both yeast and plant cells.
Interaction between P5-1-1M6 and P6M7 in yeast cells and living plant cells. (A) Interaction of P5-1-1M6 with P6M7 in transformed S. cerevisiae Y2HGold cells grown on SD/-Ade/-His/-Leu/-Trp/X-α-Gal/AbA. Segment 1, pGBKT7-53/pGADT7-T (positive control); segment 2, pGBKT7-Lam/pGADT7-T (negative control); segment 3, pGBKT7-P5-1-1M6/pGADT7-P6M7; segment 4, pGBKT7-P5-1-1M6/pGADT7 (negative control); segment 5, pGBKT7/pGADT7-P6M7 (negative control). (B) and (C) Visualization of P6M7-P5-1-1M6 interaction in N. benthamiana epidermal cells by BiFC assay. N. benthamiana leaves were co-infiltrated with pCV-nYFP-P6M7/pCV-cYFP-P5-1-1M6 (B) or pCV-nYFP-P5-1-1M6/pCV-cYFP-P6M7 (C). The white arrows indicate the positions of the cell nuclei. The results were observed 72 h after infiltration. Scale bar, 25 μm
Subcellular localization of P5-1 and P6 in N. benthamiana leaves
To gain insight into the molecular mechanism of the interaction between SRBSDV P5-1 and P6, we further examined the subcellular localization of both proteins. Constructs expressing P5-1 or P6 fused with eGFP at their C terminus (pCV-P5-1-GFP and pCV-P6-GFP) were constructed and introduced into N. benthamiana epidermal cells by Agrobacterium infiltration. GFP fluorescence was monitored by confocal microscopy at 2 dpi. In the control, the non-fused GFP was generally distributed in the cytoplasm and nucleus (Fig. 6A), but in most of the cells expressing pCV-P5-1-GFP, there were fluorescent granules of various sizes positioned at the cell periphery and fluorescence in the nucleus (Fig. 6B). In cells expressing pCV-P6-GFP, fluorescence was detected in both the cytoplasm and the nucleus. A punctate GFP signal associated with the cell periphery was also observed in infiltrated N. benthamiana epidermal cells (Fig. 6C).
Subcellular localization of SRBSDV P5-1 and P6 proteins in N. benthamiana leaf epidermal cells. GFP fluorescence in N. benthamiana leaves agroinfiltrated with pCV-GFP-N1 (A), pCV-P5-1-GFP (B) and pCV-P6-GFP (C). The white arrows indicate the positions of the cell nuclei. Representative granular bodies are marked by white triangles, and punctate bodies are marked with black triangles. The results were observed 48 h after infiltration. Scale bar, 25 μm
Intracellular localization of P5-1 and P6 in infected rice plants
Previous studies revealed that the cytopathology of SRBSDV-infected plants is typical of fijivirus infections, with virus crystals, viroplasms and tubular structures [2, 9]. Virus particles were arranged in a regular order in crystals and tubular structures, but scattered throughout the viroplasm. To determine the intracellular localization of P5-1 and P6 in infected plant cells, immunogold labeling experiments were performed using polyclonal antibodies against these proteins. Antibodies to both proteins labeled the viroplasms in infected rice cells (Fig. 7). With the P6 antibody, many immunogold particles were observed in amorphous viroplasms with higher electron density (Fig. 7B), suggesting that P6 is a major component of these viroplasms, while the smaller number of particles seen with the antibody to P5-1 suggests that this protein is a minor component (Fig. 7A). None of the structures in healthy rice plants reacted with these antibodies (data not shown).
Discussion
Viroplasms are amorphous electron-dense cytoplasmic inclusions often found in reovirus-infected cells and are thought to be structures where dsRNA is synthesized and packaged into pre-virion core particles [32–34]. There have been many studies of the composition and formation of viroplasms of animal reoviruses but only a few reports on the viroplasms of fijiviruses. In RBSDV, P6 is thought to self-interact to form punctuate VLS in the cytoplasm and to recruit viroplasm-associated protein P9-1 [13, 16–18]. MRCV P9-1 can self-interact and bind single-stranded RNA, has ATPase activity, and is thought to be associated with the formation of viroplasm structures [31, 35, 36]. A recent study has shown that SRBSDV P9-1 is a component of the viroplasm matrix [38]. Here, immunogold labeling revealed that both P5-1 and P6 of SRBSDV were localized in the viroplasm, clearly indicating that P5-1 is also a component of this structure in fijiviruses in addition to P6 and P9-1.
The region of RBSDV P6 from amino acids 395-659, which is predicted to harbour a coiled-coil structure and the ATPase domain of structural maintenance of chromosomes (SMC) proteins [17], is responsible for P6 self-interaction, heterologous interaction with P9-1, and VLS formation [17]. SRBSDV P6 has 43-62 % identity to the corresponding proteins of RBSDV and MRCV [20]. A BLAST search also showed that SRBSDV P6 contains the ATPase domain of SMC proteins, suggesting that these fijivirus proteins are homologous. In transient expression systems, fluorescence appeared as granular structures in the cytoplasm, whether P6 was expressed alone or co-expressed with P5-1, supporting the view that P6 is involved in formation of VLS. This is confirmed by the immunogold labeling of ultrathin sections showing that SRBSDV P6 was specifically enriched within the viroplasm of infected rice cells. However, the reconstituted YFP-fluorescence, formed by co-expression of the minimal truncated mutants of SRBSDV P5-1 and P6 (Fig. 5), did not aggregate into viroplasm-like structures, suggesting that something more than the interaction between the minimal interactive regions is necessary for VLS formation.
We also noted that both P5-1 and P6 were found in nuclei in addition to the cytoplasm when we studied GFP fusion proteins in the leaves of N. benthamiana (Fig. 6). The reconstituted YFP fluorescence formed by the minimal interactive truncated mutants of SRBSDV P5-1 and P6 was also observed in both the nucleus and cytoplasm (Fig. 5). However, the reconstituted YFP-fluorescent granules formed when the entire P5-1 and P6 were co-expressed in the leaves of N. benthamiana were not found in nuclei (Fig. 2), suggesting that the interaction may affect the subcellular localization of the proteins. Co-localization experiments indicated that the interaction between RBSDV P6 and P9-1 proteins can change the distribution pattern of P9-1 in onion cells [17]. It would therefore be interesting to determine whether this change of localization is a universal phenomenon during viroplasm formation in fijiviruses. In addition, interaction between P6 and P9-1 of SRBSDV was also found in a yeast two-hybrid (Y2H) assay, but it was not as strong as the interaction between P6 and P5-1 (data not shown), supporting the conclusion that fijiviral P9-1 is involved in viroplasm formation [17, 31, 35, 36, 38]. Further research on the relationships among P5-1, P6, and P9-1 would be very helpful to provide a deeper insight into viroplasm formation in fijivirus infections.
References
Zhang HM, Yang J, Chen JP, Adams MJ (2008) A black-streaked dwarf disease on rice in China is caused by a novel fijivirus. Arch Virol 153:1893–1898
Zhou GH, Wen JJ, Cai DJ, Li P, Xu DL, Zhang SG (2008) Southern rice black-streaked dwarf virus: A new proposed Fijivirus species in the family Reoviridae. Chin Sci Bull 53:3677–3685
Boccardo G, Milne RG (1984) Plant reovirus group. CMI/AAB, descriptions of plant viruses, No. 294. Association of Applied Biologists, Wellesbourne, Warwick
Attoui H, Mertens PPC, Becnel J, Belaganahalli S, Bergoin M et al (2012) Family Reoviridae. In: King AMQ, Adams MJ, Carstens EJ, Lefkowitz EJ (eds) Virus taxonomy: ninth report of the international committee on taxonomy of viruses. Elsevier Academic Press, London, pp 541–637
Luisoni E, Lovisolo O, Kitagawa Y, Shikata E (1973) Serological relationship between maize rough dwarf virus and rice black streaked dwarf virus. Virology 52:281–283
Zhang HM, Chen JP, Lei JL, Adams MJ (2001) Sequence analysis shows that a dwarfing disease on rice, wheat and maize in China is caused by rice black-streaked dwarf virus. Eur J Plant Pathol 107:563–567
Zhang HM, Chen JP, Adams MJ (2001) Molecular characterization of segments 1 to 6 of rice black-streaked dwarf virus from China provides the complete genome. Arch Virol 146:2331–2339
Marzachi C, Boccardo G, Milne R, Isogai M, Uyeda I (1995) Genome structure and variability of fijiviruses. Semin Virol 6:103–108
Hoang AT, Zhang HM, Yang J, Chen JP, Hébrard E, Zhou GH, Vien NV, Cheng JA (2011) Identification, characterization and distribution of southern rice black-streaked dwarf virus in Vietnam. Plant Dis 95:1063–1069
Li YZ, Cao Y, Zhou Q, Guo HM, Ou GC (2012) The efficiency of southern rice black-streaked dwarf virus transmission by the vector Sogatella furcifera to different host plant species. J Integr Agric 11:621–627
Arneodo JD, Guzman FA, Conci LR, Laguna IG, Truol GA (2002) Transmission features of Mal de Rio Cuarto virus in wheat by its planthopper vector Delphacodes kuscheli. Ann Appl Biol 141:195–200
Wang ZH, Fang SG, Xu JL, Sun LY, Li DW, Yu JL (2003) Sequence analysis of the complete genome of rice black-streaked dwarf virus isolated from maize with rough dwarf disease. Virus Genes 27:163–168
Isogai M, Uyeda I, Lee BC (1998) Detection and assignment of proteins encoded by rice black streaked dwarf fijivirus S7, S8, S9 and S10. J Gen Virol 79:1487–1494
Liu HJ, Wei CH, Zhong YH, Li Y (2007) Rice black-streaked dwarf virus minor core protein P8 is dimeric protein and represses transcription in tobacco protoplasts. FEBS Lett 581:2534–2540
Liu HJ, Wei CH, Zhong YW, Li Y (2007) Rice black-streaked dwarf virus outer capsid protein P10 has self-interactions and forms oligomeric complexes in solution. Virus Res 127:34–42
Zhang CZ, Liu YY, Liu LY, Lou ZY, Zhang HY, Miao HQ, Hu XB, Pang YP, Qiu BS (2008) Rice black streaked dwarf virus P9–1, an α-helical protein, self-interacts and forms viroplasms in vivo. J Gen Virol 89:1770–1776
Wang Q, Tao T, Zhang YJ, Wu WQ, Li DW, Yu JL, Han CG (2011) Rice black-streaked dwarf virus P6 self-interacts to form punctate, viroplasm-like structures in the cytoplasm and recruits viroplasm-associated protein P9–1. Virol J 8:24
Akita F, Higashiura A, Shimizu T, Pu Y, Suzuki M, Uehara-Ichiki T, Sasaya T, Kanamaru S, Arisaka F, Tsukihara T, Nakagawa A, Omura T (2012) Crystallographic analysis reveals octamerization of viroplasm matrix protein P9–1 of rice black streaked dwarf virus. J Virol 86:746–756
Zhou GH, Xu DL, Li HP (2004) Identification of rice black streaked dwarf virus infecting rice in Guangdong. In: Peng YL (ed) Proceedings of the conference on Chinese plant pathology. Agricultural Scientech Press, Beijing, pp 210–212
Wang Q, Yang J, Zhou GH, Zhang HM, Chen JP, Adams MJ (2010) The complete genome sequence of two isolates of southern rice black-streaked dwarf virus, a new member of the genus Fijivirus. J Phytopathol 158:733–737
Zhang LD, Wang ZH, Wang XB, Zhang WH, Li DW, Han CG, Zhai YF, Yu JL (2005) Two virus-encoded RNA silencing suppressors, P14 of beet necrotic yellow vein virus and S6 of rice black streak dwarf virus. Chin Sci Bull 50:305–310
Liu Y, Jia DS, Chen HY, Chen Q, Xie LH, Wu ZJ, Wei TY (2011) The P7–1 protein of southern rice black-streaked dwarf virus, a fijivirus, induces the formation of tubular structures in insect cells. Arch Virol 156:1729–1736
Lu YH, Zhang JF, Xiong RY, Xu QF, Zhou YJ (2011) Identification of an RNA silencing suppressor encoded by southern rice black-streaked dwarf virus S6. Sci Agric Sin 44:2909–2917
Fang S, Wang Z, Han C, Li D, Yu J (2007) Genomic segment 6 of rice black-streaked dwarf virus encodes for a viral non-structural protein. Acta Agric Boreali Sinica 22:5–8
Ueda S, Masuta C, Uyeda I (1997) Hypothesis on particle structure and assembly of rice dwarf phytoreovirus: interactions among multiple structural proteins. J Gen Virol 78:3135–3140
Chowdhury SR, Savithri HS (2011) Interaction of Sesbania mosaic virus movement protein with the coat protein-implications for viral spread. FEBS Lett 278:257–272
Huh SU, Kim MJ, Ham BK, Paek KH (2011) A zinc finger protein Tsip1 controls cucumber mosaic virus infection by interacting with the replication complex on vacuolar membranes of the tobacco plant. New Phytol 191:746–762
Lu Y, Yan F, Guo W, Zheng H, Lin L, Peng J, Adams MJ, Chen J (2011) Garlic virus X 11-kDa protein granules move within the cytoplasm and traffic a host protein normally found in the nucleolus. Mol Plant Pathol 12:666–676
Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1:2019–2025
Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, Harter K, Kudla J (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40:428–438
Maroniche GA, Mongelli VC, Peralta AV, Distéfano AJ, Llauger G, Taboga OA, Hopp EH, del Vas M (2010) Functional and biochemical properties of Mal de Río Cuarto virus (Fijivirus, Reoviridae) P9–1 viroplasm protein show further similarities to animal reovirus counterparts. Virus Res 152:96–103
Silvestri LS, Taraporewala ZF, Patton JT (2004) Rotavirus replication: plus-sense templates for double-stranded RNA synthesis are made in viroplasms. J Virol 78:7763–7774
Netherton C, Moffat K, Brooks E, Wileman T (2007) A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv Virus Res 70:101–182
Netherton CL, Wileman T (2011) Virus factories, double membrane vesicles and viroplasm generated in animal cells. Curr Opin Virol 1:381–387
Guzmán FA, Arneodo JD, Pons AB, Truol GA, Luque AV, Conci LR (2010) Immunodetection and subcellular localization of Mal de Río Cuarto virus P9–1 protein in infected plant and insect host cells. Virus Genes 41:111–117
Maroniche GA, Mongelli VC, Llauger G, Alfonso V, Taboga O, del Vas M (2012) In vivo subcellular localization of Mal de Río Cuarto virus (MRCV) non-structural proteins in insect cells reveals their putative functions. Virology 430:81–89
Gietz RD, Woods RA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350:87–96
Jia D, Chen H, Zheng A, Chen Q, Liu Q, Xie L, Wu Z, Wei T (2012) Development of an insect vector cell culture and RNA interference system to investigate the functional role of fijivirus replication protein. J Virol 86:5800–5807
Acknowledgments
This work was funded by the China 973 Program (2006CB708209), 863 program (2007AA10Z414), National Science and Technology Support Program (2012BAD19B03), the International Science and Technology Cooperation Project (2007DFB30350), the Special Fund for Agro-scientific Research in the Public Interest of China (201003031), the Zhejiang Provincial Science and Technology Project (2010C12027), and the Zhejiang Provincial Foundation for Natural Science (Z305165 and Y3090657). We thank Professor M. J. Adams, Rothamsted Research, Harpenden, UK, for help in correcting the English of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
J. Li and J. Xue contributed equally to this work.
Rights and permissions
About this article
Cite this article
Li, J., Xue, J., Zhang, HM. et al. Interactions between the P6 and P5-1 proteins of southern rice black-streaked dwarf fijivirus in yeast and plant cells. Arch Virol 158, 1649–1659 (2013). https://doi.org/10.1007/s00705-013-1660-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-013-1660-4