Abstract
Klebsiella pneumoniae produces 3-hydroxypropionic acid (3-HP) from glycerol with oxidation of 3-hydroxypropionaldehyde (3-HPA) to 3-HP in a reaction catalyzed by aldehyde dehydrogenase (ALDH). In the present study, two putative ALDHs of K. pneumoniae, YneI and YdcW were identified and characterized. Recombinant YneI was specifically active on 3-HPA and preferred NAD+ as a cofactor, whereas YdcW exhibited broad substrate specificity and preferred NADP+ as a cofactor. Overexpression of ALDHs in the glycerol oxidative pathway-deficient mutant K. pneumoniae AK resulted in a significant increase in 3-HP production upon shake-flask culture. The final titers of 3-HP were 2.4 and 1.8 g L−1 by recombinants overexpressing YneI and YdcW, respectively. Deletion of the ALDH gene from K. pneumoniae did not affect the extent of 3-HP synthesis, implying non-specific activity of ALDHs on 3-HPA. The ALDHs might play major roles in detoxifying the aldehyde generated in glycerol metabolism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
3-Hydroxypropionic acid (3-HP) is a valuable C3 building block, and is used as an intermediate in the synthesis of many commercially valuable chemicals employed in the production of adhesives, fibers, and resins. Despite the industrial importance of 3-HP, it is available commercially as an aqueous solution from only a few suppliers because of the high cost of production and the low yields obtained [1, 2]. Glycerol is an attractive substrate for development of a low-cost fermentation route to 3-HP because glycerol is a major by-product of biodiesel manufacturing and is, therefore, available in large quantities [3].
Two models of 3-HP biosynthesis from glycerol have been suggested in Klebsiella pneumoniae. In the coenzyme A (CoA)-dependent pathway, 3-hydroxypropionaldehyde (3-HPA) is converted to 3-HP by a CoA-dependent aldehyde dehydrogenase (PduP) via a series reactions. In the CoA-independent pathway, 3-HPA is directly converted to 3-HP by the action of aldehyde dehydrogenase (ALDH). The CoA-dependent pathway involving PduP has been described in previous study [4, 5], but ALDH involved in CoA-independent pathway has never been characterized.
To explore ALDH involved in the CoA-independent pathway, putative 176 dehydrogenase genes identified from genome sequences of K. pneumoniae were overexpressed and, as the result, three ALDHs, termed AldHk, YdcW and YneI, were revealed to increase production of 3HP. We have previously reported on the role of AldHk enhancing 3-HP production [6]. In here, two other ALDHs (YdcW and YneI) were characterized and gene effects on 3-HP biosynthesis were also investigated.
Materials and methods
Bacterial strains, plasmids and media
Klebsiella pneumoniae Cu, a derivative of strain ATCC 700721, was obtained by curing of cryptic plasmids by ethidium bromide. The K. pneumoniae AK strain (Cu Δ[orfY-dhaT-orfW-orfX-dhaR-dhaD::AprR]) has been previously described [7]. E. coli DH5α was used for DNA manipulation. The plasmid pGEM-T Easy (Promega, Madison, WI) was employed for cloning, whereas pET28a (Novagen, Darmstadt, Germany) and pBR322 were used for expression of genes in E. coli and K. pneumoniae, respectively. Microbial cells were grown in LB medium (yeast extract [Difco, Sparks, MD, USA], 0.5 %; Bacto-tryptone [Difco], 1.0 %; and NaCl, 1.0 % [all w/v]) supplemented with appropriate antibiotics (ampicillin [50 μg mL−1] and/or tetracycline [10 μg mL−1 for E. coli and 50 μg mL−1 for K. pneumoniae]). The pIJ773 vector was the source of the apramycin-resistance gene, and the temperature-sensitive plasmid pKD46 was employed when homologous recombination was required [8].
Chemical and reagents
T4 DNA ligase and restriction enzymes were obtained from Takara Bio. Inc. (Otsu, Shiga, Japan). Coomassie Brilliant blue R-250 staining solution was purchased from Noble Biosciences Inc. (Suweon, Kyounggi, Korea). N,N,N′,N′-tetramethylethylenediamine (TEMED) and 30 % (w/v) acrylamide/bis-acrylamide were the products of Biosesang Inc. (Seongnam, Kyounggi, Korea). Ammonium persulfate was obtained from Bio-Rad Laboratories, Inc. (Richmond, CA, USA). Ethidium bromide was the product of Promega. The 3-HP standard was purchased from Tokyo Chemical Industry Co. (Chuo, Tokyo, Japan). Other chemicals were the products of the Sigma Chemical Company (St. Louis, MO, USA).
Construction of plasmids for expression of ALDHs in E. coli
To construct a plasmid permitting expression of YneI and YdcW in E. coli, the relevant genes were amplified from chromosomal DNA of K. pneumoniae using appropriate primers (YneI-F: 5′-ccatggatgatgaatttatctgcaacg-3′; YneI-R: 5′-ctcgaggcgacggtctttccaga-3′; YdcW-F: 5′-ccatggatgaaacaggataatgctatg-3′; YdcW-R: 5′-ctcgaggctatgcttaaccatcacat-3′; the bases in bold indicate NcoI and XhoI sites, respectively). The PCR conditions featured initial denaturation at 95 °C for 5 min, followed by 30 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 90 s, with a final hold at 72 °C for 7 min. The PCR products were cloned into pGEM-T-Easy, and clones were subjected to nucleotide sequencing to confirm the absence of any error. After digestion with NcoI and XhoI, DNA fragments were ligated into the corresponding restriction sites of pET28a, yielding pET-yneI and pET-ydcW, respectively.
Construction of plasmids for expression of ALDHs in K. pneumoniae
The yneI (GenBank accession no. ABR77066) and ydcW (GenBank accession no. ABR77355) genes were amplified from chromosomal DNA of K. pneumoniae using appropriate primers (YneI-F: 5′-tctagaatgatgaatttatctgcaacgca-3′; YneI-R: 5′-ctcgaggcgacggtctttccaga-3′; YdcW-F: 5′-tctagaatgaaacaggataatgctatgca-3′; YdcW-R: 5′-actagtctcgagttagcgacggtctttccaga-3′). To introduce the PlacZ promoter upstream of these genes, promoter of PlacZ was amplified from pBluescript by PCR. DNA fragments were cloned into the pGEM-T-Easy vector, and clones were subjected to sequencing to confirm the absence of any error. The gene fragments, including the ALDH genes, were next inserted between the corresponding restriction sites downstream of the lacZ promoter sequence. Subsequently, pVOT (pBR- PlacZ -dhaT- PlacZ-orfWX), pGEM-PlacZ-yneI, and pGEM-PlacZ-ydcW were cut with XhoI, and ligated, yielding plasmids pBR-PlacZ-dhaT-PlacZ-yneI-PlacZ-orfWX (pVOTYI) and pBR-PlacZ-dhaT-PlacZ-ydcW-PlacZ-orfWX (pVOTYW). Plasmid pVOT, described in a previous report [7], includes the orfW gene encoding a factor reactivating DhaB. Final plasmids were transformed into the K. pneumoniae AK strain by electroporation.
Expression and purification of AldHs in/from E. coli
Recombinants of E. coli BL21 (DE3) pLysS clone harboring pET-yneI and pET-ydcW were grown to mid-exponential phase at 37 °C, with aeration, in 250 mL shake flasks, until the A 600 value attained 0.4–0.6. Expression of YneI and YdcW was induced by addition of IPTG to 0.5 mM, followed by incubation for 4 h at 37 °C. Cells were harvested by centrifugation at 4,278g for 10 min at 4 °C. Each cell pellet was washed twice in 50 mM potassium phosphate buffer (pH 7.0) and suspended in 40 mL lysis buffer (pH 8.0) containing 50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole. Next, the cells were sonicated and the resulting solutions centrifuged at 18,510g for 20 min. Each supernatant was loaded onto a Ni2+-nitrilotriacetic acid (NTA) chromatography column equilibrated with 5 mL lysis buffer. After washing (using a buffer containing 50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole), ALDHs linked to the His6 tag were eluted in elution buffer containing 50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole. Protein quantification was achieved using the Bradford protein assay reagent, employing BSA as a standard. After boiling for 5 min, SDS-PAGE was performed on 12 % (w/v) polyacrylamide gel. Proteins were stained with Coomassie Brilliant Blue R-250.
Enzyme activity assay
ALDH activity was measured by the method of Leal et al. [9], with slight modifications. The reaction mixture contained 50 mM potassium phosphate buffer (pH 8.0), 4 mM NAD+ (or NADP+), 2 mM of an aldehyde substrate, and a protein source (37.0 μg mL−1 of soluble cell lysate or 43.5 μg mL−1 of a purified ALDH); incubation proceeded at 37 °C for 10 min. Enzyme activity was determined by measuring the amount of NADH (or NADPH) produced from NAD+ (or NADP+), by absorbance at 340 nm. One unit (U) of enzyme activity was defined as the amount of enzyme producing 1 μmol NADH per min. All activity measurements were performed in triplicate.
Substrate specificity was examined at 37 °C and pH 8.0 using various aldehydes including 3-hydroxypropionaldehyde (3-HPA), propionaldehyde (PA), butyraldehyde (BA), valeraldehyde (VA), isovaleraldehyde (IVA) and furaldehyde (FA), in the presence of NAD+ or NADP+ as cofactor.
Cultivation of K. pneumoniae strains in glycerol-containing medium
Klebsiella pneumoniae strains were cultivated in medium containing 20 g L−1 glycerol, 30 mM potassium phosphate buffer (pH 7.0), 1 g L−1 yeast extract, 2 g L−1 (NH4)2SO4, 0.2 g L−1 MgSO4, 0.002 g L−1 CaCl2·2H2O, 1 mL L−1 Fe solution (5 g L−1 FeSO4·7H2O and 4 mL HCl [37 %, w/v]), and 1 mL L−1 of trace element solution (70 mg L−1 ZnCl2, 100 mg L−1 MnCl2·4H2O, 60 mg L−1 H3BO3, 200 mg L−1 CoCl2·4H2O, 20 mg L−1 CuCl2 2H2O, 25 mg L−1 NiCl2·6H2O, 35 mg L−1 Na2MoO4·2H2O, and 4 mL HCl [37 %, w/v]) [10].
When K. pneumoniae strains were grown in a 5-L bioreactor (Kobiotech Co. Ltd, Incheon, Korea), seed cells were cultivated in a 250-mL flask containing 50 mL of the preculture medium described above for 12 h at 120 rpm, and were next inoculated into the bioreactor, containing the same medium, at 2 % (v/v) with IPTG induction (0.5 mM). Batch cultivation was conducted at 37 °C, with shaking at 200 rpm, at pH 7.0. Air (0.5 vvm) was supplied to the stirred reactor containing 2 L of fermentation medium. All results reflect data obtained from three independent experiments. Average values are shown.
Analytical methods
The concentrations of 3-HP, glycerol, and other metabolites in culture broth were determined by high-performance liquid chromatography (Agilent System 1200; Santa Clara, CA); the system was equipped with a refractive index detector and an ion-exchange column (300 × 78 mm; Rezex ROA-organic acid; Phenomenex, Torrance, CA). The mobile phase was 0.0025 mol L−1 H2SO4 and the flow rate was 0.5 mL min−1. The column and cell temperatures were 65 and 45 °C, respectively [4].
Results and discussion
Sequence analysis of ALDHs
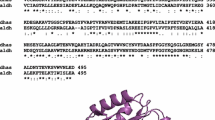
YneI contained 462 amino acids and was 76, 74, 75, and 87 % homologous to YneI of E. coli (EHX11321), YneI of Salmonella enterica (NP_460484), YneI of Citrobacter youngae (ZP_06352898), and Ald of Enterobacter aerogenes (YP_004593910), respectively (Fig. 1). Residues putatively forming the active site of an ALDH, Asn-137, Glu-234, Gly-265, and Cys-268, and NAD(P)+ binding sites were evident in YneI.
Alignment, emphasizing homology, of YneI with other aldehyde dehydrogenases: YneI of Escherichia coli (EHX11321), YneI of Salmonella enterica (NP_460484), YneI of Citrobacter youngae (ZP_06352898), and Ald of Enterobacter aerogenes (YP_004593910.1). The putative active sites (triangles) of the aldehyde dehydrogenases featuring Asn-137, Glu-234, Gly-265, and Cys-268; and the NAD(P)+ binding sites (underlined) (amino acid residues 133–137, 145, 160, 162, 163, 210–213, 216, 219–220, 234–236, 268, 365, 367, 393, 431) are shown
YdcW contained 481 amino acids and 85, 84, 92, and 97 % homologous to YdcW of E. coli (YP_001462716), YdcW of S. enterica (YP_002114622), Ald of E. aerogenes (YP_004594212), and Ald of K. variicola (YP_003439305), respectively (Fig. 2). The figure shows the catalytic residues (triangles) of Asn-155, Glu-252, Gly-283, and Cys-286; the substrate-binding sites (circles) of Asp-285 and Leu-444, and the NAD(P)+ binding sites (underline) (151, 152, 154, 178, 180, 181, 210, 215, 229–231, 234, 237, 252, 254, 286, 333, 384).
Alignment, emphasizing homology, of YdcW with aldehyde dehydrogenases YdcW of Escherichia coli (YP_001462716), YdcW of Salmonella enterica (YP_002114622), Ald of Enterobacter aerogenes (YP_004594212), and Ald of Klebsiella variicola (YP_003439305). The putative catalytic residues (triangles) of Asn-155, Glu-252, Gly-283, and Cys-286; the substrate-binding sites (circles) of Asp-285 and Leu-444; and the NAD(P)+ binding sites (underlined) (amino acid residues 151, 152, 154, 178, 180, 181, 210, 215, 229–231, 234, 237, 252, 254, 286, 333, and 384) are shown
Expression and purification of K. pneumoniae ALDHs in/from E. coli
To explore ALDH activities, recombinant proteins were purified from E. coli BL21 harboring pET28a-based clones. SDS-PAGE analysis of cytoplasmic fractions of such strains confirmed that recombinant proteins approximately 55 kDa in size were prominently expressed. No such band was observed in the cytoplasmic fraction of control cells harboring vector pET28a. His6-tagged recombinant ALDHs in cytoplasmic fractions of E. coli were purified by Ni–NTA affinity column chromatography (Fig. 3).
Functional analysis of ALDHs produced in E. coli
The specific activities of purified YneI and YdcW were 962 and 644 U mg−1 proteins, respectively, when 3-HPA was the substrate and NAD+ the cofactor (Table 1). Substrate specificity was examined using several aldehydes including 3-HPA, PA, BA, VA, IVA, and FA, in the presence of NAD+ and NADP+ as cofactors. YneI was active on 3-HPA specifically and utilized both NAD+ and NADP+ as cofactor, but with a preference for NAD+ (Fig. 4a). YdcW was active on all aldehydes tested with exception of FA. The highest activity was evident when PA served as substrate. YdcW utilized both NAD+ and NADP+ as cofactors but preferred NADP+ (Fig. 4b).
Substrate and cofactor specificity of YneI (a) and YdcW (b). 3HPA, 3-hydroxypropionaldehyde; PA, propionaldehyde; BA, butyraldehyde; VA, valeraldehyde; IVA, isovaleraldehyde; FA, furaldehyde. Data are expressed as mean ± SD. Three independent experiments were made for each end-point. Statistical differences were determined by the student’s t test. There were no statistically significant differences
Increased 3-HP production upon overexpression of ALDHs in K. pneumoniae AK
When ALDHs were overexpressed in K. pneumoniae, 3-HP production remarkably enhanced, compared to that of the control strain (the basal level was 0.6 g L−1) upon shake-flask culture over 48 h. The concentrations of 3-HP yielded by the recombinant strains were 2.4 g L−1 when YneI was expressed and 1.8 g L−1 when YdcW was synthesized (Table 2). Batch fermentation was conducted in a 5-L bioreactor. The recombinant strain harboring pVOTYI consumed all added glycerol (20 g L−1) by 20 h of culture and the maximal 3-HP production was 3.9 g L−1 at 30 h (Fig. 5b). The recombinant strain harboring pVOTYW consumed glycerol at a slower rate than did the control (measured at 24 h; Fig. 5a) and the maximal 3-HP production was 2.2 g L−1 at 30 h (Fig. 5c). K. pneumoniae first dehydrates glycerol via the action of a coenzyme B12-dependent glycerol dehydratase (DhaB) to yield 3-HPA, which is next oxidized to 3-HP by NAD+-dependent ALDH. As YdcW prefers NADP+ as cofactor, the enzyme may not efficiently catalyze the synthesis of 3-HP from 3-HPA.
Probable roles of ALDHs on glycerol metabolism in K. pneumoniae
It has been suggested that 3-HP is synthesized from glycerol, in K. pneumoniae, via two distinct routes. In the CoA-dependent pathway, 3-HPA is converted to 3-HP in a series of reactions involving catalysis by PduP and in the CoA-independent pathway, 3-HPA is directly converted into 3-HP by an ALDH. Various ALDHs have been screened with a view toward improving 3-HP production. However, this CoA-independent pathway featuring direct oxidation of 3-HPA to 3-HP has not yet been well-characterized in native organisms. Deletion of aldHk from K. pneumoniae did not affect the extent of 3-HP synthesis from glycerol [6].
To examine the role played by YneI and YdcW in 3-HP production, yneI and ydcW were precisely deleted from K. pneumoniae using a homologous recombination method [11]. Southern blotting of the wild-type and mutant strain probes specific for the genes of interest confirm that deletions had occurred and the mutants were stable. The wild-type and deletion mutant strains were subjected to batch fermentation in a 5-L bioreactor supplemented with glycerol as carbon source. All strains attained maximum optical density levels (at A 600 nm) above 5.4 and consumed almost all added glycerol (20 g L−1) by 9 h of culture. The maximal 3-HP productions noted were approximately 2 g L−1. No significant difference was evident among wild-type and mutant strains (data not shown).
In previous reports, we investigated a CoA-dependent pathway of K. pneumoniae in which 3-HPA is metabolized via the action of PduP (a CoA-dependent ALDH) to yield 3-HP. As 3-HP was still produced by a ΔpduP mutant, we speculated that a CoA-independent pathway featuring an additional ALDH might participate in 3-HP synthesis from glycerol [4]. Aldehydes vary in length and in characteristics of the alkyl chains but all are toxic because of high chemical reactivity [12]. ALDHs play major roles in detoxifying aldehydes that are generated both endogenously and exogenously. Some ALDHs oxidize only a few aldehydes; others exhibit broad substrate specificities [13]. Therefore, K. pneumoniae may produce 3-HP from glycerol via a pathway featuring the activity of a non-specific ALDH (or several such enzymes).
Some reports on the non-specific ALDHs increasing 3-HP production by overexpression have appeared. Available ALDHs include ALD4 of Saccharomyces cerevisiae; ALDH2 of Homo sapiens; AldA, AldB, AldH, and YdcW of Escherichia coli; KGSADH of Azospirillum brasiliense; AldA, AldB, PuuC, AldH, YdcW, EtuE, FeaB, GabD, BadH, and PduP of K. pneumoniae; AdhB of Zymomonas mobilis; and AldH and PduQ of Lactobacillus collinoides [14–18].
Conclusion
In the present study, we identified and characterized two putative ALDHs involved in 3HP production from glycerol in K. pneumoniae. Overexpression of the ALDHs obviously increased 3-HP production in K. pneumoniae. However, deletion of the gene from K. pneumoniae did not affect the extent of 3-HP synthesis from glycerol, indicating non-specific activity of ALDHs on 3-HPA. The ALDHs probably play major roles in detoxifying the aldehyde generated in glycerol metabolism. In addition, overexpression of genes would contribute to efficient production of 3-HP from glycerol.
References
Jiang X, Meng X, Xian M (2009) Biosynthetic pathways for 3-hydroxypropionic acid production. Appl Microbiol Biotechnol 82:995–1003
Andreessen B, Steinbüchel A (2010) Biosynthesis and biodegradation of 3-hydroxypropionate-containing polyesters. Appl Environ Microbiol 76:4919–4925
Da Silva GP, Mack M, Contiero J (2009) Glycerol: a promising and abundant carbon source for industrial microbiology. Biotechnol Adv 27:30–39
Luo LH, Kim CH, Heo SY, Oh BR, Hong WK, Kim S, Kim DH, Seo JW (2012) Production of 3-hydroxypropionic acid through propionaldehyde dehydrogenase PduP mediated biosynthetic pathway in Klebsiella pneumoniae. Bioresour Technol 103:1–6
Luo LH, Seo JW, Baek JO, Oh BR, Heo SY, Hong WK, Kim DH, Kim CH (2011) Identification and characterization of the propanediol utilization protein PduP of Lactobacillus reuteri for 3-hydroxypropionic acid production from glycerol. Appl Microbiol Biotechnol 89:697–703
Luo LH, Seo JW, Oh BR, Seo PS, Heo SY, Hong WK, Kim DH, Kim CH (2011) Stimulation of reductive glycerol metabolism by overexpression of an aldehyde dehydrogenase in a recombinant Klebsiella pneumoniae strain defective in the oxidative pathway. J Ind Microbiol Biotechnol 38:991–999
Seo MY, Seo JW, Heo SY, Baek JO, Rairakhwada D, Oh BR, Seo PS, Choi MH, Kim CH (2009) Elimination of by-product formation during production of 1,3-propanediol in Klebsiella pneumoniae by inactivation of glycerol oxidative pathway. Appl Microbiol Biotechnol 84:527–534
Doublet B, Douard G, Targant H, Meunier D, Madec JY, Cloeckaert A (2008) Antibiotic marker modifications of lambda Red and FLP helper plasmids, pKD46 and pCP20, for inactivation of chromosomal genes using PCR products in multidrug-resistant strains. J Microbiol Methods 75:359–360
Leal NA, Havemann GD, Bobik TA (2003) PduP is a coenzyme-A-acylating propionaldehyde dehydrogenase associated with the polyhedral bodies involved in B12-dependent 1,2-propanediol degradation by a Salmonella enterica serovar Typhimurium LT12. Arch Microbiol 80:353–361
Oh BR, Seo JW, Choi MH, Kim CH (2008) Optimization of culture conditions for 1,3-propanediol production from crude glycerol by Klebsiella pneumoniae using response surface methodology. Biotechnol Bioprocess Eng 13:524–532
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:640–645
Wei Y, Lin M, Oliver DJ, Schnable PS (2009) The roles of aldehyde dehydrogenases (ALDHs) in the PDH bypass of Arabidopsis. BMC Biochem 10:7
Sophos NA, Vasiliou V (2003) Aldehyde dehydrogenase gene superfamily: the 2002 update. Chem Biol Interact 143–144:5–22
Huang Y, Li Z, Shimizu K, Ye Q (2012) Simultaneous production of 3-hydroxypropionic acid and 1,3-propanediol from glycerol by a recombinant strain of Klebsiella pneumoniae. Bioresour Technol 103:351–359
Rathnasingh C, Raj SM, Jo JE, Park S (2009) Development and evaluation of efficient recombinant Escherichia coli strains for the production of 3-hydroxypropionic acid from glycerol. Biotechnol Bioeng 104:729–739
Zhu JG, Ji XJ, Huang H, Du J, Li S, Ding YY (2009) Production of 3-hydroxypropionic acid by recombinant Klebsiella pneumoniae based on aeration and ORP controlled strategy. Korean J Chem Eng 26:1679–1685
Jo JE, Raj SM, Rathnasingh C, Selvakumar E, Jung WC, Park S (2008) Cloning, expression, and characterization of an aldehyde dehydrogenase from Escherichia coli K-12 that utilizes 3-hydroxypropionaldehyde as a substrate. Appl Microbiol Biotechnol 81:51–60
Suthers PF, Cameron DC (2001) Production of 3-hydroxypropionic acid in recombinant organisms. PCT WO 01-16346
Acknowledgments
The authors are very grateful to Prof. Sunghoon Park for the kind gift of 3-hydroxypropionaldehyde. This study was supported by the Ministry of Education, Science and Technology, Korea.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Luo, L.H., Seo, JW., Heo, SY. et al. Identification and characterization of Klebsiella pneumoniae aldehyde dehydrogenases increasing production of 3-hydroxypropionic acid from glycerol. Bioprocess Biosyst Eng 36, 1319–1326 (2013). https://doi.org/10.1007/s00449-012-0880-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-012-0880-4