Abstract
Angiotensin-II acts at its type-1 receptor (AT1R) in the brain to regulate body fluid homeostasis, sympathetic outflow and blood pressure. However, the role of the angiotensin type-2 receptor (AT2R) in the neural control of these processes has received far less attention, largely because of limited ability to effectively localize these receptors at a cellular level in the brain. The present studies combine the use of a bacterial artificial chromosome transgenic AT2R-enhanced green fluorescent protein (eGFP) reporter mouse with recent advances in in situ hybridization (ISH) to circumvent this obstacle. Dual immunohistochemistry (IHC)/ISH studies conducted in AT2R-eGFP reporter mice found that eGFP and AT2R mRNA were highly co-localized within the brain. Qualitative analysis of eGFP immunoreactivity in the brain then revealed localization to neurons within nuclei that regulate blood pressure, metabolism, and fluid balance (e.g., NTS and median preoptic nucleus [MnPO]), as well as limbic and cortical areas known to impact stress responding and mood. Subsequently, dual IHC/ISH studies uncovered the phenotype of specific populations of AT2R-eGFP cells. For example, within the NTS, AT2R-eGFP neurons primarily express glutamic acid decarboxylase-1 (80.3 ± 2.8 %), while a smaller subset express vesicular glutamate transporter-2 (18.2 ± 2.9 %) or AT1R (8.7 ± 1.0 %). No co-localization was observed with tyrosine hydroxylase in the NTS. Although AT2R-eGFP neurons were not observed within the paraventricular nucleus (PVN) of the hypothalamus, eGFP immunoreactivity is localized to efferents terminating in the PVN and within GABAergic neurons surrounding this nucleus. These studies demonstrate that central AT2R are positioned to regulate blood pressure, metabolism, and stress responses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The renin angiotensin system (RAS) is best known as an endocrine system that regulates cardiovascular function and hydromineral balance. The potent vasoconstrictor and sodium conserving actions of angiotensin-II (Ang-II) at its type-1 receptor (AT1R) are widely studied and drugs that target either the synthesis of Ang-II or its action at the AT1R have been utilized as therapeutics for cardiovascular pathologies for decades. Over recent years, additional complexities of the RAS have come to light and have revealed additional therapeutic potential of targeting this system. It is now recognized that the RAS is not only an endocrine system, but also acts as a paracrine and autocrine system (Bader and Ganten 2008; Cassis et al. 2008; Cuadra et al. 2010). That is, all of the components of the RAS are found in a number of specific tissues, such as the brain and adipose, and Ang-II is thought to exert tissue-specific actions at these sites. Second, the RAS is now also established to impact numerous homeostatic systems (e.g., those that regulate metabolism and glucose homeostasis), in addition to those that regulate hydromineral balance and cardiovascular function. Finally, it is also now appreciated that Ang-II and other angiotensin peptides [e.g., angiotensin (1–7)] can act at diverse receptors, such as its type-2 receptor (AT2R) and the Mas receptor, to exert functional effects that are often in opposition to those produced by Ang-II acting at AT1R (AbdAlla et al. 2001; Kostenis et al. 2005; Santos et al. 2008, 2003).
Although the existence of the AT2R has long been acknowledged, its physiological role in adulthood has received far less attention than that of its AT1R counterpart. This likely stems from a number of reports that have indicated that AT2R expression predominates during development, while the expression of AT1R predominates during adulthood (Grady et al. 1991; Ichiki and Inagami 1995; Nuyt et al. 1999; Yu et al. 2010). Nonetheless, there are lines of evidence indicating that the brains of adult rodents densely express functional AT2Rs (Gao et al. 2012; Hauser et al. 1998; Lenkei et al. 1996, 1997) and that AT2R activation, in many instances, counteracts AT1R activation (Sumners et al. 2013). Of particular relevance, it is established that chronic activation of the AT1R can have deleterious consequences on cardiovascular function (Shi et al. 2010; Ye et al. 2002), emotional state (Krause et al. 2011; Saavedra et al. 2005) and metabolic function (de Kloet et al. 2010), while more recently, it has been recognized that activation of AT2R may protect against these effects (Bosnyak et al. 2010; Danyel et al. 2013; Gao et al. 2011, 2014; Jing et al. 2012; Ohinata et al. 2009; Okuyama et al. 1999; Qi et al. 2012; Siragy et al. 2000; Sumners et al. 2013). This has led to the hypothesis that the balance between AT1R and AT2R activation governs the responses of many tissues to Ang-II, suggesting that AT2R may serve as an important new therapeutic avenue for a number of centrally mediated pathologies. That said, determining the precise molecular mechanism(s) by which brain AT2R regulates physiology has been complicated by limitations with regard to available techniques for AT2R localization (autoradiography and immunostaining), in particular on a cellular level within the brain.
In the present manuscript, we have circumvented these obstacles by making use of recent advances in molecular and genetic approaches to assess the localization of AT2R. These new methodologies provide not only improved specificity, but also improved sensitivity for detecting the AT2R. As a consequence, the present characterization of AT2R expression provides a novel view of the location of this receptor in the brain. It not only replicates studies conducted decades ago via receptor binding assays and radioactive in situ hybridization (ISH), but also reveals the expression of AT2R in additional nuclei that are critical for the regulation of cardiovascular function, energy metabolism, and responses to stress. Furthermore, using these techniques, the present data reveal the phenotype of many of these AT2R-containing cells in the central nervous system thereby uncovering some key neural circuits in which AT2R are positioned to regulate cardiovascular, metabolic, and stress responses.
Materials and methods
Animals
Experiments were conducted in outbred male mice on a [FVB/N]/[Crl:CD1(ICR)] mixed background. Mice were 10–12 weeks old at the initiation of the experiments and were maintained in a temperature and humidity-controlled room on a 12:12-h light–dark cycle with food and water available ad libitum. The AT2R reporter mouse line was initially generated by in vitro fertilization of FVB/NJ mice with Tg(AT2R-eGFP)IP72Gsat mouse sperm (on a FVB/NTac x CD1(ICR) background) obtained from the Mutant Mouse Regional Resource Center. The details of the construct can be found at the vendor’s website (http://www.mmrrc.org/catalog/sds.php?mmrrc_id=30278). Briefly, an enhanced green fluorescent protein (eGFP) reporter gene and subsequent polyadenylation sequence were inserted into the AT2R bacterial artificial chromosome (BAC) clone at the start codon of the first coding exon of the AT2R gene. This construct was then used to produce the transgenic mouse and results in eGFP expression driven by all of the regulatory sequences of the AT2R BAC gene. Mice used for the studies were maintained on a FVB/N × CD1 mixed background and were hemizygous for the transgene. In a subset of control studies, ISH was performed on the wild-type littermates of the AT2R reporter mouse. These mice were, therefore, on the same mixed background strain. Brains obtained from AT2R knockout (KO; AT2R-/Y) mice served as negative controls for the ISH studies. These KO mice were on a C57BL/6 background and a detailed description of their development can be found in Hein et al. (1995).
Tissue collection and sectioning
At 10–12 weeks of age, mice were anesthetized with pentobarbital and perfused transcardially with 0.15 M NaCl followed by 4 % paraformaldehyde. Brains were then post-fixed for 3–4 h, after which they were stored in 30 % sucrose until sectioned using a Leica CM3050 S cryostat (Leica, Buffalo Grove, Illinois). For ISH experiments, perfused mouse brains were sectioned at 20 µm into 6 serial sections and immediately mounted onto SuperFrost Plus Gold Microscope Slides. After air-drying at room temperature for 20–30 min, slides were stored at −80 °C until further processing. All solutions were prepared with DEPC-treated water and filtered using a 0.22 µm filter and tissue collection and sectioning were performed in RNase-free conditions. For immunohistochemistry (IHC) studies, mouse brains were sectioned at 30 µm into four serial sections and stored in cryoprotective solution at −20 °C, until further processing.
In situ hybridization (RNAscope)
Single and double fluorescent RNAscope ISH studies were performed on brain tissue collected from AT2R-eGFP mice, as well as on their wild-type littermates as per the manufacturer’s instructions; however, the pretreatment procedure was optimized to provide visualization of mRNA transcripts in the presence of preserved AT2R-eGFP protein. Specifically, after allowing tissue sections to dry at 25 °C for 30 min, they were incubated with Pretreatment 4 (a protease) and then underwent the RNAscope Multiplex Fluorescent ISH protocol. Specific details of the RNAscope ISH technique are outlined on the vendor’s website (www.acdbio.com). Briefly, target probes are designed using the proprietary ACD RNAscope Probe Design pipeline, and contain 20 short double-Z oligonucleotide probe pairs that are gene specific. For amplification and visualization, both Z-probes must bind to the mRNA of interest. RNAscope detection reagents are then sequentially hybridized to amplify the signal and subsequently color label the individual mRNA transcripts. For these experiments, the color label was assigned to either FAR RED (Excitation 647 nm; Emission 690 ± 10 nm) or ORANGE (Excitation 550 nm; Emission 580 ± 10 nm). Using this technique, each punctate dot represents a single mRNA target molecule.
For the analysis of AT1R, AT2R, and eGFP mRNA localization to AT2R-eGFP-positive cells, brain regions of interest (ROIs) included the following: the nucleus of the solitary tract (NTS), the dorsal motor nucleus of the vagus (DMNV), the area postrema (AP), the medial prefrontal cortex (mPFC), and the median preoptic nucleus (MnPO). Each slide contained 4–8 sections, depending on the size of the sections. For each ROI, the following probe combinations were utilized: (1) Negative control probe, (2) Positive control probe, (3) AT2R and eGFP, and (4) AT1R. Additional negative controls for the probes included brain sections collected from KO mice that lack AT2R. One series of sections through the hindbrains of these KO mice were hybridized with the AT2R probe and revealed no positive label. Two additional series of sections were hybridized with the negative and positive control probes, to determine the exposure time and image processing necessary to provide optimal visualization of RNA signal and also to control for possible RNA degradation. Probes for glutamic acid decarboxylase-1 (Gad1) and vesicular glutamate transporter-2 (vGlut2) were used to determine the percentage of AT2R-eGFP neurons in the NTS that are GABAergic or glutamatergic, respectively. In a separate control experiment, dual ISH for Gad1 and vGlut2 was performed on AT2R-eGFP sections to assess the extent of overlap between the two probes.
Immunohistochemistry
Table 1 lists the primary antibodies, their vendors and the dilutions utilized. All primary antibodies were characterized by the manufacturers and in previously published studies (de Kloet et al. 2014; Gautron et al. 2013; Jessberger et al. 2008; Kádár et al. 2010; Krause et al. 2011; Langlet et al. 2013; Mousa et al. 2011). All secondary antibodies were purchased from Jackson Immunoresearch, raised in donkey and used at a 1:500 dilution. In general, the IHC procedures were as follows. Brain sections were removed from cryoprotectant solution and rinsed five times for 5 min in 50 mM KPBS and then incubated in blocking solution (2 % normal donkey serum and 0.2 % Triton X in 50 mM KPBS) for 2 h at 25 °C. This was followed by incubation with the primary antibody in blocking solution for 18 h at 4 °C. Sections were again rinsed five times for 5 min in 50 mM KPBS and then incubated in the secondary antibody for 2 h at 25 °C. After a final series of rinses (5 × 5 min), sections were mounted onto slides, allowed to air dry and then cover-slipped using polyvinyl alcohol mounting medium. For double-label IHC, sections were incubated with both primary antibodies simultaneously and, subsequently, both secondary antibodies simultaneously. For anti-human neuronal protein (anti-HuC/D) staining of Elav family members HuC, HuD, and Hel-N, leading to the specific labeling of neurons in mice, an additional protease digestion step, using pretreatment 4 (20 min at 25 °C) from the RNAscope ISH kit was completed prior to initiation of the IHC protocol. Importantly, all qualitative IHC studies were performed in at least four separate mice.
Image capture and processing
All images were captured and processed using Axiovision 4.8.2 software and a Zeiss AxioImager fluorescent Apotome microscope. For RNAscope ISH and dual IHC/RNAscope ISH, z-stacks of the proteins and transcripts of interest were captured at 40× magnification throughout the ROIs using neuroanatomical landmarks found in a mouse brain atlas (Franklin and Paxinos 2008). In all cases, z-steps were set at 0.5 µm, with an average of 20 optical sections per image. For each experiment, sections hybridized with the positive control probes were used to determine the exposure time and image processing required to provide optimal visualization of RNA signal. These same parameters were then used for visualization of Gad1, vGlut2, eGFP, AT2R, and AT1aR mRNA signal in experimental sections, to assess background fluorescence in sections hybridized with negative control probe (DapB) and to determine the specificity of the probes using tissue obtained from KO mice. Importantly, using these exposure times and image processing parameters there was minimal or no fluorescence in sections hybridized with the negative control probe and in sections obtained from KO mice.
To generate the results reported in Table 2, as well as the low-power images of the entire coronal sections from the mice depicted in Fig. 1, images throughout each coronal section were captured at 2.5× magnification. Subsequently, images were imported into ImageJ and the MosaicJ plugin was utilized to assemble mosaics of each coronal section. For higher power images (5× − 40×) of single, double, and triple-label IHC, exposure time was adjusted using the best fit feature in Axiovision to provide optimal visualization. All final figures were then prepared using Adobe Photoshop 7.0 where the brightness and contrast was adjusted to provide optimal visualization.
AT2R-eGFP-positive cells contain AT2R mRNA. Projection images through the (a–c) medial prefrontal cortex [mPFC; +1.70 mm rostral to bregma], (d–f) nucleus of the solitary tract [NTS; −7.20 mm caudal to bregma], and (g–i) dorsal motor nucleus of the vagus [DMNV; −7.20 mm caudal to bregma] of AT2R-eGFP reporter mice depicting (a, d, g) eGFP immunoreactivity in green, (b, e, h) AT2R-mRNA in red or magenta and (c, f, i) the merged image also including DAPI in blue. Scale bars 20 µm
Analysis
Mosaics of entire coronal sections through four separate mouse brains were analyzed to determine the number of eGFP-positive cell bodies and or the presence of eGFP fibers or terminals within brain regions important for cardiovascular, metabolic, endocrine, and behavioral responses to stress. Criteria for scoring the number of AT2R-positive cell bodies were as follows: 0 = no cells within the ROI, 1 = 1–25 % of the area occupied by cell bodies, 2 = 26–50 % of the area occupied by cell bodies, 3 = 51–75 % of the area occupied by eGFP-positive cell bodies, and 4 = 76–100 % of the area occupied by eGFP-positive cell bodies. All sections were analyzed by two separate investigators and mean score for each brain region was calculated and rounded to the nearest whole number. Brain regions that received a score between 0 and 0.5 are listed as <1. The presence of fibers or terminals was assessed in the same sections and are scored as either + (indicating that there are fibers/terminals present within the ROI) or – (indicating that no fibers/terminals were observed within the ROI).
Analysis of co-localization of mRNA transcripts with eGFP fluorescence was performed on selected brain regions in four separate AT2R-eGFP mouse brains. 40× magnification z-stacks of ROIs were used for the determination of the percentage of eGFP neurons that contain AT2R, eGFP, AT1aR, Gad1, and Vglut2 mRNA. An average of 2–8 z-stacks was captured for each ROI, depending on the rostrocaudal length of the particular ROI. eGFP neurons were considered to contain the RNA of interest if at least three visible transcripts, defined as an individual punctate dot, were observed within the volume of the eGFP fluorescence. Data are reported as the percentage of AT2R-eGFP cells that contain the RNA for each gene within each ROI.
To determine appositions between eGFP nerve terminals and the neurons within the paraventricular nucleus (PVN), 40× z-stacks (with an average of 20 optical Section (0.5 µm/section)) was assessed. Appositions were verified by a lack of separation between dual eGFP-positive boutons and the neuronal-specific marker (i.e., HuC/D).
Results
Co-localization of AT2R mRNA and eGFP protein
To assess the extent to which eGFP expression is indicative of AT2R expression, RNAscope ISH for AT2R mRNA and eGFP mRNA coupled with eGFP IHC was performed on selected brain regions determined to contain eGFP-positive cells. AT2R-eGFP cells containing mRNA for both eGFP and AT2R were observed in all of the brain nuclei assessed, which included the following: the mPFC, MnPO, NTS, AP, and DMNV. Notably, in the brain regions assessed, 100 ± 0 % of the cells that fluoresced green also contained mRNA for eGFP and 98 ± 0.18 % contained mRNA for AT2R. Conversely, AT2R mRNA was undetectable in the striatum, a region that was predetermined to lack AT2R-eGFP cell bodies and fibers/terminals (data not shown). Figure 1 contains representative images of eGFP protein (green) and AT2R mRNA (red) localized to the same cells within the mPFC (Fig. 1a–c; n = 4), NTS (Fig. 1d–f; n = 4) and DMNV (Fig. 1g–i; n = 4). To control for the possibility that the AT2R-eGFP transgene was modulating AT2R mRNA expression, RNAscope ISH in combination with IHC for the neuronal marker, HuC/D was performed on brain sections throughout the NTS and DMNV collected from wild-type mice. Qualitative assessment of these sections, revealed a similar distribution of AT2R transcripts within the NTS (Fig. 2a–c) and DMNV (Fig. 2d–f). RNAscope ISH was performed on the NTS of AT2R KO mice and revealed no positive signal when hybridized with the AT2R probe (Fig. 2g–i), but extensive labeling when hybridized with the positive control probe (Ubc1; Fig. 2j), corroborating the specificity of this ISH technique. Finally, Fig. 3 contains a representative dual IHC image highlighting the degree of overlap between native eGFP fluorescence and eGFP IHC.
AT2R mRNA expression in wild-type and AT2R KO mice. Projection images through (a–c) the NTS and (d–f) the DMNV of wild-type mice depicting (a, d) HuC/D-immunoreactivity, (b, e) AT2R-mRNA and (c, f) the merged images. g–i Projection images through the DMNV of an AT2R-KO mouse depicting lack of AT2R-mRNA (green) localization to HuC/D-containing neurons (red). j Merged projection image depicting the presence of the positive control mRNA (Ubc3) within the same area of the DMNV of the KO mouse. Scale bars 50 µm. All images were captured at −7.20 mm caudal to bregma as in Fig. 1 and using the same exposure time and brightness/contrast adjustment as tissue from AT2R-eGFP mice
AT2R-eGFP immunoreactivity in the AT2R-eGFP reporter. a Low magnification image through the NTS and AP of an AT2R-eGFP reporter mouse with the native GFP fluorescence in green and eGFP immunoreactivity in magenta. b–c Projection images of the NTS and depicting (b) AT2R-eGFP (green), eGFP immunoreactivity (magenta) and (d) the merged image. CC central canal, AP area postrema, NTS nucleus of the solitary tract. Scale bars 100 µm
Distribution of AT2R-eGFP in brain nuclei that regulate cardiovascular function, metabolism, and stress responding
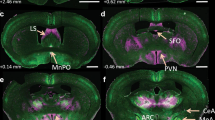
The primary goal of these studies was to evaluate whether AT2R are positioned to impact the neural circuits that regulate cardiovascular function, metabolism, and stress responding. Table 2 encompasses a survey of AT2R-eGFP expression throughout specific forebrain and hindbrain nuclei that are known to regulate these processes. Figure 4 depicts representative low-power images of key brain regions that contain AT2R-eGFP cells. In particular, high levels of AT2R-eGFP-positive cell bodies were observed in the mPFC, MnPO, portions of the amygdala, NTS, and the AP.
Localization of eGFP immunoreactivity throughout the brain of AT2R-eGFP reporter mice. a–h Representative coronal sections through selected brain regions of the AT2R-eGFP reporter mouse line. The number in the lower left of each image indicates the approximate distance rostral/caudal from bregma, in accordance with the mouse brain atlas (Franklin and Paxinos 2008). mPFC medial prefrontal cortex, MnPO median preoptic nucleus, LS lateral septum, AH anterior hypothalamus, PVN paraventricular nucleus of the hypothalamus, MeA medial amygdala, CeA central amygdala, NA nucleus ambiguus, NTS nucleus of the solitary tract, HG hypoglossal nucleus, 10 N dorsal motor nucleus of the vagus, AP area postrema. Scale bars 1 mm
Within the forebrain, the mPFC AT2R-eGFP neurons were localized to both the infralimbic and prelimbic portions at a density of approximately 26–50 % of the surface area of the nuclei. Amygdalar AT2R-eGFP neurons were primarily restricted to the central and medial nuclei (at a density of approximately 26–50 %), while no AT2R-eGFP neurons were observed within the basolateral nucleus. Furthermore, both the bed nucleus of the stria terminalis (BNST) and the lateral septum contained AT2R-eGFP cell bodies at a density of 1–25 %. Within the hypothalamus, in addition to a concentrated population of AT2R-eGFP cells within the MnPO (~50–75 % of the surface area being occupied by AT2R-eGFP cell bodies), AT2R were also localized to the anterior, lateral, dorsomedial, and posterior hypothalamic nuclei (Table 2) at an average density of ~1–25 %. Scattered or no AT2R-eGFP cell bodies were observed in other hypothalamic nuclei assessed, including the PVN. The forebrain circumventricular organs also contained few AT2R-eGFP neurons, with negligible amounts localizing to the subfornical organ and a density of 1–25 % being contained within the organum vasculosum of the lamina terminalis.
Several mid- and hindbrain nuclei were also populated with AT2R-eGFP neurons. AT2R-eGFP cell bodies occupied approximately 26–50 % of the surface area of the DMNV, NTS, and hypoglossal nuclei and 1–25 % of parabrachial and nucleus ambiguus. Other nuclei assessed included the dorsal raphe, locus coeruleus, inferior olive, rostroventrolateral medulla (RVLM) and caudal ventrolateral medulla (CVLM).
AT2R-eGFP expression is localized to neurons
Based on the morphology of the AT2R positive cells (see Figs. 1, 2, 3, 4), we hypothesized that AT2R-eGFP cells were mostly neuronal. To test this hypothesis, we focused on a select few brain regions that contained AT2R-eGFP and co-stained for neuronal (HuC/D), astroglial (GFAP), or microglial (Iba-1) markers. Figures 5 and 6 contain images of co-staining for eGFP and these cell-type-specific markers through the NTS, AP, and DMNV of the AT2R-eGFP mice. In all brain sections assessed, 100 % of the AT2R-eGFP cells exhibited positive staining for the neuronal-specific marker HuC/D (see Fig. 5), while no co-staining was observed between Iba-1 or GFAP (see Fig. 6).
AT2R-eGFP cells express the neuronal marker, HuC/D. a Low magnification image through the NTS, AP, and DMNV of an AT2R-eGFP reporter mouse co-labeled for AT2R-eGFP (green) and HuC/D (red). b–c Projection images of the NTS depicting b HuC/D (red), c AT2R-eGFP (green), and d the merged image. CC central canal, AP area postrema. Scale bars 100 µm
AT2R-eGFP cells do not co-localize with cells expressing microglial (Iba-1) or astroglial (GFAP) markers in the NTS, AP, or DMNV. Coronal sections through the AP, NTS, and DMNV of an AT2R-eGFP reporter mouse co-stained for eGFP and either (a–d) Iba-1or (e–h) GFAP. a Low magnification image through the NTS, AP, and DMNV of an AT2R-eGFP reporter mouse co-labeled for AT2R-eGFP (green) and Iba-1 (red). b–d Projection images of the NTS depicting b Iba-1 (red), c AT2R-eGFP (green), and d the merged image. e Low magnification image through the NTS, AP, and DMNV of an AT2R-eGFP reporter mouse co-labeled for AT2R-eGFP (green) and GFAP (red). f–h Projection images of the NTS depicting f GFAP (red), g AT2R-eGFP (green), and h the merged image. Scale bars 100 µm
Co-localization of AT1R mRNA with AT2R-eGFP neurons
Several lines of evidence suggest that AT2R activation counteracts AT1R-mediated actions via opposing intracellular signaling cascades; however, direct evidence localizing AT1R and AT2R to the same cells within specific cardiovascular control centers of the brain is limited. In the present studies, we performed an analysis of the localization of AT1aR mRNA to AT2R-eGFP-positive cells within selected brain regions (i.e., the PVN, MnPO, NTS, and AP) that contain AT2R-eGFP neurons and/or were also previously determined to contain AT1R (Hauser et al. 1998). Figure 7 includes representative images of AT1R mRNA and AT2R-eGFP fluorescence in the MnPO, PVN, NTS, and AP. In the brain regions assessed, a small percentage of AT2R-eGFP neurons did contain mRNA for AT1aR; however, the majority of AT2R-positive cells did not contain AT1R transcripts. Specifically, the percentage of AT2R-eGFP neurons that are positive for AT1R mRNA within these brain regions was as follows: MnPO = 17.1 ± 3.6 %, NTS = 8.7 ± 1.0 %, AP = 6.87 ± 1.85 % (n = 4). In all of the brains assessed (n = 4), there were no AT1aR transcripts observed in the AT2R-eGFP neurons that surround the PVN. The implication is that, at least in these brain regions, AT1R and AT2R are localized primarily to separate sets of neurons.
The co-localization of AT1aR mRNA and AT2R-eGFP immunoreactivity. a Low magnification image of a coronal section through the NTS (−7.76 mm caudal to bregma) of an AT2R-eGFP mouse. High magnification projection images through the NTS depicting b eGFP immunoreactivity in green, c AT1aR mRNA ISH in red, and d the merged image. e Low magnification image of a coronal section through the AP (−7.48 mm caudal to bregma) of an AT2R-eGFP mouse. High magnification projection images through the AP depicting f eGFP immunoreactivity in green, g AT1aR mRNA ISH in red, and h the merged image. i Low magnification image of a coronal section through the PVN (−0.82 mm caudal to bregma) of an AT2R-eGFP mouse. High magnification projection images through the PVN depicting j eGFP immunoreactivity in green, k AT1aR mRNA ISH in red, and l the merged image. m Low magnification image of a coronal section through the MnPO (+0.26 mm rostral to bregma) of an AT2R-eGFP mouse. High magnification projection images through the MnPO depicting n eGFP immunoreactivity in green, o AT1aR mRNA ISH in red, and (p) the merged image. Arrows denote co-labeled cells. 3v third cerebral ventricle, ac anterior commissure. Scale bars 200 µm (a, e, i, m), 50 µm (d, l, p) or 20 µm (h)
AT2R-eGFP expression is localized to GABA and ChAT, but not TH, neurons in the hindbrain
Several hindbrain nuclei are important for the regulation of cardiovascular function, metabolism, and stress responding. In particular, the NTS, the DMNV, and the nucleus ambiguus are nuclei that regulate these systems and express AT2R-eGFP and AT2R mRNA. We conducted experiments to determine the phenotype of AT2R-eGFP neurons within these hindbrain nuclei, so as to inform on potential mechanism(s) by which these AT2R may impact these systems. Figure 8 contains dual IHC/ISH results indicating the percentage of NTS neurons that express Gad1 mRNA or vGlut2 mRNA. As highlighted in Fig. 8, the majority of AT2R-eGFP neurons contain Gad1 mRNA (see Fig. 8a–d, i), rather than vGlut2 mRNA, indicating that they are primarily GABAergic. However, a small group of neurons located adjacent to the caudal AP are predominantly glutamatergic (see Fig. 8e–i). Importantly, dual ISH for Gad1 and vGlut2 indicates that Gad1 and vGlut2 mRNAs are localized to separate neurons in the NTS, thereby validating the utility of the RNAscope ISH for these mRNAs (Fig. 9). Figure 10 contains representative IHC results indicating that AT2R-eGFP is not co-localized with TH within the NTS. This was confirmed by viewing TH positive cells at multiple levels of the NTS (data not shown); however, there are numerous AT2R-eGFP cells within the DMNV and nucleus ambiguus that co-label for ChAT.
Co-localization of AT2R-eGFP and vGlut2 or Gad1 mRNA in the nucleus of the solitary tract (NTS). a Low magnification image of a coronal section through the NTS (−7.48 mm caudal to bregma) of an AT2R-eGFP mouse. High magnification projection images through the NTS depicting b eGFP immunoreactivity in green, c Gad1 mRNA ISH in magenta, and d the merged image. e Low magnification image of a coronal section through the NTS (−7.76 mm caudal to bregma) of an AT2R-eGFP mouse. High magnification projection images through the AP depicting f eGFP immunoreactivity in green, g vGlut2 mRNA ISH in magenta, and h the merged image. i Bar graph indicating the average percentage of AT2R-eGFP neurons in the NTS that contain either Gad1 mRNA or vGlut2 mRNA between −7.76 and −6.96 mm caudal to bregma (n = 4, bars represent SEM). Scale bars 200 µm (a, e) or 50 µm (b–d and f–i)
Dual multiplex ISH for vGlut2 and Gad1 mRNA with AT2R-eGFP IHC in NTS. High magnification projection images through the NTS (−7.76 mm caudal to bregma) of an AT2R-eGFP mouse depicting a eGFP immunoreactivity in green, b Gad1 mRNA ISH in cyan, c vGlut2 mRNA in magenta, and d the merged image. Scale bars 50 µm
AT2R-eGFP expression in GABAergic neurons surrounding the PVN and on terminals that contact AVP neurons
Numerous lines of evidence demonstrate that AT2R activation impacts the PVN; however, the present study indicates scant presence of AT2R-eGFP-positive cell bodies within the PVN. This point is illustrated well by Fig. 11, which also highlights the lack of co-localization between AT2R-eGFP cell bodies and arginine vasopressin (AVP)-containing PVN neurons. Despite this lack of co-localization, a higher power view indicates that there are an abundance of AT2R-eGFP-positive neuronal fibers/terminals localized to the PVN that come in close contact with these AVP neurons (see Fig. 11). While it is clear that the PVN is devoid of AT2R-eGFP neuronal cell bodies, it is apparent from Fig. 11 that there are sparse AT2R-eGFP neurons surrounding the PVN, and the representative images in Fig. 12 demonstrate that these neurons contain Gad1 mRNA, and thus are GABAergic.
AT2R-eGFP fibers/terminals are in close proximity to arginine vasopressin (AVP) neurons of the PVN. a Low magnification image through the PVN of an AT2R-eGFP reporter mouse depicting the lack of co-staining between (AVP; red) and eGFP (green), scale bar 100 µm. b High magnification projection image highlighting the presence of AT2R-eGFP fibers/terminals green in close proximity to AVP neurons in the PVN, scale bar 50 µm. c High magnification projection image highlighting the apparent lack of separation between AT2R/eGFP terminals/fibers and AVP neurons of the PVN, scale bar 10 µm
AT2R-eGFP cells surrounding the PVN express Gad1. a Low magnification image through the PVN (−1.06 mm caudal to bregma) of an AT2R-eGFP reporter mouse co-labeled for AT2R-eGFP (green) and Gad1 mRNA (red). b–d Projection images of neurons surrounding the PVN depicting b Gad1 mRNA (red), c AT2R-eGFP (green), and d the merged image. 3v third ventricle. Scale bars a 100 µm and b–d 50 µm
Discussion
In recent years, the potential therapeutic value of targeting the AT2R for various neurogenic pathologies has gained significant attention. Of particular relevance, AT2R are thought to be involved in a number of brain functions, including cognition (Gallo-Payet et al. 2011; Jing et al. 2012), limiting damage during stroke (Joseph et al. 2014; McCarthy et al. 2012) and in blood pressure regulation (Gao et al. 2011; Li et al. 2003). That said, the progression of research in these areas has been slow, due to technological issues that have limited the ability to effectively localize AT2R at a cellular level within the brain. The present study circumvents these issues by making use of the increased sensitivity and resolution of recent advances in genetic techniques in mice and ISH to provide an in depth analysis of AT2R localization within selected mouse brain nuclei. In the present study, we used an AT2R-eGFP BAC-transgenic mouse line to uncover the distribution of AT2R in the adult mouse brain. The major findings of the present study are that AT2R are well positioned to influence brain nuclei controlling cardiovascular function, metabolism, and stress responding. Furthermore, using this approach, we have been able to determine, for the first time, the neurotransmitter phenotype of key populations of AT2R-eGFP neurons within select cardiovascular control centers of the brain and have also demonstrated the potential for intracellular AT2R-AT1aR interactions by evaluating AT1aR/AT2R-eGFP co-localization. Lastly, the present data also indicate that under normal conditions, the expression of AT2R within the brain is limited to neurons and that microglia and astrocytes do not exhibit detectable levels of this receptor.
A novel approach to localize AT2R within the brain
Until this point, elucidation of the mechanisms by which activation of brain AT2R exerts functional effects has been hindered by a number of factors: the unreliability of antibodies for angiotensin receptors (Benicky et al. 2012; Hafko et al. 2013; Herrera et al. 2013), the low degree of sensitivity and resolution of traditional receptor autoradiography and ISH approaches, and the fact that receptor mRNAs are often expressed at a lower level than other mRNAs has made the use of ISH for the detection of receptors challenging. An important contribution of the present studies was, therefore, to assess the utility of a newly generated BAC-transgenic AT2R-eGFP reporter mouse line and of recent advances in ISH to examine the cellular localization of AT2R within the brain. Initial experiments conducted in the AT2R-eGFP BAC-transgenic mouse line revealed a high degree of co-localization between cells that display eGFP immunoreactivity, with those that contain mRNA for AT2R, thereby validating this mouse line for the investigation of AT2R localization within the brain. Although the quantity of visible AT2R transcripts within these GFP cells was, in some instances, low relative to some other mRNAs examined (e.g., Gad1), mammalian proteins are generally orders of magnitude more abundant than their mRNAs (Schwanhausser et al. 2011). As a consequence, even an apparently low number of mRNAs within a given cell can have functional relevance at the level of protein expression.
AT2R localization within the brain
It has long been assumed that the expression and function of AT2R predominates during development, while that of AT1R prevails during adulthood (Nuyt et al. 1999; Yu et al. 2010). As a consequence, the role of AT2R in adults has often been overlooked. Nonetheless, there are studies that have revealed AT2R expression and binding in adult rodents. These studies were primarily conducted in the rat (Johren et al. 1995; Lenkei et al. 1996, 1997; Millan et al. 1991) and documented AT2R expression in many areas that are consistent with the present study. Such studies have also been conducted in the mouse, which is of particular relevance for the current manuscript. In this regard, Hauser et al. (1998) documented AT2R binding in many mouse forebrain and hindbrain nuclei, such as the amygdala and NTS. Our mRNA and eGFP expression results are in many instances, consistent with these findings; however, there are some key differences. For example, the present studies revealed that AT2R-eGFP cells and mRNA were abundant within the MnPO, while Hauser et al. (1998) did not observe AT2R binding within this region. Importantly, rather than these discrepancies pointing to a deficiency in either the present or the former study, the combined interpretation of these data may inform on the cellular localization of AT2R. That is, it is possible that AT2R are localized exclusively to the terminals of MnPO neurons, rather than their cell bodies. This localization would lead to the presence of AT2R-eGFP cell bodies in the MnPO (as reported here), but no AT2R binding [as documented in Hauser et al. (1998)]. Another cause for discrepancies between the present and former data may arise from the higher level of resolution of the methodologies used in the present studies, thereby revealing AT2R expression in some additional brain regions. Importantly, the increased resolution of the techniques used in the current study, have allowed for an in-depth analysis of the phenotype of AT2R positive cells and provide insight into how activation of AT2R may impact brain function.
In addition, a number of studies have examined AT2R immunoreactivity within the brain (Coleman et al. 2009; Premer et al. 2013; Reagan et al. 1994). However, the controversial nature of available angiotensin receptor antibodies for IHC (Benicky et al. 2012; Hafko et al. 2013; Herrera et al. 2013) has led to waning in the confidence in the results of these studies. Nonetheless, it is of direct relevance to consider these previous results in the context of the present studies, and this is done throughout the subsequent sections of the discussion.
AT2R-eGFP cells are neuronal
The present data indicate that, at least under basal conditions and within the brain regions assessed, AT2R-eGFP cells are neuronal. AT2R-eGFP cells within the brain not only exhibit neuronal morphology, but they also display extensive immunoreactivity for the neuronal marker, HuC/D. Conversely, no immunoreactivity for microglial or astroglial markers was observed in AT2R-eGFP cells under these conditions. Consistent with these observations, Lenkei et al. (1996) previously determined that in the lateral septum AT2R mRNA is not detected in GFAP containing cells (Lenkei et al. 1996). Furthermore, although Saavedra and colleagues found that the AT2R receptor ligand CGP42112, binds to microglia/macrophages, they also determined that Ang-II does not bind microglia/macrophages in the brain (Saavedra and Pavel 2006). The implication is that this particular AT2R ligand also binds to a non-AT2R site in these cells (Egidy et al. 1997; Saavedra and Pavel 2006).
Although we did not observe AT2R-eGFP co-localization with Iba-1 or GFAP in the present study, there are lines of evidence that AT2R activation impacts non-neuronal cells within the brain (McCarthy et al. 2012, 2014; Miyoshi et al. 2008; Rodriguez-Pallares et al. 2008). In this regard, AT2R mRNA and protein have been localized to cultured rat microglia and astrocytes (Miyoshi et al. 2008; Rodriguez-Pallares et al. 2008), which contrast our in vivo results in mice. Furthermore, in vivo, AT2R agonism using Compound 21 (C21) enhances microglial activation when given prior to stroke (McCarthy et al. 2014). These previous findings coupled with the present results suggest that the impact of AT2R agonism on non-neuronal cells within the brain is likely an indirect consequence of neuronal AT2R activation. However, the present studies do not rule out the possibility that other cell- types within the brain begin to express the receptor in certain circumstances; perhaps in response to an injury or insult. There is some evidence that this may be the case (Fuchtbauer et al. 2010). For example, studies using an AT2R antibody suggest that there is an increase in co-localization between AT2R and the microglial marker lectin in the site of ischemia induced by middle cerebral artery occlusion (Wu et al. 2013). The approaches utilized in the current study may be used to elucidate AT2R plasticity during pathological conditions.
The hindbrain
The present studies revealed that AT2R-eGFP-positive neurons and fibers/terminals are densely localized to the NTS, DMNV, and NA, while only fibers/terminals are present in the RVLM. In agreement with these findings, there is recent Western blot evidence that AT2R are densely expressed in the whole brainstem of adult mice (Gao et al. 2012) and rats (Gao et al. 2011; Yu et al. 2010). Although AT2R have traditionally been thought to function predominantly in development (Nuyt et al. 1999; von Bohlen und Halbach et al. 2001; Yu et al. 2010), these previous studies determined that the levels of AT2R in adult rodent hindbrains exceed those of brainstems collected from fetal rodents (Gao et al. 2012, 2011), suggesting the possibility of a physiological role for AT2R in adulthood.
The nucleus of the solitary tract
The NTS is a hindbrain nucleus that densely expresses AT2R-eGFP neurons and plays a pivotal role in the neural regulation of cardiovascular function, metabolism, and stress responding. The intermediate third of the NTS, which is populated with AT2R-eGFP neurons, is considered a locus for the regulation of cardiovascular function (Guyenet 2006). It is the principle site for the termination of baroreceptor afferent fibers, and thus mediates the inhibitory actions of baroreceptors on sympathetic outflow, thereby participating in the acute regulation of blood pressure. During various pathological conditions (e.g., metabolic challenges (Guimaraes et al. 2014)), there is a ‘resetting’ of the baroreflex that is mediated by altered activity of the NTS that culminates in the defense of another (higher or lower) level of blood pressure. In addition to its prominent role in the regulation of cardiovascular function, the NTS also has many distinct roles in the regulation of metabolism and stress responses. (Hayes et al. 2009; Travers et al. 1987; Scheuer 2010; Zhang et al. 2010).
Another key contribution of this study is an in-depth analysis of the distribution and phenotype of AT2R-eGFP neurons in the NTS. AT2R-eGFP neurons within the NTS do not display tyrosine hydroxylase (TH) immunoreactivity. Moreover, minimal co-localization between AT2R-eGFP and vGlut2 was observed, with the exception of a small group of neurons adjacent to the caudal-most portion of the AP. Rather, the majority of AT2R neurons within the NTS are GABAergic (i.e., they express Gad1 mRNA). This is of importance because within the NTS, GABA is known to have potent effects on cardiovascular function (Masubuchi et al. 2004; Tsukamoto and Sved 1993). GABA receptor agonists elicit pressor responses when injected into the NTS that are exacerbated in rats that are subjected to hypertensive stimuli, such as a high salt diet or DOCA/salt hypertension (Tsukamoto and Sved 1993; Masubuchi et al. 2004; Vitela and Mifflin 2001). Conversely, GABAB receptor antagonism (CGP-35348) prompts greater reductions in blood pressure in rodents subjected to hypertensive stimuli (Masubuchi et al. 2004). In Spontaneously Hypertensive Rats, GABAA or GABAB blockade into the NTS increases sympathetic baroreflex range and gain, suggesting that GABAergic mechanisms may be tonically reducing sympathetic baroreflex range and gain in this model of hypertension. Although the present studies indicate that AT2R are likely localized to GABAergic neurons within this region, the impact of AT2R activation on their neuronal activity is yet to be determined. That being said, it has been documented that angiotensin receptor binding and expression are sensitive to hypertensive stimuli (Plunkett and Saavedra 1985) and that manipulation of the RAS within the NTS regulates the baroreflex and blood pressure (Eshima et al. 2000; Polson et al. 2007; Shan et al. 2013; Wang et al. 2007), as well as indices of GABA signaling (Zhang et al. 2009).
The dorsal motor nucleus of the vagus and nucleus ambiguus
Another novel finding of the current study is that AT2R-eGFP is co-localized predominantly with choline acetyl transferase (ChAT) immunoreactivity within the DMNV and the NA. The high degree of co-localization with ChAT is noteworthy because cholinergic vagal efferent preganglionic axons arise from the NA and the DMNV and these nuclei both send projections to neurons within the cardiac ganglia. As such, these nuclei are critical for parasympathetic cardiac regulation and increased vagal input to the heart causes bradycardia. Furthermore, the DMNV is widely recognized to impact metabolic function via vagal input to the pancreas and various organs of the gastrointestinal tract (Mussa and Verberne 2013) and Ang-II has previously been found to excite DMNV neurons (Mo et al. 1992). It is also clear from our data that not all of the ChAT cells in the DMNV and NA are positive for AT2R. Thus, it is possible that the AT2R-containing ChAT neurons are a subset that serves only one of the above-mentioned functions, rather than both.
The ventrolateral medulla
In the present studies, we did not observe AT2R-eGFP-positive cell bodies in the RVLM and CVLM. Despite the absence of AT2R-eGFP neurons in the RVLM/CVLM, there are several previous reports of AT2R actions within these areas. For example, there is western blot evidence for AT2R protein in the RVLM which is reduced in congestive heart failure (Gao et al. 2008) and it has been proposed that reduced AT2R signaling within the RVLM may be involved in sympathetic over activity that is characteristic of congestive heart failure (Gao et al. 2008). In further support of a functional role for AT2R within the RVLM, microinjection of the AT2R agonist CGP42112 into this region decreases blood pressure and sympathetic outflow, effects that contrast AT1R activation within this area (Gao et al. 2008). Overexpression of AT2R in the RVLM decreases blood pressure in normal rats (Gao et al. 2008), while knockdown of AT2R causes elevations in blood pressure (Wang et al. 2004). Although we do not observe many AT2R-eGFP cell bodies within these areas, it is important to note that these areas are densely populated with AT2R-eGFP-positive terminals/fibers. It is possible that rather than being localized to cell bodies within the RVLM, AT2R are expressed presynaptically on nerve terminals and that activation of these AT2R, may influence neurotransmitters released from these terminals.
The forebrain
A number of forebrain nuclei that regulate cardiovascular function, metabolism, and stress responding are also populated with AT2R-eGFP neurons, with a particular abundance being localized to the mPFC, the MnPO, as well as portions of the amygdala and hypothalamus.
The medial preoptic area
The medial preoptic area contains a collection of subnuclei (e.g., the MnPO) that are implicated in the control of body fluid homeostasis and blood pressure, particularly in response to hyperosmotic stimuli. Of the brain nuclei assessed in the present study, the MnPO has the greatest abundance of AT2R-eGFP neurons. As discussed earlier, while these findings contradict previous data, which demonstrated no AT2R binding within this nucleus (Hauser et al. 1998), they might suggest that AT2R are localized to the terminals of MnPO neurons, rather than their soma. The present studies also suggest that AT2R-eGFP efferent terminals are within the PVN of the hypothalamus and it is intriguing to hypothesize that AT2R-eGFP neurons of the MnPO may be projecting to PVN neurons and impacting their activity.
The paraventricular nucleus of the hypothalamus
The PVN is a critical control center for numerous homeostatic functions, including blood pressure regulation, energy balance, and stress responding. Further, numerous studies, including our own, have indicated that Ang-II actions at its AT1R within this nucleus are important contributors to these processes (Chen et al. 2011; de Kloet et al. 2013, de Kloet et al. 2014; Northcott et al. 2010). Another important finding of the current manuscript is that, for the most part, AT2R-eGFP neurons do not reside within the PVN. As expected based on the minimal localization of AT2R within the PVN, AT2R-eGFP is not localized to PVN AVP neurons; however, there is an abundance of AT2R-eGFP-positive terminals/fibers in the PVN. This is in agreement with a number of ISH studies conducted in mice and rats (Lenkei et al. 1996, 1997); however, it contrasts immunohistochemical studies (Coleman et al. 2009; Reagan et al. 1994) which have documented AT2R binding and immunoreactivity within the PVN. This discrepancy could be the result of species differences between mice and rats, as the previous studies were predominantly conducted in rats, while the present studies are exclusively conducted in mice. However, it is also possible that although there are no AT2R-eGFP-positive neurons in the PVN, AT2R are localized to nerve terminals within this region. This could account for the above-mentioned AT2R binding and immunoreactivity within the PVN, but not for the lack of detection of AT2R mRNA (Lenkei et al. 1996, 1997) or AT2R-eGFP cell bodies, as determined in the present studies. Consistent with this notion, many of the regions we found to contain AT2R-eGFP neurons, including the MnPO and BNST are known to project to the PVN (Choi et al. 2007; Stocker and Toney 2005) and, therefore, may be the source of these AT2R-eGFP terminals within this nucleus.
Other hypothalamic nuclei
Other hypothalamic nuclei that contain AT2R-eGFP neurons include the anterior (AH), posterior (PH), and dorsomedial hypothalamic (DMH) nuclei. Of particular interest, the DMH plays an important role in the regulation of blood pressure, energy balance and stress responses. For example, there is evidence for a DMH-NTS connection that mediates cardiovascular changes (i.e., change in heart rate variability and baroreflex) associated with social defeat-induced anxiety (Sevoz-Couche et al. 2013).
Amygdala
The amygdala is an important component of the limbic system that contains a collection of subnuclei, some of which are populated with AT2R-eGFP neurons. In particular, AT2R-eGFP neurons are distributed throughout portions of the central (CeA) and medial (MeA) nuclei, but are absent within the basolateral nucleus. These AT2R-eGFP-containing areas of the amygdala are involved in various processes, such as memory, emotional reactions, hydromineral balance, and metabolism (Petrovich et al. 2009; Schulkin et al. 1989; Zardetto-Smith et al. 1994). Both the CeA and MeA are implicated in behavioral and cardiovascular responses associated with fear and anxiety (Saha et al. 2005). Although the RAS is known to influence the function of amygdalar neurons (Breigeiron et al. 2002; Cecconello et al. 2010; Heshmatian et al. 2007; Llano Lopez et al. 2012; von Bohlen und Halbach et al. 2001), the contribution of AT2R to these effects is not clear. Activation of the RAS within the amygdala has been found to impact blood pressure (Heshmatian et al. 2007), hydromineral balance (Sakai et al. 2000; Yan et al. 2014), and sexual behavior (MeA) (Breigeiron et al. 2002; Cecconello et al. 2010). Pharmacological inhibition of amygdalar AT1R via bilateral infusion of losartan is anxiolytic in acutely stressed rats when tested in the elevated plus maze (Llano Lopez et al. 2012). The presence of AT2R within this area brings to light the possibility that an increase in AT2R binding upon losartan administration is contributing to the anxiolytic effect of the drug. Consistent with a developmental effect of AT2R within this area, genetic deletion of AT2R increases cell number in the amygdala (von Bohlen und Halbach et al. 2001).
The bed nucleus of the stria terminalis and lateral septum
The BNST is another limbic brain region that contains AT2R-eGFP neurons and fibers/terminals and is known to participate in cardiovascular, endocrine, and behavioral response to stress (Choi et al. 2007; Ciriello et al. 2013; Crestani et al. 2013). It serves as a relay between other limbic areas of the forebrain and the hypothalamus and brainstem (Crestani et al. 2013) and Ang-II has been found to impact neuronal activity within the BNST via an AT1R-dependent mechanism (Lienard et al. 1996). Consistent with our finding that AT2R-eGFP neurons are localized to the BNST, Hauser et al. observed AT2R binding within this region (Hauser et al. 1998). Previous studies have also observed that angiotensin-II receptor binding sites in the ventral portion of the BNST are reduced upon interruption of the medial forebrain bundle, while lesion of cell bodies within this area using a neurotoxin did not alter binding (Grove et al. 1998). The implication is that Ang-II receptors are likely located on nerve terminals in this region that have their cell bodies in the brainstem.
The lateral septum is also involved in autonomic and behavioral responses to stressful stimuli (Reis et al. 2011) and contains AT2R-eGFP neurons. A lateral septum to MeA pathway has been found to regulate cardiovascular homeostasis (Scopinho et al. 2012). Furthermore, the lateral septum is a part of a pressor pathway that originates in the cingulate cortex (Fernandes et al. 2005).
Ventral medial prefrontal cortex
The mPFC is involved in cardiovascular modulation (Fernandes et al. 2003, 2007; Tavares et al. 2004), metabolic and behavioral control, particularly in response to stressful stimuli (McKlveen et al. 2013; Muller-Ribeiro et al. 2012). Both the infralimbic and prelimbic portions of the ventral mPFC are involved in cardiovascular and behavioral responses to stressful stimuli, and contain a particular abundance of AT2R-eGFP neurons. While these areas are connected to portions of the amygdala as well as hindbrain and thoracic (Bacon and Smith 1993) cardiovascular regulatory pathways, there is no available evidence for effects of Ang-II via AT2R at these sites.
AT1R/AT2R interactions
Another contribution of the present manuscript is an analysis of the co-expression of AT1aR and AT2R in neuronal populations within selected brain nuclei. For the most part, AT1a mRNA and AT2R-eGFP were localized to distinct cells within the brain regions examined; however, a small percentage of neurons examined did contain both receptors. Several lines of evidence have led to the hypothesis that stimulation of AT2R opposes AT1R activation via triggering opposing intracellular signaling cascades (Sumners et al. 1991). It has, therefore, been speculated that the balance between AT1R and AT2R contributes to the extent by which the RAS impacts various homeostatic systems. Our results suggest that in some instances AT1R and AT2R may counter-regulate each other by activating opposing intracellular signaling cascades within the same neurons; however, it appears that these subtypes of angiotensin receptors more frequently influence the function of separate neurons that reside in the same brain nuclei.
Conclusions: implications for the central control of cardiovascular, metabolic, and stress homeostasis
Based on the localization observed in this study, it is reasonable to hypothesize that activation of AT2R within the brain would influence cardiovascular function, energy homeostasis and stress responding. Although the literature is replete with data suggesting that Ang-II can act via AT1R to negatively impact these processes, there are far less studies examining the potential role of AT2R within the brain. Nonetheless, there is both genetic and pharmacological evidence that complement these neuroanatomical studies and suggest that there are indeed functional AT2R in the adult brain that exert protective effects for many aspects of physiology. In regards to cardiovascular function, a number of studies have revealed a cardioprotective role for brain AT2R. AT2R null mice have elevated blood pressure relative to controls, while Ang-II given intracerebroventricularly evokes a larger increase in blood pressure in AT2R KO mice (Ichiki et al. 1995; Li et al. 2003), implying that AT2R plays an inhibitory role in Ang-II-induced blood pressure elevation. Furthermore, direct activation of central AT2R using C21 decreases BP and NE excretion in the urine (Gao et al. 2011) and improves arterial baroreflex sensitivity in rats with heart failure (Gao et al. 2014). There is also some evidence for beneficial actions of central AT2R on metabolic function. Rats that overexpress AT2R have a lower body weight (Peters et al. 2012). Further, AT2R activation using C21 improves glucose tolerance (Shao et al. 2014) and has direct effects on the pancreatic islets to increase insulin biosynthesis and secretion (Shao et al. 2013). The present studies also reveal that AT2R are distributed within many brain regions that regulate stress responding and mood and there are some published studies supporting the notion that AT2R activation may serve as a therapeutic for stress-related pathologies. AT2R KO mice exhibit increased anxiety-like behavior compared to wild-type mice (Okuyama et al. 1999) and AT2R also act in opposition to AT1R, in regard to the cardiovascular responses to stress (Israel et al. 2000a, b). It has been found that upon administration of an AT1R antagonist, AT2R signaling becomes augmented. To this end, administration of an AT1R antagonist and then subsequent exposure to foot shock stress, leads to a consistent vasodepressor response that is mediated by AT2R, as it is blocked administration of an AT2R antagonist (Israel et al. 2000b). Collectively, the current localization of AT2R provides an important and novel look at the distribution of AT2R within brain regions that regulate cardiovascular, metabolic, and behavioral responses to psychological or physiological stressors, thereby opening doors for future investigation of the functional role of these receptors in the neural control of these processes.
References
AbdAlla S, Lother H, Abdel-tawab AM, Quitterer U (2001) The angiotensin II AT2 receptor is an AT1 receptor antagonist. J Biol Chem 276:39721–39726. doi:10.1074/jbc.M105253200
Bacon SJ, Smith AD (1993) A monosynaptic pathway from an identified vasomotor centre in the medial prefrontal cortex to an autonomic area in the thoracic spinal cord. Neuroscience 54:719–728
Bader M, Ganten D (2008) Update on tissue renin-angiotensin systems. J Mol Med 86:615–621
Benicky J, Hafko R, Sanchez-Lemus E, Aguilera G, Saavedra JM (2012) Six commercially available angiotensin II AT1 receptor antibodies are non-specific. Cell Mol Neurobiol 32:1353–1365. doi:10.1007/s10571-012-9862-y
Bosnyak S, Welungoda IK, Hallberg A, Alterman M, Widdop RE, Jones ES (2010) Stimulation of angiotensin AT2 receptors by the non-peptide agonist, Compound 21, evokes vasodepressor effects in conscious spontaneously hypertensive rats. Br J Pharmacol 159:709–716. doi:10.1111/j.1476-5381.2009.00575.x
Breigeiron MK, Morris M, Lucion AB, Sanvitto GL (2002) Effects of angiotensin II microinjected into medial amygdala on male sexual behavior in rats. Horm Behav 41:267–274. doi:10.1006/hbeh.2002.1771
Cassis LA, Police SB, Yiannikouris F, Thatcher SE (2008) Local adipose tissue renin-angiotensin system. Curr Hypertens Rep 10:93–98
Cecconello AL, Raineki C, Sebben V, Lucion AB, Sanvitto GL (2010) Effect of acute stress on sexual behavior in female rats: participation of the central angiotensinergic system. Behav Brain Res 207:429–433. doi:10.1016/j.bbr.2009.10.026
Chen AD, Zhang SJ, Yuan N, Xu Y, De W, Gao XY, Zhu GQ (2011) Angiotensin AT1 receptors in paraventricular nucleus contribute to sympathetic activation and enhanced cardiac sympathetic afferent reflex in renovascular hypertensive rats. Exp Physiol 96:94–103. doi:10.1113/expphysiol.2010.054353
Choi DC, Furay AR, Evanson NK, Ostrander MM, Ulrich-Lai YM, Herman JP (2007) Bed nucleus of the stria terminalis subregions differentially regulate hypothalamic-pituitary-adrenal axis activity: implications for the integration of limbic inputs. J Neurosci 27:2025–2034. doi:10.1523/jneurosci.4301-06.2007
Ciriello J, Caverson MM, Li Z (2013) Effects of hypocretin and norepinephrine interaction in bed nucleus of the stria terminalis on arterial pressure. Neuroscience 255:278–291. doi:10.1016/j.neuroscience.2013.09.032
Coleman CG, Anrather J, Iadecola C, Pickel VM (2009) Angiotensin II type 2 receptors have a major somatodendritic distribution in vasopressin-containing neurons in the mouse hypothalamic paraventricular nucleus. Neuroscience 163:129–142. doi:10.1016/j.neuroscience.2009.06.032
Crestani CC, Alves FH, Gomes FV, Resstel LB, Correa FM, Herman JP (2013) Mechanisms in the bed nucleus of the stria terminalis involved in control of autonomic and neuroendocrine functions: a review. Curr Neuropharmacol 11:141–159. doi:10.2174/1570159X11311020002
Cuadra AE, Shan Z, Sumners C, Raizada MK (2010) A current view of brain renin-angiotensin system: is the (pro)renin receptor the missing link? Pharmacol Ther 125:27–38
Danyel LA, Schmerler P, Paulis L, Unger T, Steckelings UM (2013) Impact of AT2-receptor stimulation on vascular biology, kidney function, and blood pressure. Integr Blood Press Control 6:153–161. doi:10.2147/IBPC.S34425
de Kloet AD, Krause EG, Woods SC (2010) The renin angiotensin system and the metabolic syndrome. Physiol Behav 100:525–534. doi:10.1016/j.physbeh.2010.03.018
de Kloet AD et al (2013) Angiotensin type 1a receptors in the paraventricular nucleus of the hypothalamus protect against diet-induced obesity. J Neurosci 33:4825–4833. doi:10.1523/JNEUROSCI.3806-12.2013
de Kloet AD, Pioquinto DJ, Nguyen D, Wang L, Smith JA, Hiller H (2014) CS Obesity induces neuroinflammation mediated by altered expression of the renin-angiotensin system in mouse forebrain nuclei. Physiol Behav 136:31–38
Egidy G, Friedman J, Viswanathan M, Wahl LM, Saavedra JM (1997) CGP-42112 partially activates human monocytes and reduces their stimulation by lipopolysaccharides. Am J Physiol 273:C826–C833
Eshima K, Hirooka Y, Shigematsu H, Matsuo I, Koike G, Sakai K, Takeshita A (2000) Angiotensin in the nucleus tractus solitarii contributes to neurogenic hypertension caused by chronic nitric oxide synthase inhibition. Hypertension 36:259–263
Fernandes KB, Crippa GE, Tavares RF, Antunes-Rodrigues J, Correa FM (2003) Mechanisms involved in the pressor response to noradrenaline injection into the cingulate cortex of unanesthetized rats. Neuropharmacology 44:757–763
Fernandes KB, Tavares RF, Correa FM (2005) The lateral septal area is involved in the pressor pathway activated by microinjection of norepinephrine into the rat brain cingulate cortex. Neuropharmacology 49:564–571. doi:10.1016/j.neuropharm.2005.04.025
Fernandes KBP, Tavares RF, Pelosi GG, Corrêa FMA (2007) The paraventricular nucleus of hypothalamus mediates the pressor response to noradrenergic stimulation of the medial prefrontal cortex in unanesthetized rats. Neurosci Lett 426:101–105. doi:10.1016/j.neulet.2007.08.063
Franklin KBJ, Paxinos G (2008) The mouse brain: in stereotaxic coordinates, 3rd edn. Elsevier, New York
Fuchtbauer L, Toft-Hansen H, Khorooshi R, Owens T (2010) Expression of astrocytic type 2 angiotensin receptor in central nervous system inflammation correlates with blood-brain barrier breakdown. J Mol Neurosci 42:89–98. doi:10.1007/s12031-010-9371-8
Gallo-Payet N, Guimond MO, Bilodeau L, Wallinder C, Alterman M, Hallberg A (2011) Angiotensin II, a neuropeptide at the frontier between endocrinology and neuroscience: is there a link between the angiotensin ii type 2 receptor and Alzheimer’s disease? Front Endocrinol 2:17. doi:10.3389/fendo.2011.00017
Gao J, Chao J, Parbhu KJ, Yu L, Xiao L, Gao F, Gao L (2012) Ontogeny of angiotensin type 2 and type 1 receptor expression in mice. J Renin Angiotensin Aldosterone Syst 13:341–352. doi:10.1177/1470320312443720
Gao J, Zhang H, Le KD, Chao J, Gao L (2011) Activation of central angiotensin type 2 receptors suppresses norepinephrine excretion and blood pressure in conscious rats. Am J Hypertens 24:724–730. doi:10.1038/ajh.2011.33
Gao J, Zucker IH, Gao L (2014) Activation of central angiotensin type 2 receptors by compound 21 improves arterial baroreflex sensitivity in rats with heart failure. Am J Hypertens. doi:10.1093/ajh/hpu044
Gao L, Wang W, Li H, Sumners C, Zucker IH (2008) Effects of angiotensin type 2 receptor overexpression in the rostral ventrolateral medulla on blood pressure and urine excretion in normal rats. Hypertension 51:521–527. doi:10.1161/HYPERTENSIONAHA.107.101717
Gautron L, Rutkowski JM, Burton MD, Wei W, Wan Y, Elmquist JK (2013) Neuronal and nonneuronal cholinergic structures in the mouse gastrointestinal tract and spleen. J Comp Neurol 521:3741–3767. doi:10.1002/cne.23376
Grady EF, Sechi LA, Griffin CA, Schambelan M, Kalinyak JE (1991) Expression of AT2 receptors in the developing rat fetus. J Clin Invest 88:921–933. doi:10.1172/jci115395
Grove KL, Speth RC, Palmer AA, Ganong WF, Steele MK (1998) Angiotensin II receptor binding sites in the ventral portion of the bed nucleus of the stria terminalis are reduced by interruption of the medial forebrain bundle. Brain Res 809:5–11
Guimaraes PS, Huber DA, Campagnole-Santos MJ, Schreihofer AM (2014) Development of attenuated baroreflexes in obese Zucker rats coincides with impaired activation of nucleus tractus solitarius. Am J Physiol Regul Integr Comp Physiol 306:R681–R692. doi:10.1152/ajpregu.00537.2013
Guyenet PG (2006) The sympathetic control of blood pressure nature reviews. Neuroscience 7:335–346. doi:10.1038/nrn1902
Hafko R, Villapol S, Nostramo R, Symes A, Sabban EL, Inagami T, Saavedra JM (2013) Commercially available angiotensin II At(2) receptor antibodies are nonspecific. PloS One 8:e69234. doi:10.1371/journal.pone.0069234
Hauser W, Johren O, Saavedra JM (1998) Characterization and distribution of angiotensin II receptor subtypes in the mouse brain. Eur J Pharmacol 348:101–114
Hayes MR, Bradley L, Grill HJ (2009) Endogenous hindbrain glucagon-like peptide-1 receptor activation contributes to the control of food intake by mediating gastric satiation signaling. Endocrinology 150:2654–2659. doi:10.1210/en.2008-1479
Hein L, Barsh GS, Pratt RE, Dzau VJ, Kobilka BK (1995) Behavioural and cardiovascular effects of disrupting the angiotensin II type-2 receptor gene in mice. Nature 377:744–747
Herrera M, Sparks MA, Alfonso-Pecchio AR, Harrison-Bernard LM, Coffman TM (2013) Lack of specificity of commercial antibodies leads to misidentification of angiotensin type 1 receptor protein. Hypertension 61:253–258. doi:10.1161/HYPERTENSIONAHA.112.203679
Heshmatian B, Parviz M, Karimian SM, Keshavarz M, Sohanaki H (2007) Cardiovascular response to renin substrate microinjection into the central nucleus of the amygdala of rats. Neuroreport 18:675–678. doi:10.1097/WNR.0b013e3280ba49d8
Ichiki T, Inagami T (1995) Expression, genomic organization, and transcription of the mouse angiotensin II type 2 receptor gene. Circ Res 76:693–700
Ichiki T et al (1995) Effects on blood pressure and exploratory behaviour of mice lacking angiotensin II type-2 receptor. Nature 377:748–750. doi:10.1038/377748a0
Israel A, Cierco M, Sosa B (2000a) Angiotensin AT(2) receptors mediate vasodepressor response to footshock in rats. Role of kinins, nitric oxide and prostaglandins. Eur J Pharmacol 394:103–108
Israel A, Sosa B, Gutierez CI (2000b) Brain AT(2) receptor mediate vasodepressor response to footshocks: role of kinins and nitric oxide. Brain Res Bull 51:339–343
Jessberger S, Toni N, Clemenson GD Jr, Ray J, Gage FH (2008) Directed differentiation of hippocampal stem/progenitor cells in the adult brain. Nat Neurosci 11:888–893. doi:10.1038/nn.2148
Jing F et al (2012) Direct stimulation of angiotensin II type 2 receptor enhances spatial memory. J Cereb Blood Flow Metab 32:248–255. doi:10.1038/jcbfm.2011.133
Johren O, Inagami T, Saavedra JM (1995) AT1A, AT1B, and AT2 angiotensin II receptor subtype gene expression in rat brain. Neuroreport 6:2549–2552
Joseph JP et al (2014) The angiotensin type 2 receptor agonist Compound 21 elicits cerebroprotection in endothelin-1 induced ischemic stroke. Neuropharmacology 81:134–141. doi:10.1016/j.neuropharm.2014.01.044
Kádár A et al (2010) Distribution of hypophysiotropic thyrotropin-releasing hormone (TRH)-synthesizing neurons in the hypothalamic paraventricular nucleus of the mouse. J Comp Neurol 518:3948–3961. doi:10.1002/cne.22432
Kostenis E et al (2005) G-protein-coupled receptor Mas is a physiological antagonist of the angiotensin II type 1 receptor. Circulation 111:1806–1813
Krause EG et al (2011) Blood-borne angiotensin II acts in the brain to influence behavioral and endocrine responses to psychogenic stress. J Neurosci 31:15009–15015. doi:10.1523/JNEUROSCI.0892-11.2011
Langlet F, Mullier A, Bouret SG, Prevot V, Dehouck B (2013) Tanycyte-like cells form a blood—cerebrospinal fluid barrier in the circumventricular organs of the mouse brain. J Comp Neurol 521:3389–3405. doi:10.1002/cne.23355
Lenkei Z, Palkovits M, Corvol P, Llorens-Cortes C (1996) Distribution of angiotensin II type-2 receptor (AT2) mRNA expression in the adult rat brain. J Comp Neurol 373:322–339. doi:10.1002/(SICI)1096-9861(19960923)373:3<322:AID-CNE2>3.0.CO;2-4
Lenkei Z, Palkovits M, Corvol P, Llorens-Cortès C (1997) Expression of Angiotensin Type-1 (AT1) and Type-2 (AT2) Receptor mRNAs in the adult rat brain: a functional neuroanatomical review. Front Neuroendocrinol 18:383
Li Z, Iwai M, Wu L, Shiuchi T, Jinno T, Cui TX, Horiuchi M (2003) Role of AT2 receptor in the brain in regulation of blood pressure and water intake. Am J Physiol Heart Circ Physiol 284:H116–H121. doi:10.1152/ajpheart.00515.2002
Lienard F, Thornton SN, Martial FP, Mousseau MC, Galaverna O, Meile MJ, Nicolaidis S (1996) Effects of DOCA pretreatment on neuronal sensitivity and cell responsiveness to angiotensin II, in the bed nucleus of the stria terminalis in the rat. Regul Pept 66:59–63
Llano Lopez LH et al (2012) Anxiolytic-like effect of losartan injected into amygdala of the acutely stressed rats. Pharmacol Rep 64:54–63
Masubuchi Y, Tsukamoto K, Isogai O, Yajima Y, Ito S, Saito S, Uchiyama T (2004) Effect of a high-salt diet on gamma-aminobutyric acid-mediated responses in the nucleus tractus solitarius of Sprague-Dawley rats. Brain Res Bull 64:221–226. doi:10.1016/j.brainresbull.2004.07.009
McCarthy CA, Vinh A, Broughton BR, Sobey CG, Callaway JK, Widdop RE (2012) Angiotensin II type 2 receptor stimulation initiated after stroke causes neuroprotection in conscious rats. Hypertension 60:1531–1537. doi:10.1161/HYPERTENSIONAHA.112.199646
McCarthy CA, Vinh A, Miller AA, Hallberg A, Alterman M, Callaway JK, Widdop RE (2014) Direct angiotensin AT2 receptor stimulation using a novel AT2 receptor agonist, compound 21 evokes neuroprotection in conscious hypertensive rats. PloS One 9:e95762. doi:10.1371/journal.pone.0095762
McKlveen JM, Myers B, Flak JN, Bundzikova J, Solomon MB, Seroogy KB, Herman JP (2013) Role of prefrontal cortex glucocorticoid receptors in stress and emotion. Biol Psychiatry 74:672–679. doi:10.1016/j.biopsych.2013.03.024
Millan MA, Jacobowitz DM, Aguilera G, Catt KJ (1991) Differential distribution of AT1 and AT2 angiotensin II receptor subtypes in the rat brain during development. Proc Natl Acad Sci USA 88:11440–11444
Miyoshi M, Miyano K, Moriyama N, Taniguchi M, Watanabe T (2008) Angiotensin type 1 receptor antagonist inhibits lipopolysaccharide-induced stimulation of rat microglial cells by suppressing nuclear factor kappaB and activator protein-1 activation. Eur J Neurosci 27:343–351. doi:10.1111/j.1460-9568.2007.06014.x
Mo ZL, Katafuchi T, Muratani H, Hori T (1992) Effects of vasopressin and angiotensin II on neurones in the rat dorsal motor nucleus of the vagus, in vitro. J Physiol 458:561–577
Mousa SA, Shaqura M, Schäper J, Treskatsch S, Habazettl H, Schäfer M, Abdul-Khaliq H (2011) Developmental expression of δ-opioid receptors during maturation of the parasympathetic, sympathetic, and sensory innervations of the neonatal heart: Early targets for opioid regulation of autonomic control. J Comp Neurol 519:957–971. doi:10.1002/cne.22560
Muller-Ribeiro FC, Zaretsky DV, Zaretskaia MV, Santos RA, DiMicco JA, Fontes MA (2012) Contribution of infralimbic cortex in the cardiovascular response to acute stress. Am J Physiol Regul Integr Comp Physiol 303:R639–R650. doi:10.1152/ajpregu.00573.2011
Mussa BM, Verberne AJ (2013) The dorsal motor nucleus of the vagus and regulation of pancreatic secretory function. Exp Physiol 98:25–37. doi:10.1113/expphysiol.2012.066472
Northcott CA, Watts S, Chen Y, Morris M, Chen A, Haywood JR (2010) Adenoviral inhibition of AT1a receptors in the paraventricular nucleus inhibits acute increases in mean arterial blood pressure in the rat. Am J Physiol Regul Integr Comp Physiol 299:R1202–R1211. doi:10.1152/ajpregu.00764.2009
Nuyt AM, Lenkei Z, Palkovits M, Corvol P, Llorens-Cortes C (1999) Ontogeny of angiotensin II type 2 receptor mRNA expression in fetal and neonatal rat brain. J Comp Neurol 407:193–206
Ohinata K, Fujiwata Y, Shingo F, Masaru I, Masatsugu H, Yoshikawa M (2009) Orally administered novokinin, an angiotensin AT2 receptor agonist, suppresses food intake via prostaglandin E2-dependent mechanism in mice. Peptides 30:1105–1108. doi:10.1016/j.peptides.2009.03.003
Okuyama S, Sakagawa T, Chaki S, Imagawa Y, Ichiki T, Inagami T (1999) Anxiety-like behavior in mice lacking the angiotensin II type-2 receptor. Brain Res 821:150–159
Peters B et al (2012) A new transgenic rat model overexpressing the angiotensin II type 2 receptor provides evidence for inhibition of cell proliferation in the outer adrenal cortex. Am J Physiol Endocrinol Metab 302:E1044–E1054. doi:10.1152/ajpendo.00080.2011
Petrovich GD, Ross CA, Mody P, Holland PC, Gallagher M (2009) Central, but not basolateral, amygdala is critical for control of feeding by aversive learned cues. J Neurosci 29:15205–15212. doi:10.1523/JNEUROSCI.3656-09.2009
Plunkett LM, Saavedra JM (1985) Increased angiotensin II binding affinity in the nucleus tractus solitarius of spontaneously hypertensive rats. Proc Natl Acad Sci USA 82:7721–7724
Polson JW, Dampney RA, Boscan P, Pickering AE, Paton JF (2007) Differential baroreflex control of sympathetic drive by angiotensin II in the nucleus tractus solitarii. Am J Physiol Regul Integr Comp Physiol 293:R1954–R1960. doi:10.1152/ajpregu.00041.2007
Premer C, Lamondin C, Mitzey A, Speth RC, Brownfield MS (2013) Immunohistochemical localization of AT1a, AT1b, and AT2 angiotensin II receptor subtypes in the rat adrenal pituitary, and brain with a perspective commentary. Int J Hypertens 2013:175428. doi:10.1155/2013/175428
Qi Y et al (2012) Moderate cardiac-selective overexpression of angiotensin II type 2 receptor protects cardiac functions from ischaemic injury. Exp Physiol 97:89–101. doi:10.1113/expphysiol.2011.060673
Reagan LP, Flanagan-Cato LM, Yee DK, Ma LY, Sakai RR, Fluharty SJ (1994) Immunohistochemical mapping of angiotensin type 2 (AT2) receptors in rat brain. Brain Res 662:45–59
Reis DG, Scopinho AA, Guimaraes FS, Correa FM, Resstel LB (2011) Behavioral and autonomic responses to acute restraint stress are segregated within the lateral septal area of rats. PloS One 6:e23171. doi:10.1371/journal.pone.0023171
Rodriguez-Pallares J, Rey P, Parga JA, Munoz A, Guerra MJ, Labandeira-Garcia JL (2008) Brain angiotensin enhances dopaminergic cell death via microglial activation and NADPH-derived ROS. Neurobiol Dis 31:58–73. doi:10.1016/j.nbd.2008.03.003
Saavedra J, Pavel J (2006) The discovery of a novel macrophage binding site. Cell Mol Neurobiol 26:507–524. doi:10.1007/s10571-006-9044-x
Saavedra JM, Ando H, Armando I, Baiardi G, Bregonzio C, Juorio A, Macova M (2005) Anti-stress and anti-anxiety effects of centrally acting angiotensin II AT1 receptor antagonists. Regul Pept 128:227–238
Saha S, Drinkhill MJ, Moore JP, Batten TF (2005) Central nucleus of amygdala projections to rostral ventrolateral medulla neurones activated by decreased blood pressure. Eur J Neurosci 21:1921–1930. doi:10.1111/j.1460-9568.2005.04023.x
Sakai RR, McEwen BS, Fluharty SJ, Ma LY (2000) The amygdala: site of genomic and nongenomic arousal of aldosterone-induced sodium intake. Kidney Int 57:1337–1345. doi:10.1046/j.1523-1755.2000.00972.x
Santos RA, Ferreira AJ, Simoes ESAC (2008) Recent advances in the angiotensin-converting enzyme 2-angiotensin(1-7)-Mas axis. Exp Physiol 93:519–527
Santos RA et al (2003) Angiotensin-(1-7) is an endogenous ligand for the G protein-coupled receptor Mas. Proc Natl Acad Sci USA 100:8258–8263
Scheuer DA (2010) Regulation of the stress response in rats by central actions of glucocorticoids. Exp Physiol 95:26–31. doi:10.1113/expphysiol.2008.045971
Schulkin J, Marini J, Epstein AN (1989) A role for the medial region of the amygdala in mineralocorticoid-induced salt hunger. Behav Neurosci 103:179–185
Schwanhausser B et al (2011) Global quantification of mammalian gene expression control. Nature 473:337–342. doi:10.1038/nature10098
Scopinho AA, Aguiar DC, Resstel LB, Guimaraes FS, Correa FM (2012) Brain pathways involved in the modulatory effects of noradrenaline in lateral septal area on cardiovascular responses. Cell Mol Neurobiol 32:1147–1157. doi:10.1007/s10571-012-9840-4
Sevoz-Couche C, Brouillard C, Camus F, Laude D, De Boer SF, Becker C, Benoliel JJ (2013) Involvement of the dorsomedial hypothalamus and the nucleus tractus solitarii in chronic cardiovascular changes associated with anxiety in rats. J Physiol 591:1871–1887. doi:10.1113/jphysiol.2012.247791
Shan Z et al (2013) Chronic knockdown of the nucleus of the solitary tract AT1 receptors increases blood inflammatory-endothelial progenitor cell ratio and exacerbates hypertension in the spontaneously hypertensive rat. Hypertension 61:1328–1333. doi:10.1161/HYPERTENSIONAHA.111.00156
Shao C, Yu L, Gao L (2014) Activation of angiotensin type 2 receptors partially ameliorates streptozotocin-induced diabetes in male rats by islet protection. Endocrinology 155:793–804. doi:10.1210/en.2013-1601
Shao C, Zucker IH, Gao L (2013) Angiotensin type 2 receptor in pancreatic islets of adult rats: a novel insulinotropic mediator. Am J Physiol Endocrinol Metab 305:E1281–E1291. doi:10.1152/ajpendo.00286.2013
Shi P et al (2010) Brain microglial cytokines in neurogenic hypertension. Hypertension 56:297–303
Siragy HM, de Gasparo M, Carey RM (2000) Angiotensin type 2 receptor mediates valsartan-induced hypotension in conscious rats. Hypertension 35:1074–1077
Stocker SD, Toney GM (2005) Median preoptic neurones projecting to the hypothalamic paraventricular nucleus respond to osmotic, circulating Ang II and baroreceptor input in the rat. J Physiol 568:599–615. doi:10.1113/jphysiol.2005.094425
Sumners C, Horiuchi M, Widdop RE, McCarthy C, Unger T, Steckelings UM (2013) Protective arms of the renin-angiotensin-system in neurological disease. Clin Exp Pharmacol Physiol 40:580–588. doi:10.1111/1440-1681.12137
Sumners C, Tang W, Zelezna B, Raizada MK (1991) Angiotensin II receptor subtypes are coupled with distinct signal-transduction mechanisms in neurons and astrocytes from rat brain. Proc Natl Acad Sci USA 88:7567–7571
Tavares RF, Antunes-Rodrigues J, de Aguiar Correa FM (2004) Pressor effects of electrical stimulation of medial prefrontal cortex in unanesthetized rats. J Neurosci Res 77:613–620. doi:10.1002/jnr.20195
Travers JB, Travers SP, Norgren R (1987) Gustatory neural processing in the hindbrain. Ann Rev Neurosci 10:595–632. doi:10.1146/annurev.ne.10.030187.003115
Tsukamoto K, Sved AF (1993) Enhanced gamma-aminobutyric acid-mediated responses in nucleus tractus solitarius of hypertensive rats. Hypertension 22:819–825
Vitela M, Mifflin SW (2001) γ-Aminobutyric acidb receptor-mediated responses in the nucleus tractus solitarius are altered in acute and chronic hypertension. Hypertension 37:619–622. doi:10.1161/01.hyp.37.2.619
Von Bohlen und Halbach O, Walther T, Bader M, Albrecht D (2001) Genetic deletion of angiotensin AT2 receptor leads to increased cell numbers in different brain structures of mice. Regul Pept 99:209–216
Wang H, Gallinat S, Li HW, Sumners C, Raizada MK, Katovich MJ (2004) Elevated blood pressure in normotensive rats produced by ‘knockdown’ of the angiotensin type 2 receptor. Exp Physiol 89:313–322. doi:10.1113/expphysiol.2004.027359
Wang WZ, Gao L, Pan YX, Zucker IH, Wang W (2007) AT1 receptors in the nucleus tractus solitarii mediate the interaction between the baroreflex and the cardiac sympathetic afferent reflex in anesthetized rats. Am J Physiol Regul Integr Comp Physiol 292:R1137–R1145. doi:10.1152/ajpregu.00590.2006
Wu CY et al (2013) Expression of angiotensin II and its receptors in activated microglia in experimentally induced cerebral ischemia in the adult rats. Mol Cell Biochem 382:47–58. doi:10.1007/s11010-013-1717-4
Yan JB et al (2014) Natriorexigenic effect of DAMGO is decreased by blocking AT1 receptors in the central nucleus of the amygdala. Neuroscience 262:9–20. doi:10.1016/j.neuroscience.2013.12.046
Ye S, Zhong H, Duong VN, Campese VM (2002) Losartan reduces central and peripheral sympathetic nerve activity in a rat model of neurogenic hypertension. Hypertension 39:1101–1106. doi:10.1161/01.hyp.0000018590.26853.c7
Yu L, Zheng M, Wang W, Rozanski GJ, Zucker IH, Gao L (2010) Developmental changes in AT1 and AT2 receptor-protein expression in rats. J Renin Angiotensin Aldosterone Syst 11:214–221. doi:10.1177/1470320310379065
Zardetto-Smith AM, Beltz TG, Johnson AK (1994) Role of the central nucleus of the amygdala and bed nucleus of the stria terminalis in experimentally-induced salt appetite. Brain research 645:123–134
Zhang Q, Yao F, O’Rourke ST, Qian SY, Sun C (2009) Angiotensin II enhances GABA(B) receptor-mediated responses and expression in nucleus tractus solitarii of rats. Am J Physiol Heart Circ Physiol 297:H1837–H1844. doi:10.1152/ajpheart.00354.2009
Zhang R, Jankord R, Flak JN, Solomon MB, D’Alessio DA, Herman JP (2010) Role of glucocorticoids in tuning hindbrain stress integration. J Neurosci 30:14907–14914. doi:10.1523/JNEUROSCI.0522-10.2010
Acknowledgments
This work was supported by NIH grants HL-076803 (CS), HL-093186 (CS/DS), HL-096830 (EGK), T32-HL-083810 (ADdK), and F32-HL-116074 (ADdK).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
de Kloet, A.D., Wang, L., Ludin, J.A. et al. Reporter mouse strain provides a novel look at angiotensin type-2 receptor distribution in the central nervous system. Brain Struct Funct 221, 891–912 (2016). https://doi.org/10.1007/s00429-014-0943-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00429-014-0943-1