Abstract
In Klebsiella pneumoniae, aldehyde dehydrogenases (ALDH) convert 3-hydroxypropionaldehyde (3-HPA) into 3-hydroxypropionic acid (3-HP). Although ALDHs can increase the production of 3-HP in K. pneumoniae, the substrate specificity of ALDH homologues from other microorganisms toward 3-HPA is less documented. Here we report that DhaS, a putative ALDH from Bacillus subtilis, shows high specificity toward 3-HPA when heterologously expressed in K. pneumoniae. Using NAD+ as a cofactor, DhaS exhibited higher catalytic activity (2.3 U mg−1) and lower K m value (0.4 mmol l−1) toward 3-HPA than that toward other aldehydes. Under shake-flask conditions, the recombinant strain produced 2.1 g 3-HP l−1 in 24 h, which is 3.9-fold of that in a control harboring a blank vector. Under non-optimized bioreactor conditions, the recombinant strain produced 18 g 3-HP l−1 and 1,3-propanediol (1,3-PDO) at 27 g l−1 in 24 h. The overall conversion rate from glycerol to 3-HP and 1,3-PDO reached 59.4 mol mol−1. Homology modeling of DhaS illustrates substrate specificity and NAD+-binding site. DhaS is thus a 3-HPA-specific enzyme useful for production of 3-HP.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
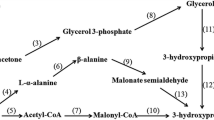
3-Hydroxypropionic acid (3-HP) has attracted significant attention because it can be converted into a variety of commercially-valuable compounds, including acrylic acid, 3-hydroxypropionaldehyde (3-HPA), 1,3-propanediol (1,3-PDO) and malonic acid (Werpy et al. Werpy and Petersen 2004; Valdehuesa et al. 2013; Kumar et al. 2013). Klebsiella pneumoniae is a promising 3-HP-synthesizing strain due to active cell proliferation and high capacity to metabolize glycerol (Huang et al. 2013). Under anaerobic conditions, glycerol dissimilation in K. pneumoniae is mediated by the dha regulon which involves parallel oxidation and reduction pathways (Forage and Lin Forge and Lin 1982). In the glycerol oxidation pathway, glycerol is converted into dihydroxyacetone phosphate, which subsequently enters glycolic pathway. In the glycerol reduction pathway, glycerol is converted into 3-HPA by glycerol dehydratase (dhaB) (Forage and Foster 1982). Next, 3-HPA is catalyzed into 1,3-PDO by 1,3-propanediol oxidoreductase (dhaT) (Forage and Lin 1982) and 3-HP by aldehyde dehydrogenases (ALDH) (Ashok et al. 2011) respectively.
3-HP production has been achieved by overexpression of ALDHs, including ALD4 from Saccharomyces cerevisiae (Li et al. 2013), KGSADH from Azospirillum brasilense (Rathnasingh et al. 2009), and isozymes PuuC, YdcW and YneI from K. pneumoniae (Raj et al. 2010; Luo et al. 2013). However, the yield of 3-HP is too low for commercialization. The reasons behind include but are not limited to the following: (i) massive consumption of NAD+, which slows cell growth and thereby hinders 3-HP production; (ii) low specificity of ALDHs toward 3-HPA, that is, other aldehydes beyond 3-HPA in cytoplasm were also catalyzed into corresponding acids by ALDHs, leading to metabolic perturbation and the decrease of 3-HP biosynthesis. In fact, in wild-type strains, ALDHs usually demonstrate low activity to maintain cofactor balance (Ko et al. 2012). Based on these information, we speculate that in addition to catalytic activity, the ALDH specificity toward 3-HPA might be crucial for overproduction of 3-HP.
In this study, DhaS, a putative ALDH from B. subtilis, was heterologously expressed in K. pneumoniae to produce 3-HP. Both the in vitro activities toward different aldehydes and the specificity toward 3-HPA were examined. Homology modeling and sequence analysis of DhaS were carried out to predict its spatial structure. Shake-flask and bioreactor cultivation in micro-anaerobic conditions were performed to comprehensively evaluate the efficiency of DhaS for the production of 3-HP.
Materials and methods
Strains, plasmids and reagents
Klebsiella pneumoniae DSM 2026 and Bacillus subtilis subsp. 168 were from DSMZ GmbH, Germany. The vector pET-28a (Novagen) was used in this study with minor modification. The original T7 promoter in vector pET-28a was replaced by pk, the native promoter of dhaB1 gene (GenBank: YP_001337151.1) in K. pneumoniae DSM 2026. The resulting vector was named as pET-pk (Li et al. 2013). Restriction enzymes and Taq DNA polymerase were purchased from TaKaRa (Dalian, China). Other chemicals for enzymatic activity assay, gel electrophoresis and HPLC analysis were products of Sigma. DNA synthesis and sequencing were performed by Biomed Co. Ltd., China.
Cultivation conditions
B. subtilis subsp. 168 was grown in LB medium. The medium for producing 3-HP by K. pneumoniae contained the following components (per liter): K2HPO4·3H2O, 3.4 g; KH2PO4, 1.3 g; (NH4)2SO4, 4 g; MgSO4·7H2O, 0.5 g; CaCO3, 0.1 g; yeast extract, 3 g; glycerol, 40 g; and 1.25 ml trace element solution. The trace element solution contained (per liter): FeSO4, 32 g; ZnCl2·6H2O, 2.72 g; MnCl2·4H2O, 0.68 g; CoCl2·6H2O, 1.88 g; H3BO3, 0.24 g; Na2MoO4, 0.02 g; CuCl2·2H2O, 1.88 g; and 40 ml conc. HCl. The recombinant strain was grown microaerobically in 50 ml Erlenmeyer flasks containing 20 ml medium and 50 μg kanamycin ml−1, and shaken at 150 rpm for 24 h.
Construction of the recombinants
The native pk promoter of dhaB1 gene was used to drive gene expression. The gene dhaS (GenBank: NC_000964.3) was amplified from the genomic DNA of B. subtilis by using the following primers. Forward: 5′-CGCGGATCCATGAGTTCTTTAACGAT-3′, Reverse: 5′-TCCGAGCTCTTAGTCTTCAAGGTTTACC-3′, the underlined nucleotides indicate BamHI and SacI restriction sites, respectively. The PCR procedures were: initial denaturation at 94 °C for 4 min; followed by 30 cycles of 94 °C for 1 min, 55 °C for 45 s, 72 °C for 90 s, and 72 °C for 10 min. All other molecular manipulations followed the standard protocol. The PCR product of dhaS was cloned into vector pET-pk, leading to recombinant vector pET-pk-dhaS. It was then transformed into K. pneumoniae DSM 2026, the positive recombinants were screened by LB kanamycin plate and further confirmed by sequencing.
Determination of enzyme activities
The recombinant strains were grown in fermentation medium containing kanamycin of 50 μg ml−1. After 9 h cultivation, cells were centrifuged at 10,000×g for 10 min and washed by 5 ml phosphate buffer solution (PBS, pH 7.0). 50 µl PMSF (10 mg ml−1) was used to inhibit the protease activity. The cells were sonicated and the resulting solution was centrifuged at 17,000g for 15 min. Protein concentration was determined using the Bradford reagent (BioRad). 100 µl cell-free solutions were added into 2 ml centrifugation tube containing 0.2 mmol NADH (or NADPH) and 5 mmol aldehyde, and incubated at 37 °C for 5 min. The amount of consumed NADH (or NADPH) was determined by measuring the decrease of absorbance at 340 nm. The experiment was performed in triplicate. One unit activity of ALDH was defined as the amount of enzyme used in consuming 1 μmol NADH per min.
Homology modeling of DhaS
The online Basic Local Alignment Search Tool (BLAST) was used to search suitable template structure from Protein Data Bank (PDB). Based on the crystal structure of human mitochondrial ALDH (PDB: 3inj_H) (Steinmetz et al. 1997), the DhaS model was generated using Swiss-Model software at the ExPASy website (http://swissmodel.expasy.org/). The predicted structures were evaluated by VERIFY-3D (a structure evaluation server), a program developed to verify the average distance between the backbones of superimposed proteins, thus enabling a quantitative comparison between the protein structures (Luthy et al. 1992). These models were used to identify the active site and cofactor-binding site.
3-HP production by recombinant K. pneumoniae
In shake-flask cultivations, the recombinants were grown in LB medium containing: yeast extract 5 g, NaCl 10 g, peptone 10 g, and kanamycin 50 mg. 1 % overnight culture was inoculated to the medium containing the same concentration of antibiotics. To maintain microaerobic conditions, the flasks were plugged with an O2-permeable cotton stopper and incubated at 37 °C with shaking at 150 rpm.
Fed-batch cultivation of the recombinant K. pneumoniae strain Kp(pET-pk-dhaS) was carried out at 37 °C in a 5 l bioreactor (Baoxing, China) containing 3 l fermentation medium. The fermentation conditions were the same as those previously reported (Huang et al. 2013). The strain was pre-cultivated in a 100 ml fermentation medium overnight at 37 °C and then added into bioreactor. The agitation speed was 400 rpm and the air was supplied at 1.5 vvm. pH was maintained at 7 by automatic addition of 5 M NaOH. Residual glycerol was maintained at 25 g l−1. Dissolved oxygen (DO) was monitored automatically. Samples were taken every 3 h, and metabolites, cell concentration and residual glycerol were examined.
Analytical methods
Cell concentrations were measured by using microplate reader at 600 nm. 3-HP, lactic acid and acetic acid were determined by HPLC equipped with a C18 column with the eluate being monitored at 210 nm. The mobile phase was 0.05 % phosphoric acid 0.8 ml min−1. Residual glycerol concentration was measured every 3 h by a titration method with NaIO4 (for control of glycerol). 1,3-PDO was quantitatively analyzed by HPLC equipped with a column of Aminex HPX-87H Ion Exclusion particles (300 × 7.8 mm, Bio-Rad, Hercules, CA, USA) using a differential refractive index detector. The column was maintained at 65 °C and the mobile phase was 5 mM H2SO4 (in Milli-Q water) at 0.6 ml min−1. All samples were filtered through 0.22 µm membrane filter.
Results and discussion
Characterization of the recombinants
The recombinant vectors were electro-transformed into K. pneumoniae DSM 2026. All positive recombinants were screened by a LB kanamycin plate and further confirmed by sequencing. The dhaS gene in recombinant vector pET-pk-dhaS showed 100 % sequence identity with the reported sequence in the GenBank, suggesting that it was correctly acquired. The recombinant strain Kp(pET-pk-dhaS) was used for subsequent activity assay and production of 3-HP. By using constitutive promoter pk, the enzyme DhaS was constitutively induced by dihydroxyacetone (DHA), the metabolite of glycerol oxidization.
DhaS activity and specificity
The in vitro DhaS catalytic activities toward different aldehydes were investigated. When NADP+ was the cofactor, DhaS did not show any activity toward propionaldehyde, benzaldehyde, valeraldehyde, butyraldehyde and acetaldehyde, with exception of only 0.1 U mg−1 activity toward propionaldehyde. In contrast, when NAD+ was the cofactor, the activity of DhaS toward 3-HPA reached up to 2.3 U mg−1, and the recombinant K. pneumoniae harboring vector pET-pk (as the control) showed activity of 1.3 U mg−1 which was contributed by the inherent ALDHs in K. pneumoniae. Interestingly, except 3-HPA, the activities of K. pneumoniae (pET-pk-dhaS) toward other five aldehydes were nearly equal to that of K. pneumoniae (pET-pk), implying that other aldehydes are not the preferred substrates of DhaS. Indeed, DhaS activities toward other aldehydes are much lower than that toward 3-HPA when NAD+ was the cofactor. The relative activities of DhaS toward distinct aldehydes were shown in Table 1. The K m value of DhaS for 3-HPA was 0.4 mM, which is much lower than that toward other aldehydes. Collectively, 3-HPA is an appropriate substrate of DhaS, and DhaS is an ideal enzyme for catalyzing 3-HPA into 3-HP.
Homology modeling and sequence analysis of DhaS
To better understand the spatial structure of DhaS, homology modeling and sequence analysis were performed. Homology modeling of DhaS was accomplished based on the crystal structure of human mitochondrial ALDH (3inj_H). The protein structure checking tool, Verify 3D, showed that the model of DhaS is reliable, because the statistics of non-bonded interactions analysis displayed that the overall quality factor is 87.9, which is higher than the standard (>85). Amino acid sequence alignment showed 42 % identify between the dhaS from B. subtilis and the aldH from E. coli. Despite low sequence similarity between two ALDHs, there exist conserved domains, such as Glu267 and Cys302 residues, related to the catalytic activities toward aldehydes and a glycine-rich motif G-x-G-x-x–x-G (GFGEDAG in DhaS) involved in binding NAD(P)+ (Fig. 1). Interestingly, most of NAD+-dependent ALDHs contain lysine and glutamate residues adjacent to adenosine ribose (Perozich et al. 2000). A lysine residue, also conserved in NADP+-dependent ALDHs, interacts with the adenosine ribose in NAD+, or the 2′-phosphate in NADP+. Unlike the lysine residue, the glutamate residue likely participates in the exclusion of NADP+. In view of putative structure of DhaS, Lys191 and Glu194 residues adjacent to glycine motif are postulated to bind NAD+. Overall, 3D-structure model of DhaS revealed that NAD+ is the appropriate cofactor.
Sequence analysis and homology modeling of DhaS. a Amino acid sequence alignment between DhaS from B. subtilis and AldH from E. coli K12 (GenBank AAA23428.1). Arrows indicate the putative active site catalyzing aldehydes into acids. The glycine motif, G-X-G-X–X-X-G, is the NAD(P)+-binding site. b The predicted 3D structure of DhaS. Cys 302 and Glu267 residues highlight the active site. Lys191 and Glu194 residues in glycine motif indicate cofactor-binding site
Flask cultivation of the recombinant K. pneumoniae expressing dhaS
Since DhaS showed catalytic activity for in vitro conversion of 3-HPA into 3-HP, its in vivo performance was investigated. For shake-flask cultivation, 40 g glycerol was initially added to the medium. After 24 h cultivation, more than 20 g glycerol l−1 was left in the broth of the strain K. Pneumoniae (pET-pk-dhaS), while less than 10 g glycerol l−1 was left in that of wild-type K. pneumoniae (Kp-WT) (Fig. 2a). In accordance with glycerol consumption, the recombinant strain Kp(pET-pk-dhaS) grew more slowly than wild-type K. pneumoniae (Fig. 2b).
Shake-flask cultivation of the strains. a Residual glycerol, b cell concentration, c lactic acid, d acetic acid, e 3-hydroxypropionic acid, f 1,3-propanediol. Kp-WT: Wild-type K. pneumoniae; Kp(pET-pk): Recombinant K. pneumoniae harboring blank vector pET-pk; Kp(pET-pk-dhaS): Recombinant K. pneumoniae harboring vector pET-pk-dhaS. Error bars represent standard deviation from three independent experiments
The decreased cell growth and glycerol consumption may be largely ascribed to plasmid replication which overburdened the cells. To determine the influences of DhaS overexpression on glycerol allocation, the two major byproducts, lactic acid and acetic acid, were measured. In the strain Kp(pET-pk-dhaS), the production of lactic acid was slightly affected compared to that in wild-type, however, acetic acid reached up to 1.5 g l−1 at 24 h, which was 2.5-fold of that in wild-type strain (Fig. 2c, d). Strain Kp(pET-pk-dhaS) yielded 2.1 g 3-HP l−1 at 24 h, which was 3.9-fold of that in control harboring a blank vector. In addition, Kp(pET-pk-dhaS) yielded 7 g 1,3-PDO l−1 at 24 h, while wild-type strain produced 14 g 1,3-PDO l−1. The overall conversion rate from glycerol to 3-HP and 1,3-PDO of strain Kp(pET-pk-dhaS) was 59.4 mol mol−1, indicating that glycerol was efficiently utilized (Table 2).
Bioreactor cultivation of the recombinant K. pneumoniae expressing dhaS
To further investigate the performance of Kp(pET-pk-dhaS), fed-batch fermentation was carried out in a 5 l bioreactor containing 3 l medium. Under O2-sufficient conditions, the recombinant strain Kp(pET-pk-dhaS) produced 18.5 g 3-HP l−1 and 27 g 1,3-PDO l−1 in 24 h (Fig. 3). Although air supply was efficient for aerobic fermentation, the dissolved O2 (DO) dropped dramatically to zero at the initial 1 h and remained unchanged during the entire fermentation process. Along with the low level of DO, lactic acid reached 36.1 g l−1. The overall conversion rate from glycerol to 3-HP and 1,3-PDO of the strain Kp(pET-pk-dhaS) was 37.1 mol mol−1 (Table 3), which is lower than that in the shake-flask condition. The decreased glycerol conversion rate may be ascribed to the environmental differences between shake-flask and bioreactor. Nevertheless, DhaS is shown to be a 3-HPA-specific enzyme and promising for conversion of 3-HPA into 3-HP.
Although DhaS shows strict specificity toward 3-HPA and NAD+, the current 3-HP yield is far from commercialization. The reasons behind may be multifaceted. One reason is cofactor imbalance. 3-HP biosynthesis is positively correlated with cell growth, and there exists a competition between 3-HP production and cell growth for NAD+. Even though K. pneumoniae can regenerate NAD+ by the formation of lactic acid, whereby lactate dehydrogenase converts NADH into NAD+ (Laval et al. 1984), the amount of NAD+ may not be sufficient for overproduction of 3-HP. In addition to cofactor imbalance, 3-HP toxicity to the cell may be the second reason for hindering 3-HP accumulation. In fact, excessive 3-HP impeded cell growth which in turn retarded the formation of 3-HP. The third reason hindering 3-HP accumulation is the low activity of ALDH toward 3-HPA. Therefore, we suggest the enhancement of catalytic activity via directed evolution but without compromising substrate specificity. In summary, high 3-HP yield depends on cofactor availability, cell tolerance, enzymatic properties, or beyond.
Conclusion
A variety of aldehydes are catalyzed into corresponding acids by ALDHs and 3-HP is one of the enzymatic products of ALDHs. To preferentially produce 3-HP, a 3-HPA-specific enzyme is required. In this study, DhaS, an ALDH from B. subtilis, was shown to be a 3-HPA-specific enzyme. In a recombinant K. pneumoniae expressing dhaS, DhaS exhibited higher activity (2.3 U mg−1) toward 3-HPA than toward other aldehydes, indicating that 3-HPA is the preferred substrate of DhaS (Table 1). Furthermore, a predicted model of DhaS demonstrated the spatial structure for binding 3-HPA and NAD+ (Fig. 1). In shake-flasks and in a 5 l bioreactor, the recombinant strain Kp(pET-pk-dhaS) produced 2.1 and 18 g l−1 of 3-HP, respectively (Figs. 2, 3). Overall these results indicate that DhaS is a promising enzyme for production of 3-HP.
References
Ashok S, Raj SM, Rathnasingh C, Park S (2011) Development of recombinant Klebsiella pneumoniae∆ dhaT strain for the co-production of 3-hydroxypropionic acid and 1,3-propanediol from glycerol. Appl Microbiol Biotechnol 90:1253–1265
Forage RG, Foster MA (1982) Glycerol fermentation in Klebsiella pneumoniae: functions of the coenzyme B12-dependent glycerol and diol dehydratases. J Bacteriol 149:413–419
Forge RG, Lin ECC (1982) dha System Mediating Aerobic and Anaerobic Dissimilation of Glycerol in Klebsiella pneumoniae NCIB 418. J Bacteriol 151:591–599
Huang Y, Li Z, Shimizu K, Ye Q (2013) Co-production of 3-hydroxypropionic acid and 1,3-propanediol by Klebseilla pneumoniae expressing aldH under microaerobic conditions. Bioresour Technol 128:505–512
Ko Y, Ashok S, Zhou S, Kumar V, Park S (2012) Aldehyde dehydrogenase activity is important to the production of 3-hydroxypropionic acid from glycerol by recombinant Klebsiella pneumoniae. Process Biochem 47:1135–1143
Kumar V, Ashok S, Park S (2013) Recent advances in biological production of 3-hydroxypropionic acid. Biotechnol Adv 31:945–961
Laval JM, Bourdillon C, Moiroux J (1984) Enzymic electrocatalysis: electrochemical regeneration of NAD+ with immobilized lactate dehydrogenase modified electrodes. J Am Chem Soc 106:4701–4706
Li Y, Su M, Ge X, Tian P (2013) Enhanced aldehyde dehydrogenase activity by regenerating NAD+ in Klebsiella pneumoniae and implications for the glycerol dissimilation pathways. Biotechnol Lett 35:1609–1615
Luthy R, Bowie JU, Eisenberg D (1992) Assessment of protein models with three-dimensional profiles. Nature 356:83–85
Luo LH, Seo JW, Heo SY, Oh BR, Kim DH, Kim CH (2013) Identification and characterization of Klebsiella pneumoniae aldehyde dehydrogenases increasing production of 3-hydroxypropionic acid from glycerol. Bioprocess Biosyst Eng 36:1319–1326
Perozich J, Kuo I, Wang BC, Boesch JS, Lindahl R, Hempel J (2000) Shifting the NAD/NADP preference in class 3 aldehyde dehydrogenase. Eur J Biochem 267:6197–6203
Raj SM, Rathnasingh C, Jung WC, Selvamumar E, Park S (2010) A novel NAD+-dependent aldehyde dehydrogenase encoded by the puuC gene of Klebsiella pneumoniae DSM 2026 that utilizes 3-hydroxypropionaldehyde as a substrate. Biotechnol Bioprocess Eng 15:131–138
Rathnasingh C, Raj SM, Jo JE, Park S (2009) Development and evaluation of efficient recombinant Escherichia coli strains for the production of 3-hydroxypropionic acid from glycerol. Biotechnol Bioeng 104:729–739
Steinmetz CG, Xie P, Weiner H, Hurley TD (1997) Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion. Struct 5:701–711
Valdehuesa KNG, Liu H, Nisola GM, Chung WJ, Lee SH, Park SJ (2013) Recent advances in the metabolic engineering of microorganisms for the production of 3-hydroxypropionic acid as C3 platform chemical. Appl Microbiol Biotechnol 97:3309–3321
Werpy T, Petersen G (2004) Top value added chemicals from biomass volume 1-Results of screening for potential candidates from sugars and synthesis gas. Department of Energy, Washington, DC
Acknowledgments
This work was supported by Grants from National Basic Research Program of China (973 Program) (2012CB725200) and National Natural Science Foundation of China (No. 21076013, 21276014).
Author information
Authors and Affiliations
Corresponding author
Additional information
Mingyue Su and Ying Li have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Su, M., Li, Y., Ge, X. et al. 3-Hydroxypropionaldehyde-specific aldehyde dehydrogenase from Bacillus subtilis catalyzes 3-hydroxypropionic acid production in Klebsiella pneumoniae . Biotechnol Lett 37, 717–724 (2015). https://doi.org/10.1007/s10529-014-1730-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-014-1730-z