Abstract
Previously, we constructed a glycerol oxidative pathway-deficient mutant strain of Klebsiella pneumoniae by inactivation of glycerol dehydrogenase (dhaD) to eliminate by-product synthesis during production of 1,3-propanediol (1,3-PD) from glycerol. Although by-product formation was successfully blocked in the resultant strain, the yield of 1,3-PD was not enhanced, probably because dhaD disruption resulted in insufficient regeneration of the cofactor NADH essential for the activity of 1,3-PD oxidoreductase (DhaT). To improve cofactor regeneration, in the present study we overexpressed an NAD+-dependent aldehyde dehydrogenase in the recombinant strain. To this end, an aldehyde dehydrogenase AldHk homologous to E. coli AldH but with NAD+-dependent propionaldehyde dehydrogenase activity was identified in K. pneumoniae. Functional analysis revealed that the substrate specificity of AldHk embraced various aldehydes including propionaldehyde, and that NAD+ was preferred over NADP+ as a cofactor. Overexpression of AldHk in the glycerol oxidative pathway-deficient mutant AK/pVOTHk resulted in a 3.6-fold increase (0.57 g l−1 to 2.07 g l−1) in the production of 3-hydroxypropionic acid (3-HP), and a 1.1-fold enhancement (8.43 g l−1 to 9.65 g l−1) of 1,3-PD synthesis, when glycerol was provided as the carbon source, compared to the levels synthesized by the control strain (AK/pVOT). Batch fermentation using AK/pVOTHk showed a significant increase (to 70%, w/w) in conversion of glycerol to the reductive metabolites, 1,3-PD and 3-HP, with no production of by-products except acetate.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Currently, there is great demand for biodiesel production from animal fat or plant oil. However, raw glycerol is a significant by-product of biodiesel production, constituting as much as 10% (w/w) of biodiesel generated [9]. This surplus of raw glycerol has not only greatly disturbed the market for glycerol, affecting both the utility of traditional production methods and price, but is also a significant environmental problem, because untreated glycerol cannot be discharged [2]. Thus, considerable research effort has been devoted to developing methods to refine glycerol (a low-cost feedstock) into industrially valuable materials such as fuels, building blocks for organic syntheses, and bioactive substances.
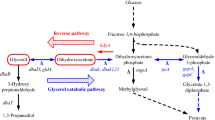
The facultative anaerobic bacterium Klebsiella pneumoniae is typical of microbes capable of fermenting glycerol as the sole source of carbon and energy, and both the genetic and biochemical aspects of the relevant metabolic pathways have been extensively studied (Fig. 1) [21, 22]. Glycerol is dehydrogenated to dihydroxyacetone (DHA) by nicotinamide adenine dinucleotide (NAD+)-dependent glycerol dehydrogenase (DhaD), and DHA is next phosphorylated to dihydroxyacetone phosphate (DHAP) by an ATP-dependent DHA kinase (DhaK). To assist in oxidative metabolism, a reductive pathway has evolved, wherein glycerol is first converted to 3-hydroxypropionaldehyde (3-HPA) by the coenzyme-B12-dependent glycerol dehydratase (DhaB), which is next reduced to 1,3-propanediol (1,3-PD) by a reduced nicotinamide adenine dinucleotide (NADH)-dependent 1,3-PD oxidoreductase (DhaT) [13].
The glycerol fermentative pathway in Klebsiella pneumoniae. The metabolic pathway reconstituted in the present study, using AldHk, is indicated by the dashed line. AldHk NAD+-dependent aldehyde dehydrogenase, DHA dihydroxyacetone, DhaB glycerol dehydratase, DhaD glycerol dehydrogenase, DhaK dihydroxyacetone kinase, DHAP dihydroxyacetone phosphate, DhaT 1,3-propanediol oxidoreductase, G-3-P glycerol-3 phosphate, PEP phosphoenol pyruvate, 3-HPA 3-hydroxypropionaldehyde
The major product of glycerol metabolism, 1,3-PD, is a valuable chemical that is used principally in polymerization with terephthalates to form polymethylene terephthalates [1, 17]. In turn, these chemicals are employed in the manufacture of textile fibers, films, and plastics. Because of the commercial importance of these products, many metabolic and process engineering endeavors have sought to enhance production of 1,3-PD from glycerol by K. pneumoniae [19]. Large levels (approximately 70% of 1,3-PD concentration) of by-products including acetate, ethanol, lactate, succinate, and 2,3-butanediol (2,3-BD) are usually associated with production of 1,3-PD from glycerol by K. pneumoniae wild-type strains, because a glycerol oxidative pathway is also operative. Indeed, the presence of 2,3-BD, a major by-product, may render it difficult to obtain high-purity 1,3-PD during downstream processing, because the boiling points of the two materials are similar. Thus, enhancement of 1,3-PD production and minimization of by-product levels are both desirable goals of metabolic engineering [16].
Previously, we constructed and analyzed mutant strains that did not form by-products during 1,3-PD synthesis. This was achieved by inactivating dhaD, which is the first gene in the oxidative branch of the K. pneumoniae glycerol metabolic pathway [20]. As expected, by-product formation, with the exception of acetate, was completely eliminated in our engineered K. pneumoniae strains. However, the yield of 1,3-PD did not increase, but rather was slightly lower in engineered strains compared to wild type. The most likely explanation is a shortage of intracellular NADH crucially required for DhaT activity when catalyzing synthesis of 1,3-PD. Inactivation of the oxidative pathway would inhibit NADH synthesis (Fig. 1). Recently, Park and colleagues [15] described an NAD+-dependent aldehyde dehydrogenase (AldH) catalyzing the synthesis of 3-hydroxypropionic acid (3-HP) from glycerol in a recombinant Escherichia coli expressing the dhaB gene of K. pneumoniae. This encouraged us to further engineer K. pneumoniae strains to overexpress an NAD-dependent propionaldehyde dehydrogenase that might permit efficient regeneration of NADH from NAD+. To this end, a homolog of AldH was identified and overexpressed in a K. pneumoniae strain, and the effect of this manipulation on reductive glycerol metabolism was examined in the present study.
Materials and methods
Strains, plasmids, and media
The K. pneumoniae AK strain (Fig. 2a; Cu Δ[orfY-dhaT-orfW-orfX-dhaR-dhaD::AprR]) has been previously described [20]. E. coli DH5α was used for DNA manipulation. The plasmid pGEM-T Easy was employed for cloning, whereas pET28a and pBR322 were used for expression of the aldHk gene in E. coli and K. pneumoniae, respectively. Microbial cells were grown in LB medium [yeast extract (Difco), 0.5%; Bactotryptone (Difco), 1.0%; and NaCl, 1.0% (all w/v)] supplemented with appropriate antibiotics [ampicillin (50 μg ml−1); and/or tetracycline (10 μg ml−1 for E. coli and 50 μg ml−1 for K. pneumoniae)].
Cloning and construction of a plasmid permitting K. pneumoniae aldHk gene expression in E. coli
The 1.5-kb aldHk gene (GenBank accession no. ABR76453) was obtained from chromosomal DNA of K. pneumoniae by PCR amplification using the primers P1 (5′-CCATGGATGATGAATTTTCAGCACCTGG-3′; the italicized letters indicate the start codon of aldHk) and P2 (5′-CTCGAGAGACTCCAGGGCAATCCAGA-3′; the italicized letters indicate the Ser-495). The PCR product was cloned into pGEM-T Easy, and clones were subjected to nucleotide sequencing to confirm the absence of any errors. AldHk DNA digested with EcoRI was ligated into the corresponding restriction sites of pET28a, yielding pET-aldHk.
Construction of a plasmid permitting aldHk expression in K. pneumoniae
A schematic representation of the strategy used to construct the pBR-dhaT-aldHk-orfWX (pVOTHk) plasmid is shown in Fig. 2b. The aldHk gene was amplified from chromosomal DNA of K. pneumoniae using the following primers: PaldHk-F (5′-TCTAGA ATGATGAATTTTCAGCACC-3′, the underlined and italicized letters indicate an XbaI site and the start codon of aldHk, respectively) and PaldHk-R (5′-GGATCC GTTAAC TCAAGACTCCAGGGCAATCC-3’, italicized and underlined letters indicate the BamHI and HpaI sites, respectively; the bold letters refer to a stop codon). To introduce the lacZ promoter upstream of aldHk, the gene was also amplified from pBluescript by PCR using the following primers: PlacZ-aldHk-F (5′-CTCGAGGCGCAACGCAATTAATG-3′) and PlacZ-aldHk-R (5′-GGATCC TCTAGAAGCTGTTTCCTGTGT-3′, italicized and underlined letters indicate the BamHI and XbaI sites, respectively). The DNA fragments were cloned into the pGEM-T Easy vector, and clones were subjected to sequencing to confirm the absence of any errors. The XbaI-BamHI fragment including the aldHk gene was next inserted into the corresponding restriction sites downstream of the lacZ promoter sequence. Subsequently, pVOT (pBR-dhaT-orfWX) and pGEM-lacZ-aldHk were restricted by NotI, followed by Klenow treatment and HpaI cleavage, and ligated with pVOT digested with XhoI and treated with the Klenow treatment, yielding plasmid pVOTHk. Plasmid pVOT, described in a previous study [20], includes the orfW gene encoding a reactivation factor of DhaB. All plasmids were transformed into K. pneumoniae strains by electroporation.
Heterologous expression and purification of K. pneumoniae AldHk in E. coli
An E. coli BL21 (DE3) pLysS clone harboring pET-aldHk was grown to mid-exponential phase at 37°C with aeration until the A 600 value attained 0.4–0.6. The expression of aldHk was induced by addition of isopropyl-β-d-thiogalactoside (IPTG) to 0.1 mM followed by incubation for 3 h at 37°C. An E. coli clone harboring pET28a was used as a control. Soluble and insoluble fractions were obtained by centrifugation at 13,000g for 20 min, and AldHk was purified from the soluble fraction by Ni2+-nitrilotriacetic acid (NTA) affinity chromatography.
Cultivation of recombinant K. pneumoniae strains
K. pneumoniae strains were cultivated in 250-ml round-bottomed flasks in 50-ml amounts of defined medium containing 20 g l−1 glycerol, 0.1 M potassium phosphate buffer (pH 7.0), 1 g l−1 yeast extract, 2 g l−1 (NH4)2SO4, 0.2 g l−1 MgSO4, 0.02 g l−1 CaCl2·2H2O, 1 ml Fe solution [5 g l−1 FeSO4·7H2O and 4 ml of HCl (37%, w/v)], and 1 ml of trace element solution [70 mg l−1 ZnCl2, 100 mg l−1 MnCl2·4H2O, 60 mg l−1 H3BO3, 200 mg l−1 CoCl2·4H2O, 20 mg l−1 CuCl2·2H2O, 25 mg l−1 NiCl2·6H2O, 35 mg l−1 Na2MoO4·2H2O, and 4 ml HCl (37%, w/v)] at 37°C and 120 rpm for 36 h. To induce gene expression, IPTG was added to 0.5 mM [6, 20].
To cultivate K. pneumoniae strains in a 5-l bioreactor, seed cells were prepared in a 250-ml flask containing 50 ml preculture medium as described above, and inoculated into the fermentor at 2% (v/v) concentration. Batch and fed-batch fermentation was conducted at 37°C, 200 rpm, and pH 7.0, with 0.5 volume per min of air, in a 5-l stirred fermentor (Kobiotech. Co., Ltd, Korea) containing 2 l of fermentation medium. Tetracycline (10 μg ml−1) and IPTG (0.5 mM) were added in the beginning of the culture cultivation.
AldHk activity assay
AldHk activity was measured according to the method of Leal et al. [11]. The reaction mixture contained 50 mM potassium phosphate buffer (pH 7.0), 1 mM dithiothreitol (DTT), 4 mM NAD+ (or NADP+), 2 mM of an aldehyde substrate, and a protein source (80 μg ml−1 for total cell lysate and 8 μg ml−1 for purified AldHk); incubation proceeded at 37°C for 10 min. Enzyme activity was determined by measuring the amount of NADH (or NADPH) produced from NAD+ (or NADP+), using absorbance readings at 340 nm. One unit (U) of enzyme activity was defined as the amount of enzyme producing 1 μmole NADH per min. The substrate specificity of AldHk was examined by using various aldehydes including propionaldehyde, butyraldehyde, valeraldehyde, isovaleraldehyde, furaldehyde, and benzaldehyde.
Metabolite analysis
The concentrations of glycerol, 3-HP, 1,3-PD, and other metabolites in culture broth were determined by high-performance liquid chromatography (1200, Agilent, CA, USA) equipped with a refractive index detector and an ion-exchange column (300 × 78 mm; Aminex HPX-87H; Bio-Rad, USA). The mobile phase was 0.005 mol l−1 H2SO4 and the flow rate 0.8 ml min−1 during elution. The column and cell temperatures were 65 and 45°C, respectively [25]. A 3-HP standard was purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan).
Gas chromatography–mass spectrometry (GC–MS) was conducted by using an Agilent 6890 GC/5973 MSD fitted with a capillary column (DB5; 0.32 mm × 30 m; J & W Scientific, USA). The temperature was programmed to increase from 70 to 300°C at a rate of 10°C min−1 and the final temperature was held for 5 min. Helium was used as the carrier gas, at a flow rate of 2 ml min−1 [5, 10, 12]. Aliquots of culture supernatants were freeze-dried and extracted in ethyl acetate. Subsequently, the fraction was heated to 70°C for 30 min after suspension in 100-μl amounts of N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA; Sigma–Aldrich, St. Louis, MO) and samples were next analyzed by GC–MS [18].
Results
Cloning and sequencing of an aldH homolog from Klebsiella pneumoniae
Sequence homology alignment analysis identified a homolog of E. coli aldH (an NAD+-dependent aldehyde dehydrogenase catalyzing production of 3-HP from 3-HPA) in the chromosomal DNA of a recombinant strain of K. pneumoniae expressing glycerol dehydratase (dhaB) [14]. The AldH homolog consisted of 496 amino acids and showed 85, 83, 82, and 82% homology with Ald of Enterobacter cloacae (ADF61777), AldH of E. coli (ACT43165), PuuC of Citrobacter rodentium (CBG87141), and AldH of Shigella flexneri (ABF03518; Fig. 3). Residues putatively forming the active site of an aldehyde dehydrogenase, Asn-168, Glu-267, Gly-299, and Cys-302, and a glycine motif involved in binding of the NAD(P)+ cofactor, were well preserved in the AldH homolog of K. pneumoniae, which was termed AldHk.
Homology alignment of AldHk with aldehyde dehydrogenases. Kp_AldHk from Klebsiella pneumoniae (ABR76435); Ec_Ald from Enterobacter cloacae (ADF61777); Ec_AldH from Escherichia coli (ACT43165); Cr_PuuC from Citrobacter rodentium (CBG87141); and Sf_AldH from Shigella flexneri (ABF03518). The putative active sites (triangles) of aldehyde hydrogenases, featuring Asn-168, Glu-267, Gly-299, and Cys-302; and glycine motifs involved in binding of the NAD(P)+ cofactor, are indicated in bold
Expression and purification of K. pneumoniae AldHk in E. coli
SDS–PAGE analysis (Fig. 4a) of the cytoplasmic fraction of recombinant E. coli harboring pET-aldHk confirmed the prominent expression of a recombinant protein of approximately 55 kDa, in line with the expected molecular weight of K. pneumoniae AldHk. No such band was found in the cytoplasmic fraction of control cells harboring vector pET28a. The His6-tagged recombinant AldHk from the cytoplasmic fraction of E. coli harboring pET-aldHk was purified by Ni–NTA affinity column chromatography (data not shown).
Functional analysis of K. pneumoniae AldHk produced in E. coli
Analysis of the cytoplasmic fraction of E. coli cells harboring pET-aldHk revealed an aldehyde dehydrogenase activity when propionaldehyde and NAD+ were employed as substrate and cofactor, respectively, but no activity was detected when supernatant from E. coli harboring control pET28a was examined (Table 1). When NADP+ was employed as a cofactor, dehydrogenase activity fell by 73.9%, indicating that AldHk prefers NAD+ to NADP+ as a cofactor (Fig. 5). In addition to propionaldehyde, AldHk was also active on butyraldehyde, valeraldehyde, isovaleraldehyde, furaldehyde, and benzaldehyde. NAD+ was the preferred cofactor for all substrates except benzaldehyde and furaldehyde.
Overexpression of AldHk in the K. pneumoniae AK strain
To examine the effect of AldHk overexpression on glycerol metabolism, plasmid pVOTHk, including the aldHk, dhaT, and orfW genes, was prepared as shown in Fig. 2b and introduced into K. pneumoniae AK. DhaT encodes the 1,3-PD oxidoreductase of the reductive glycerol pathway, whereas dhaD encodes the glycerol dehydrogenase which is the first enzyme in the oxidative glycerol pathway (Fig. 2b) [20]. SDS–PAGE analysis of the cytoplasmic fraction of recombinant K. pneumoniae AK harboring pVOTHk confirmed the prominent expression of a protein corresponding to AldHk (Fig. 4b). No such band was found in the cytoplasmic fraction of control cells harboring pVOT. Consistent with these results, the cytoplasmic protein fraction of K. pneumoniae AK strains harboring pVOTHk showed much-enhanced aldehyde dehydrogenase activity when propionaldehyde and NAD+ were used as substrate and cofactor, respectively, but no activity was detected in the cytoplasmic protein of a control K. pneumoniae AK strain harboring pVOT (Table 1).
Metabolite analysis of culture broth from recombinant K. pneumoniae AK harboring pVOTHk
Growth of the recombinant AK strain was not affected by overexpression of aldHk when glycerol was used as the sole carbon source (3.6 and 3.8 OD600 of AK strain harboring pVOT and pVOTHk, respectively). One HPLC peak was significantly elevated when culture broth of the AK strain harboring pVOTHk was analyzed (data not shown). The peak was identified by GC–MS, and the mass spectrum and proposed fragmentation mechanism are presented in Fig. 1 in the electronic supplementary material. The BSTFA derivatives displayed a common fragmentation pattern, with ion fragments at m/z = 73 derived by substitution of the active hydrogen atom with the –Si(CH3)3 (trimethylsilyl) group. In addition, fragmentation yielded peaks at m/z 147, 177, and 219, matching database characteristics of 3-HP. The yield of 3-HP was elevated 3.6-fold by overexpression of AldHk (Table 2). As with 3-HP, the yield of 1,3-PD was also enhanced upon AldHk overexpression, from 8.43 to 9.65 g l−1. However, no by-products, with the exception of acetate, were detected in medium from the recombinant AK strain.
Fermentation of the K. pneumoniae AK strain harboring pVOTHk using 5-l bioreactor
The effect of AldHk expression on reductive glycerol metabolism in the K. pneumoniae AK strain was examined by batch fermentation using a 5-l bioreactor. The recombinant AK strain harboring pVOTHk consumed all added glycerol (20 g l−1) after 20 h. Maximal 1,3-PD production was obtained at higher level of 9.2 g l−1 compared to 8.5 g l−1 by the control AK strain harboring pVOT until 18 h of growth; further increase in the incubation period resulted in slight decrease of 1,3-PD yield (to 8.1 g l−1; Fig. 6). This may reflect the fact that DhaT activity is reversible [8]. In line with this suggestion, 3-HP production increased continuously to 6.0 g l−1. In contrast, the production of 3-HP reached 1.1 g l−1 in the control strain. For both 1,3-PD and 3-HP, the conversion yield from glycerol was dramatically increased (to over 70%, w/w) in the AK strain harboring pVOTHk, compared to the control strain with pVOT (about 45%, w/w). Also, by-product formation, except for acetate, was completely blocked in the recombinant strains.
Additionally, fed-batch fermentation using K. pneumoniae AK strain harboring pVOTHk gave the maximal production of 1,3-PD (22.7 g l−1; Fig. 7). Compared to 1,3-PD, the maximal 3-HP production was slightly increased by fed-batch fermentation (6.8 g l−1), probably due to the serious toxicity of the compound.
Discussion
The fermentation of glycerol by the K. pneumoniae wild-type strain yields the valuable chemical 1,3-PD, but also generates large amounts of by-products, including lactate and 2,3-BD (to a level approximately 70% that of 1,3-PD), thus compromising efforts to obtain high-purity 1,3-PD by downstream processing. Previously, we engineered K. pneumoniae to inactivate the oxidative glycerol pathway responsible for by-product production by substitution of the glycerol dehydrogenase gene (dhaD) gene on chromosomal DNA with a gene conveying resistance to the antibiotic apramycin [20]. Although this K. pneumoniae strain (AK/pVOT) did not produce by-products, the yield of 1,3-PD was not elevated and the rate of glycerol consumption fell somewhat [20]. A possible explanation is insufficient regeneration of the cofactor NADH crucial for the activity of DhaT (Fig. 1) of the recombinant strain (AK/pVOT), because inactivation of DhaD may adversely affect the cellular balance between the cofactors NAD+ and NADH. To stimulate regeneration of NADH in the AK strain harboring pVOT, we overexpressed an NAD+-dependent aldehyde dehydrogenase in this strain. To this end, AldHk, exhibiting a dehydrogenase activity on propionaldehyde as a substrate, and preferring NAD+ to NADP+ as a cofactor, was identified and overexpressed in the AK strain, yielding strain AK/pVOTHk. This manipulation significantly stimulated the reductive glycerol pathway, enhancing synthesis of 1,3-PD and 3-HP. The fermentation using AK/pVOTHk shows no production of by-products except acetate. Thus, we have further engineered the AK strain to increase the yield of reductive metabolites, 1,3-PD and 3-HP, from glycerol. The increased production of acetate was unrelated to expression of aldHk because the high production was also found in the control strain AK/pVOT. Although the molecular basis for the increase of acetate level in the recombinant strain defective of glycerol oxidative pathway is unclear [20], it could be related to increased demand of energy production (ATP) in the mutant strain.
As described above, overexpression of AldHk significantly enhanced production of 3-HP in the AK strain. This indicates that AldHk is active on 3-HPA as a substrate, similar to its activity on propionaldehyde, although this was not directly demonstrated because 3-HPA is not commercially available. We also explored the effect of AldHk in a wild-type strain of K. pneumoniae. However, no stimulation of reductive glycerol metabolism was evident in that strain (data not shown), indicating that inactivation of the oxidative glycerol pathway is required before the effect of AldHk overexpression is apparent. In other words, an enzyme activity compensating the effect of AldHk is provided by oxidative metabolism in the wild-type strain.
To the best of our knowledge, this is the first report to identify 3-HP in the culture broth of K. pneumoniae. We also found 3-HP in the broth of a wild-type strain of K. pneumoniae grown in glycerol- but not glucose-containing medium (data not shown), suggesting that K. pneumoniae has a de novo metabolic pathway synthesizing 3-HP from glycerol. However, AldHk is unlikely to be involved in this process, as shown by a gene inactivation experiment. An aldHk mutant strain (ΔaldHk) of K. pneumoniae was constructed by substitution of the chromosomal aldHk gene with a gene encoding resistance to the antibiotic apramycin (data not shown). This mutant strain also produced 3-HP (data not shown), indicating that the metabolic pathway was still functional. It has been reported that some Lactobacillus species, such as L. reuteri and L. collinoides, can produce 3-HP from glycerol [4, 23]. Although the catalytic mechanism remains unclear, a propanediol utilization protein (PduP) has been suggested as an important component of the metabolic pathway [26]. A homolog of Lactobacillus pduP is present in chromosomal DNA of K. pneumoniae.
3-HP is regarded as a potential C3 building block and is hence ranked, by the United States Department of Energy, as one of the top 12 value-added chemicals produced from biomass [24]. Although several chemical and biological synthesis methods have been reported to date, 3-HP is produced commercially by only a few suppliers, as an aqueous solution, because of high production costs and low yield [3, 7]. Therefore, an organism with a metabolic pathway enabling the production of 3-HP via very low-cost fermentation would be valuable. Recently, Mohan Raj et al. [14] reported that a recombinant E. coli strain expressing glycerol dehydratase and aldehyde dehydrogenase genes produced 3-HP to a maximum of 31 g l−1 in fed-batch fermentation [15]. However, fermentation using recombinant E. coli requires supplementation of the medium with high-cost coenzyme B12 to activate glycerol dehydratase, because E. coli does not possess the biosynthetic pathway for coenzyme B12. By contrast, K. pneumoniae is a typical glycerol-fermenting bacterium that biosynthesizes coenzyme B12. Thus, the recombinant K. pneumoniae strain described in the present study will be useful in industrial scale-up processes, to produce these valuable chemicals from glycerol.
References
Bhatia SK, Kurian JV (2008) Biological characterization of Sorona polymer from corn-derived 1,3-propanediol. Biotechnol Lett 30:619–623. doi:10.1007/s10529-007-9607-z
Da Silva GP, Mack M, Contiero J (2009) Glycerol: a promising and abundant carbon source for industrial microbiology. Biotechnol Adv 27:30–39. doi:10.1016/j.biotechadv.2008.07.006
Della Pina C, Falletta E, Rossi M (2009) Oxidation of allyl alcohol in the presence of a gold catalyst: a route to 3-hydroxypropionic acid. Chem Sus Chem 2:57–58. doi:10.1002/cssc.200800172
Garai-Ibabe G, Ibarburu I, Berregi I, Claisse O, Lonvaud-Funel A, Irastorza A, Dueñas MT (2008) Glycerol metabolism and bitterness producing lactic acid bacteria in cider making. Int J Food Microbiol 121:253–261. doi:10.1016/j.ijfoodmicro.2007.11.004
Ghassempour A, Nojavan S, Talebpour Z, Amiri AA, Najafi NM (2004) Monitoring of the fermentation media of citric acid by the trimethylsilyl derivatives of the organic acids formed. J Agric Food Chem 52:6384–6388. doi:10.1021/jf030751v
Hao J, Lin R, Zheng Z, Sun Y, Liu D (2008) 3-Hydroxypropionaldehyde guided glycerol feeding strategy in aerobic 1,3-propanediol production by Klebsiella pneumoniae. J Ind Microbiol Biotechnol 35:1615–1624. doi:10.1007/s10295-008-0405-y
Jiang X, Meng X, Xian M (2009) Biosynthetic pathways for 3-hydroxypropionic acid production. Appl Microbiol Biotechnol 82:995–1003. doi:10.1007/s00253-009-1898-7
Johnson EA, Lin EC (1987) Klebsiella pneumoniae 1,3-propanediol NAD oxydoreductase. J Bacteriol 169:2050–2054
Johnson DT, Taconi KA (2007) The glycerin glut: options for the value-added conversion of crude glycerol resulting from biodiesel production. Environ Prog 26:338–348
Kakinuma H, Yamamoto S, Nishimuta T, Yamashita H (1986) A computer program for automated diagnosis of organic acidurias by GC/MS. J Mass Spectrom Soc Jpn 34:131–138
Leal NA, Havemann GD, Bobik TA (2003) PduP is a coenzyme-A-acylating propionaldehyde dehydrogenase associated with the polyhedral bodies involved in B12-dependent 1,2-propanediol degradation by Salmonella enterica serovar Typhimurium LT2. Arch Microbiol 180:353–361. doi:10.1007/s00203-003-0601-0
Lee SH, Park SJ, Park OJ, Cho J, Rhee JW (2009) Production of 3-hydroxypropionic acid from acrylic acid by newly isolated Rhodococcus erythropolis LG12. J Microbiol Biotechnol 19:474–481
Marçal D, Rêgo AT, Carrondo MA, Enguita FJ (2009) 1,3-Propanediol-dehydrogenase from Klebsiella pneumoniae: decameric quaternary structure and possible subunit cooperativity. J Bacteriol 191:1143–1151. doi:10.1128/JB.01077-08
Mohan Raj S, Rathnasingh C, Jo JE, Park S (2008) Production of 3-hydroxypropionic acid from glycerol by a novel recombinant Escherichia coli BL21 strain. Process Biochem 43:1440–1446
Mohan Raj S, Rathnasingh C, Jung WC, Park S (2009) Effect of process parameters on 3-hydroxypropionic acid production from glycerol using a recombinant Escherichia coli. Appl Microbiol Biotechnol 84:649–657. doi:10.1007/s00253-009-1986-8
Oh BR, Seo JW, Choi MH, Kim CH (2008) Optimization of culture conditions for 1,3-propanediol production from crude glycerol by Klebsiella pneumoniae using response surface methodology. Biotechnol Bioprocess Eng 13:524–532
Pagliaro M, Ciriminna R, Kimura H, Rossi M, Pina CD (2007) From glycerol to value-added products. Angew Chem Int Eng 46:4434–4440
Pietrogrande MC, Bacco D, Mercuriali M (2010) GC-MS analysis of low-molecular-weight dicarboxylic acids in atmospheric aerosol: comparison between silylation and esterification derivatization procedures. Anal Bioanal Chem 396:877–885. doi:10.1007/s00216-009-3212-z
Saxena RK, Anand P, Saran S, Isar J (2009) Microbial production of 1,3-propanediol: recent developments and emerging opportunities. Biotechnol Adv 27:895–913. doi:10.1016/j.biotechadv.2009.07.003
Seo MY, Seo JW, Heo SY, Baek JO, Rairakhwada D, Oh BR, Seo PS, Choi MH, Kim CH (2009) Elimination of by-product formation during production of 1,3-propanediol in Klebsiella pneumoniae by inactivation of glycerol oxidative pathway. Appl Microbiol Biotechnol 84:527–534. doi:10.1007/s00253-009-1980-1
Skraly FA, Lytle BL, Cameron DC (1998) Construction and characterization of a 1,3-propanediol operon. Appl Environ Microbio1 64:98–105
Sun J, van den Heuvel J, Soucaille P, Qu Y, Zeng AP (2003) Comparative genomic analysis of dha regulon and related genes for anaerobic glycerol metabolism in bacteria. Biotechnol Prog 19:263–272. doi:10.1021/bp025739m
Talarico TD, Casas IA, Cheng TC, Dobrodogsz WJ (1988) Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri. Antimicrob Agents Chemother 32:1854–1858
Werpy T, Holladay J, White J (2004) Top value-added chemicals from biomass. PNNL-14808. PNNL, Richland
Yang G, Tian J, Li J (2007) Fermentation of 1,3-propanediol by a lactate deficient mutant of Klebsiella oxytoca under microaerobic conditions. Appl Microbiol Biotechnol 73:1017–1024. doi:10.1007/s00253-006-0563-7
Yasuda S, Mukoyama M, Ilorikawa H, Toraya T, Morita H (2007) Process for producing 1,3-propanediol and/or 3-hydroxypropionic acid. US Patent Application US 2007/0148749 A1
Acknowledgments
This research was supported by Korea Ministry of Environment "Conversing Technology Project."
Author information
Authors and Affiliations
Corresponding author
Additional information
The authors Lian Hua Luo and Jeong-Woo Seo are co-first authors and contributed equally.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Luo, L.H., Seo, JW., Oh, BR. et al. Stimulation of reductive glycerol metabolism by overexpression of an aldehyde dehydrogenase in a recombinant Klebsiella pneumoniae strain defective in the oxidative pathway. J Ind Microbiol Biotechnol 38, 991–999 (2011). https://doi.org/10.1007/s10295-010-0872-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-010-0872-9