Abstract
Key message
This work shows that overexpression of the WUS gene from Arabidopsis enhanced the expression of embryogenic competence and triggered organogenesis from some cells of the regenerated embryo-like structures.
Abstract
Agrobacterium-mediated genetic transformation of cotton was described in the late 1980s, but is still time consuming and largely genotype dependant due to poor regeneration. To help solve this bottleneck, we over-expressed the WUSCHEL (WUS) gene, a homeobox transcription factor cloned in Arabidopsis thaliana, known to stimulate organogenesis and/or somatic embryogenesis in Arabidopsis tissues cultured in vitro. The AtWUS gene alone, and AtWUS gene fused to the GFP marker were compared to the GFP gene alone and to an empty construct used as a control. Somatic embryogenesis was improved in WUS expressed calli, as the percentage of explants giving rise to embryogenic tissues was significantly higher (×3) when WUS gene was over-expressed than in the control. An interesting result was that WUS embryogenic lines evolved in green embryo-like structures giving rise to ectopic organogenesis never observed in any of our previous transformation experiments. Using our standard in vitro culture protocol, the overexpression of AtWUS in tissues of a recalcitrant variety did not result in the production of regenerated plants. This achievement will still require the optimization of other non-genetic factors, such as the balance of exogenous phytohormones. However, our results suggest that targeted expression of the WUS gene is a promising strategy to improve gene transfer in recalcitrant cotton cultivars.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Transgenesis has been developed largely on the cotton plant, which is the most widely used source of vegetable textile fiber. Its economic importance is illustrated by the fact that 350 million people are employed in its production and manufacture. Given its economic importance and its vulnerability to insect pests, it is no surprise that cotton was one of the first crops studied with a view to conferring new agronomic traits through gene transfer. Genetically modified cotton is now widespread. Two-thirds (68 %) of the 36 million hectares under cotton cultivation were biotech in 2011 (source ISAAA: The International Service for the Acquisition of Agri-biotech Applications http://www.isaaa.org). Although results have been published on biolistic cotton tissue transformation (Finer and McMullen 1990; McCabe and Martinell 1993) and more recently by Liu and coll (Liu et al. 2011), until now the most commonly used process has been the Agrobacterium-mediated transformation and in vitro regeneration of transgenic plants. For cotton, the in vitro regeneration process is somatic embryogenesis which was first described in Gossypium klotzschiaanum (Price and Smith 1979). Following this pioneering work, several authors reported the regeneration of the cultivated cotton species Gossypium hirsutum through somatic embryogenesis (Trolinder and Goodin 1987; Gawel et al. 1986; Shoemaker et al. 1986). The Wilkins team developed highly regenerable elite Acala cotton (Mishra et al. 2003). This represents an important step towards genotype-independent regeneration, and hence, transformation. After the first reports of the production of transgenic cotton plants expressing the NPTII gene (Umbeck et al. 1987; Shoemaker et al. 1986; Firoozabady et al. 1987), the development of this technique to create new varieties expressing genes of interest (herbicidal or insect resistant) has been very rapid. Agrobacterium-mediated transformation of cotton and regeneration via somatic embryogenesis (SE) is being developed in several laboratories and private companies. Although several studies (Trolinder and Xhixian 1989; Cousins et al. 1991; Firoozabady and DeBoer 1993; Kumar et al. 1998; Sakhanokho et al. 2001, 2004; Sun et al. 2006; Zhang et al. 2009; Wu et al. 2004; Jin et al. 2006) reported the regeneration of various cultivars through somatic embryogenesis, many elite cultivars remain poorly regenerable (Mishra et al. 2003; Obembe et al. 2011). Following the first results published by Umbeck et al. (1987) and Firoozabady et al. (1987), the most efficient transformation process was developed for Coker varieties (Pannetier et al. 1997; Sunilkumar and Rathore 2001; Wilkins et al. 2004), and most transgenic cotton currently cultivated in the world came from primary transformants obtained on a Coker variety. However, the method is still time consuming mainly due to incomplete control of the regeneration process. Research into the regeneration process itself tends to be both tedious and protracted. For this reason as well as the economic premium attached to the rapid development of transgenic cotton, there have been few studies on the regeneration process and particularly on the induction of embryogenesis. The influence of media composition (mineral content and phytohormones) has been studied (Trolinder and Goodin 1988a, b). Transcriptomic approaches have been used to identify genes involved in somatic embryogenesis in cotton (Zeng et al. 2006; Wu et al. 2009; Yang et al. 2012) and more than 200 unigenes have been identified as upregulated during cotton SE (Zeng et al. 2006). It will be very challenging to identify among these genes those that regulate directly the process of somatic embryogenesis, and for evidence the recent work of Hu et al. (2011) is pioneer in this field.
Somatic embryogenesis is the biological process by which many plants can regenerate in vitro. In the past, most studies focused on hormonal regulation of this process; (Zimmerman 1993; Lazzeri et al. 1987; Jimenez 2005) and the literature abounds with articles describing different strategies for regenerating a number of species via SE (Feher et al. 2003; Verdeil et al. 2007). More recently, due to progress made in studies of zygotic embryogenesis and of the shoot apical meristem a number of genes involved in SE induction have been identified (Verdeil et al. 2007; Tahir and Stasolla 2006; Rose and Nolan 2006). More specifically, many transcription factors: LEAFY COTYLEDON1 (LEC1) BABY BOOM (BBM) and AGAMOUS-LIKE 15 (AGL15) were shown to enhance embryo formation from vegetative cells, immature microspores or zygotic embryos (Alemanno et al. 2008; Lotan et al. 1998; Boutilier et al. 2002; Harding et al. 2003). We were interested in the effects of the WUSCHEL gene from A. thaliana expressed in cotton tissue cultured in vitro.
The WUSCHEL gene (WUS), which encodes a homeodomain transcription factor, was initially identified as being required to maintain a pool of pluripotent stem cells in the shoot apical meristem (SAM) in an undifferentiated state (Endrizzi et al. 1996; Laux et al. 1996; Mayer et al. 1998). WUS expression is confined to a small group of cells in the lower part of the central zone of SAM, but it can drive signals across cell layers and is expressed non-autonomously (Mayer et al. 1998). WUS is thought to interact with CLAVATA3, a gene expressed in the underlying cell layers, by a regulatory loop controlling the size of the stem cell population, with the CLV genes repressing WUS at the transcript level, and WUS expression being sufficient to induce meristem cell identity (Brand et al. 2000; Schoof et al. 2000).
Ectopic expression of the A. thaliana WUS gene was shown to induce stem cells in vegetative tissues which can differentiate into somatic embryos without external plant hormones (Zuo et al. 2002). Other studies reported stem cell differentiation into organogenesis (Gallois et al. 2002). The ability of WUS to stimulate organogenesis and/or somatic embryogenesis appears to be dependant on the cellular context (Xu et al. 2005) or on the exogenous hormonal regime (Gallois et al. 2004). A connection was observed between WUS and cytokinins in the regulation of stem cells in the SAM (Leibfried et al. 2005). Proper apical meristem function requires the interaction between WUS and Arabidopsis response regulator (ARR) genes, which act in the negative feedback loop of cytokinin signaling, with WUS repressing the transcription of several ARRs. Thus, WUSCHEL facilitates high cytokinin activity (Shani et al. 2006). Cytokinin signals and WUS reinforce each other through multiple feedback loops (Sablowski 2009; Gordon et al. 2009). Su et al. (2009) showed a link between WUS and auxin. A correct WUS expression, regulated by a defined level of exogenous auxin concentration, is essential for somatic embryo induction. Eventhough these studies concern de novo shoot formation, several papers highlight the role of WUS during in vitro regeneration (Cary et al. 2002; Gordon et al. 2007; Atta et al. 2009). All these results on the auxin–cytokinin–WUS connection, which clarify the role of hormones during in vitro shoot regeneration, along with previously published results on the ectopic expression of WUSCHEL leading to the formation of somatic embryos, provide supplementary arguments to perform experiments to analyze the effect of WUS overexpression on the cotton regeneration process through somatic embryogenesis.
Materials and methods
Plant material
Delinted cotton seeds of the Coker 310 variety were sterilized in a bayrochlor (Bayrol) solution (0.3 % active chlorine) for 30 min, rinsed with sterile water and sown in test tubes on a half strength MS medium (Murashige and Skoog 1962) supplemented with Morel and Wetmore vitamins (Morel and Wetmore 1951) 10 g/l sucrose and solidified with agar 8 g/L. Seedlings were grown in a culture room at 29° ± 1 °C under a low light intensity (4 μmol m−2 s—1) for 5 days. Hypocotyl fragments of 4–5 mm in length were used as explants. Explants were placed on a basic medium (BM) composed of MS mineral salts (Murashige and Skoog 1962) supplemented with vitamins according to Morel and Wetmore (Morel and Wetmore 1951) and 20 g/L glucose, solidified with 4 g/L agarose (Litex, LSM 5000, Lonza Copenhagen). All the media used in our experiments had a pH adjusted at 5.8 and were autoclaved 20 min at 115 °C. For callogenesis and embryogenesis induction, 2,4-dichlorophenoxyacetic acid (2,4-D) (0.1–0.05 mg/L) and kinetin (0.1–0.05 mg/L) were added. After 2–6 months, embryogenic clusters (Fig. 1c) appeared on the calli; they were excised and subcultured on a new medium to maintain embryogenic lines. The new medium was composed of the same Basic Medium containing 30 g/L sucrose instead of glucose and was free of phytohormones. Tissues were subcultured every 4 weeks. Control and WUS-overexpressing embryogenic clusters were isolated on the same basic medium.
Somatic embryogenesis in control and WUS-overexpressing tissues: a transformed calli on hypocotyl explant, b isolated callus, c embryogenic tissues (arrows) on callus. d Embryogenic lines: d control; d1, d2, d3 successive stages of a WUS-overexpressing line, e somatic embryo development on control line (bar 0.5 cm)
Agrobacterium tumefaciens-mediated transformation
Cultures of A. tumefaciens were initiated from a single plated colony or from glycerol stocks and grown overnight at 28 °C with shaking (150 rpm) in liquid Luria–Bertani medium containing 50 mg/L kanamycin, carbenicilin and rifampicin, to mid-log phase (OD660 = 0.9–1.2). The A. tumefaciens cells were collected by centrifugation and resuspended in liquid inoculation medium same as the BM plant medium. The A. tumefaciens culture was then diluted 1/50 for explant inoculation. Hypocotyl fragments were dipped in the A. tumefaciens suspension culture for 25 min. The bacterial suspension was then removed and the explants were wiped on filter paper to remove excess bacteria. Explants were placed on callus induction medium (BM medium supplemented containing 2,4-D and kinetin at the concentration of 0.1 mg/L) at 25 °C for 48 h for co-cultivation and then subcultured on the same callus induction medium supplemented with 25 mg/L of kanamycin and 500 mg/L of cefotaxime. Subcultures were performed every 14 days. Cefotaxime concentration was decreased gradually to 250 mg/L after the second subculture, 125 mg/L after the fourth subculture. The cultures were kept at 29° ± 1 °C under 16–8 h photoperiod.
Plasmid constructs
Four constructs were made using the pGWB Gateway cloning series (Nakagawa et al. 2007): pGWB2-WUS (35S:WUS), pGWB5-WUS (35S:WUS-GFP fusion), pGWB2-GFP (35S:GFP), and pGWB1 (empty vector: no promoter, no gene). pGWB2-GFP and pGWB1 were used as controls in transformation experiments. The pENTR-WUS cDNA (provided by Pr. Laux’ laboratory, University of Freiburg, Germany) was cloned in the REGIA project (Paz-Ares and The REGIA Consortium 2002). The WUS cDNA was amplified from A. thaliana using specific primers AtWUS-1 (5′ATGGAGCCGCCACAGC) and AtWUS-2 (5′CATGTTCAGACGTAGCTC) and cloned into the pCR II TOPO blunt vector (Invitrogen). Subsequently, the WUS open reading frame (ORF) was amplified with the primers attB1AtWUS (5′AAGGGGACAAGTTTGTACAAAAAAGCAGGCTGCATGGAGCCGCCACAGC) and attB2AtWUS (5′AAGGGGACCACTTTGTACAAGAAAGCTGGGTGCCATGTTCAGACGTAGCT) which were used to amplify AtWUS and recombined the gene into pDONR201 (Invitrogen). The pENTR-GFP was obtained by PCR amplification of GFP gene from the pGWB5 vector using attB1GFP (5′GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGAGAGGATCCATGGTGAGCAA) and attB2GFP (5′GGGGACCACTTTGTACAAGAAAGCTGGGTTTTACTTGTACAGCTCGTCCATGCC) and recombination into pDONR207 (Invitrogen). The pENTR-delta used for deleting gateway cassette from pGWB1 was obtained by modification of pENTR11 (Invitrogen). The ccdB gene for negative selection was deleted from pENTR11 by SalI–XhoI restriction enzyme digestion and religation. The inserts in pENTR-WUS, pENTR-GFP and pENTR-delta were transferred by LR recombination in the destination binary vectors pGWB2, pGWB5 and pGWB1, respectively. The fragment generated by PCR, the ligated junctions and the cloned fragments were verified by sequencing in all vectors. These plasmids were transferred into the A. tumefaciens strain C58::pGV2260 (Deblaere et al. 1985) by electroporation.
RT-PCR analysis
Total RNA was isolated from 100 mg samples with the RNeasy mini kit (QIAGEN S.A., Courtabeuf, France) following the manufacturer’s instructions, and including the optional RNase-free DNase step to avoid contamination with genomic DNA. RNA was extracted from embryogenic tissues from transformed and untransformed lines. For the transformed lines, successive stages of differentiation were studied in the transformed lines (see “Results”). Reverse transcription of mRNA was carried out in a 20 μL final volume from 1 μg total RNA with the SuperScript II reverse transcriptase (Invitrogen) according to manufacturer’s instructions. PCRs contained 2 μL of cDNA, corresponding to 50 ng of total RNA, in a 50 μL final volume, 1X PCR buffer, 0.2 mM dNTP, 1.5 mM MgCl2, Taq DNA polymerase native and recombinant (Invitrogen®) 0.05 U and 0.2 μM of each primer (AtWUS-1 and AtWUS-2 see above).
Histological analysis
Embryogenesis tissues were sampled at successive stages of differentiation and fixed in 0.2 M phosphate buffer at pH 7.2, supplemented with 2 % (v/v) paraformaldehyde, 1 % (w/v) caffeine, and 1 % (v/v) glutaraldehyde in a vacuum chamber for 30 min, then overnight at 4 °C. Tissues were dehydrated in ethanol, from 70 to 100 % progressively, then impregnated with ethanol–resin (50/50) for 2 h, and finally with resin 100 % overnight at 4 °C. Samples were embedded in Technovit 7100 resin (Heraeus Kulzer GmbH, Germany). Finally, 4-μm sections were double-stained with periodic acid Schiff (PAS) (Merck) and Naphthol Blue Black (NBB) (Sigma-Aldrich) (Buffard-Morel et al. 1992), and imaged with a Leica DMRXB microscope.
Confocal and scanning electronic microscopy
The WUS–GFP signal was characterized by confocal laser scanning microscopy (Confocal LSM710 ZEISS CARL SAS) in whole mounts or fresh hand-cut 1-mm sections. Spectral analysis confirmed that the observed fluorescence corresponded to GFP and not autofluorescence. Vibratome sections of the samples expressing AtWUS-GFP were stained with 4,6-diamidino-2-phenylindole dihydrochloride (DAPI) at 1 g/L in PBS for 15 min, then rinsed and mounted in water. For scanning electronic microscopy, fresh tissues were cooled at −33 °C by a Deben Coolstage and observed with a Hirox SH-1500 benchtop SEM.
Determination of auxin content
Auxin (IAA) content was measured in cotton callus. The procedure used homogenized frozen tissue. Samples were weighted and 10 ng of a standard 13C6-IAA (Cambridge Isotope Laboratory Inc.) was initially added as internal tracers for recovery and analytical purposes used to quantify AIA as described previously (Denancé et al. 2012).
Statistical analyses
Induction of somatic embryogenesis was quantified through a logistic regression with the R software environment for statistical computing and graphics (www.r-project.org/).
Comparison of IAA content in calli expressing WUS and in control calli was performed with a non-parametric test (Kruskal and Wallis) using Rcmdr package of R software (http://cran.r-project.org/web/packages/Rcmdr/).
Results
We have analyzed the effect of WUSCHEL overexpression during regeneration via somatic embryogenesis, from the appearance of embryogenic tissues through the development of somatic embryos. AtWUS-expressing tissues were compared to controls with regard to developmental and morphological characteristics. Figure 1 summarizes the successive steps of the regeneration process for both control and AtWUS-expressing calli.
WUSCHEL enhances the induction of somatic embryogenesis
In six independent experiments, each including ~100 explants per construct, we observed a significant increase in the percentage of embryogenic lines in calli where WUS was overexpressed (Table 1). AtWUS overexpressors yielded 3–4 times more embryogenic lines than control GFP overexpressors. Variability between experiments probably reflects the heterogeneity of the starting material. For example, experiments 1 and 2 were done with a different seed batch than experiment 3.
The positive effect of AtWUS overexpression was confirmed in another experiment where pGWB1 was used as the second control (repetition 7 in Table 1). This result shows that, the GFP gene has no detrimental effect on somatic embryogenesis. The percentage of explants giving rise to embryogenic tissues was statistically higher when the fusion AtWUS-GFP is overexpressed compared to AtWUS. In some cases, we have observed the appearance of embryogenic calli directly on the explant expressing the fusion AtWUS-GFP (Fig. 2). Compared to the GFP control, AtWUS overexpressor calli produced many more embryogenic cell clusters (Fig. 3a, b). The clusters contained dark blue-stained active cells, indicating a high rate of soluble proteins, with a thick cell wall, a big nucleus and a single nucleolus (Fig. 3b′). These cytological features are common to embryogenic cells in general whatever the species (Michaux-Ferrière and Schwendiman 1992).
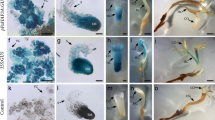
Histological examination (NBB staining) of WUS-induced embryogenic line (EmbS) and embryo-like structures (ES). a Control GFP-expressing EmbS, b WUS-expressing EmbS with multiple embryogenic cell clusters (arrows), b′ detail of embryogenic cells, c WUS-expressing ES showing ectopic development of bud-like formation, d detail showing that ectopic formations originated from the peripheral zone. Bar a, b 150 μm; c, d 500 μm
Embryogenic lines overexpressing AtWUS show specific morphological and histological features
The macroscopic morphology of embryogenic lines overexpressing GFP was identical to that of lines we previously transformed with GUS gene or various genes of interest (more than five different types). The tissues overexpressing GFP have passed through the classical stages leading to somatic embryogenesis and plantlet regeneration, typically observed in cotton (Fig. 1). Different stages of differentiation are usually found in a single cotton embryogenic line and the embryogenic lines can simultaneously proliferate and form embryos during several years on hormone-free medium (Fig. 1d). Accordingly, control embryogenic lines, including those overexpressing GFP, produced several types of structures: aggregates of embryogenic cells, pro-embryos and embryos at different stages of development (Fig. 1d, e). Embryogenic lines overexpressing AtWUS differed from this typical scenario and produced tissues going through three distinct stages of differentiation. The first stage (S1) corresponded to embryogenic aggregates same as control at the time of appearance but that rapidly evolved in clusters, initially forming small pale yellow round structures (Fig. 1d1), then growing into large round structures distinguished as the second stage (S2) (Fig. 1d2). In the third stage (S3), these structures evolved into even larger green masses, usually not observed during cotton regeneration (Fig. 1d3). At stage 3 and beyond, these structures can exhibit characteristics of somatic embryos with a well-organized root pole (Fig. 4c). We conclude that the formation of abnormal embryo-like structures results from the constitutive expression of AtWUS.
Tissues in embryogenic lines overexpressing either AtWUS or GFP were conducted. Similar to control embryogenic tissues, the AtWUS-induced S3 embryo-like structures can be maintained on a hormone-free medium, through multiplication of similar adventitious S3 embryos produced by “budding” (Fig. 1d3). More than 100 embryogenic lines were thereby subcultured with a stable phenotype for over 2 years.
AtWUS ectopic expression promotes the formation of leaf-like structures
Despite the fact that embryoids over expressing WUSCHEL have never gone into differentiation of shoots, ectopic leaf-like structures were produced by these embryoid formations (Figs. 3c, 4). This phenomenon has never been observed on control somatic embryos. Histological examination showed that these ectopic leafy structures arise from the peripheral zone of the embryo-like structures where cells actively divide (Figs. 3c, d, 5e, f). The leaf-like formations developed on embryos expressing AtWUS alone as well as AtWUS-GFP. In the latter case, the corresponding GFP fluorescence was detected in the nucleus as expected for the AtWUS fusion protein. Confocal imaging of S3 embryogenic-like formations expressing AtWUS-GFP revealed that signal was highest in the globular formations arising at the peripheral zone (Fig. 5c, d). These formations are constituted of active meristematic cells as shown by NBB coloration (Fig. 5a, b). This observation suggests that the expression of AtWUS leads to the reactivation of cells giving rise to organogenic structures.
Embryo-like formations depend on AtWUS expression
Some of the embryogenic lines transformed with the AtWUS or GFP-AtWUS transgenes did not show a developmental pattern giving rise to embryo-like structures. Instead, they only grew as proliferating tissues without any embryonic differentiation, remaining at stage S1. Therefore, we tested the WUSCHEL expression in this type of tissue as well as in the S3 structures. RT-PCRs have been done with tissues exhibiting globular green masses (Fig. 1d3) and cultures of undifferentiated WUS lines. All tissues were sampled after the same delay of 1 year in in vitro culture. AtWUS expression was only detected in embryogenic S3 structures (Fig. 6). The results indicate that transformed embryogenic tissue not evolving into S2 and S3 embryo-like structures do not express AtWUS, confirming that S3 formations and ectopic leaf-like structures result from overexpression of AtWUS.
Analysis of WUS ectopic expression in transgenic tissues. a RT-PCR analysis of WUS transcript level. S1 cultures at stage 1, S3 cultures at stage 3; C RNA control to show that the amplification is not due to residual DNA; C+ positive WUS control. Bottom fragment corresponds to amplification of the small ribosomal subunit (SSU) cDNA. b Macroscopic views of the cultures, at stages 1 and 3, analyzed by RT-PCR
WUSCHEL did not interact with endogenous IAA content
Endogenous IAA levels were measured in AtWUS overexpressing embryogenic callus and in GFP overexpressors, for two independent experiments with ten samples. No differences were seen for IAA content between AtWUS (1.68 pg.mg−1) and GFP (1.31 pg mg−1) expressing lines (Kruskal–Wallis rank sum test, p value = 0.32).
Discussion
With this work, we wished to test whether genes that stimulate organogenesis or somatic embryogenesis in A. thaliana promote the same developmental programs in cotton tissues and can induce the regeneration process starting with explants from recalcitrant cultivars.
In our hands and with our standard in vitro culture protocol, the overexpression of AtWUS in tissues of a recalcitrant variety (a CIRAD-IRAD variety, Irma96 + 97) did not result in the production of regenerated plants. This achievement will still require the optimization of other non-genetic factors, such as the balance of exogenous phytohormones and the composition of in vitro culture media. Nevertheless, we showed that AtWUS overexpression in an in vitro routinely used genotype improved somatic embryogenesis and induced organogenesis on embryo-like structures cultured on a hormone-free medium.
The positive effect of AtWUS on somatic embryogenesis was observed in all experiments, when overexpressed by itself or as a translational fusion with GFP. However, the fraction of explants giving rise to embryogenic tissues was higher with AtWUS-GFP, possibly because the fusion product may be more stable.
Embryogenic lines overexpressing AtWUS evolved following a quite different scenario from the one we usually observe in cotton. In the classical scenario, different stages of differentiation are usually found in a single embryogenic line: as soon as embryogenic tissues are observed on calli they can be isolated on a hormone-free medium. On this medium, the embryogenic culture can proliferate for years by regular subcultures. Two phenomena are observed simultaneously: proliferation of embryogenic clumps and somatic embryo differentiation.
In a AtWUS overexpressing context and without exogenous phytohormones, three distinct stages were observed leading to large embryo-like green masses, highly differentiated and supporting adventitious organogenesis. Histological examinations showed that, in contrast to control lines, AtWUS overexpressing lines start to differentiate very actively. Highly active zones, defined as clusters of embryogenic cells, appeared only in the WUS tissues. In the third stage of development, the highly active cells were located in the peripheral zone of the embryo-like structures. The active zones overlap with AtWUS expression as confocal examination of lines expressing AtWUS-GFP fusion showed fluorescence in the corresponding zones. These peripheral active zones give rise to adventive bud-like formations. AtWUS overexpression triggered the cell totipotency in these tissues and lead to new meristems. Xu et al. (2005) observed similar results where in their system, ectopic flower meristems were initiated from the differentiated cortex cells. The reason why AtWUS expression is limited or at least stronger to the peripheral zone, while under the 35S promoter it is supposed to be overexpressed in all tissues, is unknown. AtWUS expression is probably defined by the distribution of CLV3 signaling peptides as CLAVATA and WUS are known to be regulated in a feedback loop (Schoof et al. 2000; Brand et al. 2000).
Considering the new knowledge in hormonal control of shoot stem cell niche in interaction of transcription factors (Leibfried et al. 2005; Zhao et al. 2010), we can say that cell differentiation observed in WUS over-expressors is due to an interaction of WUS with phytohormones. In our conditions, WUSCHEL did not alter IAA activity as no differences were seen for IAA content between WUS and GFP-expressing lines whilst on the other hand WUS expression facilitates high cytokinin activity in the SAM (Leibfried et al. 2005; Shani et al. 2006).
We demonstrate in this work that AtWUS overexpression dramatically promotes the production of embryogenic lines and could potentially improved regeneration. We tried to regenerate plants from these tissues obtained in every transformation experiments performed and could potentially improve plant regeneration. We have regenerated few plants from AtWUS overexpressing tissues from our experiments. These plants had present a strong WUS phenotype (Kieffer et al. 2006; Xu et al. 2005) with many branches and waffle-curled leaves and expressed the Arabidopsis gene at detectable levels from tissues taken at several parts of the plants (data not shown). Therefore, we speculate that the Arabidopsis WUS gene is useful to improve the regeneration/transformation process in cotton but only if it is induced at critical steps, for example, when in vitro regeneration is initiated. Separate studies focusing on the effect of AtWUS ectopic expression in pepper (Solis-Ramos et al. 2009) and coffea (Arroyo-Herrera et al. 2008) under the control of estradiol-inducible promoter resulted in no normal plant regenerated because of the leakiness of the promoter. In White spruce (Klimaszewska et al. 2010), very few and severely abnormal WUS transgenic somatic embryos developed on medium containing 17-β-estradiol and morphologically normal somatic embryos were collected only on media containing no, or very few concentrations of the inducer. In future studies, other types of inducible systems should be tested to find a reliable one that insure expression of WUS transgene only at specific steps of regeneration process.
Our results have shown that the overexpression of the WUS gene from Arabidopsis can promote the expression of embryogenic competence of dedifferentiated proliferating cells obtained on cotton hypocotyl explants. Further experiments are needed to understand interactions between endogenous hormone and WUS expression to use WUS over-expression to regenerate recalcitrant genotypes.
We can notice that our approach could lead to a promising way to obtain marker-free transgenic plants. Using an Agrobacterium binary vector carrying WUS and a gene of interest, the transformed calli would be the only one able to give rise to embryogenic lines and plants. A co-transformation method or the use of two binary vectors in a same Agrobacterium would allow avoiding the presence of “embryogenic gene” in plants through subsequent segregation.
References
Alemanno L, Devic M, Niemenak N, Sanier C, Guilleminot J, Rio M, Verdeil JL, Montoro P (2008) Characterization of leafy cotyledon1-like during embryogenesis in Theobroma cacao L. Planta 227(4):853–866. doi:10.1007/s00425-007-0662-4
Arroyo-Herrera A, Ku Gonzalez A, Canche Moo R, Quiroz-Figueroa F, Loyola-Vargas V, Rodriguez-Zapata L, Burgeff D′Hondt C, Suárez-Solís V, Castaño E (2008) Expression of WUSCHEL in Coffea canephora causes ectopic morphogenesis and increases somatic embryogenesis. Plant Cell Tissue Organ Cult 94(2):171–180. doi:10.1007/s11240-008-9401-1
Atta R, Laurens L, Boucheron-Dubuisson E, Guivarc’h A, Carnero E, Giraudat-Pautot V, Rech P, Chriqui D (2009) Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J 57(4):626–644
Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu CM, van Lammeren AA, Miki BL, Custers JB, van Lookeren Campagne MM (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14(8):1737–1749
Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in arabidopsis on a feedback loop regulated by CLV3 activity. Science 289(5479):617–619
Buffard-Morel J, Verdeil JL, Pannetier C (1992) Somatic embryogenesis of coconut (Cocos nucifera L.) from leaf explants: histological study. Can J Bot 70(4):735–741
Cary A, Che P, Howell S (2002) Developmental events and shoot apical meristem gene expression patterns during shoot development in Arabidopsis thaliana. Plant J 32(6):867–877
Cousins Y, Lyon B, Llewellyn D (1991) Transformation of an Australian cotton cultivar: prospects for cotton improvement through genetic engineering. Aust. J. Plant Physiol 18
Deblaere R, Bytebier B, De Greve H, Deboeck F, Schell J, Van Montagu M, Leemans J (1985) Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res 13(13):4777–4788
Denancé N, Ranocha P, Oria N, Barlet X, Rivière M-P, Yadeta KA, Hoffmann L, Perreau F, Clément G, Maia-Grondard A, van den Berg GCM, Savelli B, Fournier S, Aubert Y, Pelletier S, Thomma BPHJ, Molina A, Jouanin L, Marco Y, Goffner D (2012) Arabidopsis wat1 (walls are thin1)-mediated resistance to the bacterial vascular pathogen, Ralstonia solanacearum, is accompanied by cross-regulation of salicylic acid and tryptophan metabolism. Plant J 73(2):225–239. doi:10.1111/tpj.12027
Endrizzi K, Moussian B, Haecker A, Levin JZ, Laux T (1996) The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J 10(6):967–979
Feher A, Pasternak TP, Dudits D (2003) Transition of somatic plant cells to an embryogenic state. Plant Cell Tissue Organ Cult 74(3):201–228. doi:10.1023/a:1024033216561
Finer J, McMullen M (1990) Transformation of cotton (Gossypium hirsutum L.) by Agrobacterium tumefaciens and regeneration of transgenic plants. Plant Cell Rep 8(10):203–206
Firoozabady E, DeBoer DL (1993) Plant regeneration via somatic embryogenesis in many cultivars of cotton (Gossypium hirsutum L.). In vitro Cell Dev Biol Plant J Tissue Cult Assoc 29(4):166–173
Firoozabady E, Deboer DL, Murray EE, Merlo DJ, Adang MJ, Halk EL (1987) Transformation of cotton (Gossypium hirsutum L.) by Agrobacterium tumefaciens and regeneration of transgenic plants. Plant Mol Biol 10(2):105–116. doi:10.1007/BF00016148
Gallois JL, Woodward C, Reddy GV, Sablowski R (2002) Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development 129(13):3207–3217 (Unspdev0423)
Gallois JL, Nora FR, Mizukami Y, Sablowski R (2004) WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev 18(4):375–380. doi:10.1101/gad.291204
Gawel NJ, Rao AP, Robacker CD (1986) Somatic embryogenesis from leaf and petiole callus-cultures of Gossypium hirsutum L. Plant Cell Rep 5(6):457–459
Gordon S, Heisler M, Reddy G, Ohno C, Das P, Meyerowitz E (2007) Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134(19):3539–3548
Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM (2009) Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci 106(38):16529–16534
Harding EW, Tang W, Nichols KW, Fernandez DE, Perry SE (2003) Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. Plant Physiol 133(2):653–663. doi:10.1104/pp.103.023499
Hu L, Yang X, Yuan D, Zeng F, Zhang X (2011) GhHmgB3 deficiency deregulates proliferation and differentiation of cells during somatic embryogenesis in cotton. Plant Biotechnol J 9(9):1038–1048. doi:10.1111/j.1467-7652.2011.00617.x
Jimenez V (2005) Involvement of plant hormones and plant growth regulators on in vitro somatic embryogenesis. Plant Growth Regul 47(2–3):91–110. doi:10.1007/s10725-005-3478-x
Jin S, Zhang X, Nie Y, Guo X, Liang S, Zhu H (2006) Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol Plant 50(4):519–524. doi:10.1007/s10535-006-0082-5
Kieffer M, Stern Y, Cook H, Clerici E, Maulbetsch C, Laux T, Davies B (2006) Analysis of the transcription factor WUSCHEL and its functional homologue in antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell Online 18(3):560–573
Klimaszewska K, Pelletier G, Overton C, Stewart D, Rutledge RG (2010) Hormonally regulated overexpression of Arabidopsis WUS and conifer LEC1 (CHAP3A) in transgenic white spruce: implications for somatic embryo development and somatic seedling growth. Plant Cell Rep 29(7):723–734. doi:10.1007/s00299-010-0859-z
Kumar S, Sharma P, Pental D (1998) A genetic approach to in vitro regeneration of non-egenerating cotton (Gossypium hirsutum L.) cultivars. Plant Cell Rep 18(1/2):59–63
Laux T, Mayer KF, Berger J, Jürgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122(1):87–96
Lazzeri P, Hildebrand D, Collins G (1987) Soybean somatic embryogenesis—effects pf hormones and culture manipulations. Plant Cell Tissue Organ Cult 10(3):197–208. doi:10.1007/BF00037304
Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU (2005) WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438(7071):1172–1175. doi:10.1038/nature04270
Liu X, Kim YJ, Müller R, Yumul RE, Liu C, Pan Y, Cao X, Goodrich J, Chen X (2011) AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 23(10):3654–3670. doi:10.1105/tpc.111.091538
Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93(7):1195–1205
Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95(6):805–815 (pii:S0092-8674(00)81703-1)
McCabe DE, Martinell BJ (1993) Transformation of elite cotton cultivars via particle bombardment of meristems. Nat Biotechnol 11(5):596–598
Michaux-Ferrière N, Schwendiman J (1992) Histology of somatic embryogenesis. In: Dattée Y, Dumas C, Gallais A (eds) Reproductive biology and plant breeding, pp 247–259. ISBN 3-540-54641-3
Mishra R, Wang H-Y, Yadav N, Wilkins T (2003) Development of a highly regenerable elite Acala cotton (Gossypium hirsutum cv; Maxxa)—a step towards genotype-independent regeneration. Plant Tissue Organ Cult 73(1):21–35
Morel G, Wetmore R (1951) Tissue culture of monocotyledons. Am J Bot 38(2):138–140
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15(3):473–497
Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, Watanabe Y, Nakamura K, Kimura T, Ishiguro S (2007) Improved gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71(8):2095–2100
Obembe OO, Khan T, Popoola JO (2011) Use of somatic embryogenesis as a vehicle for cotton transformation. J Med Plants Res 5(17):4009–4020
Pannetier C, Giband M, Couzi P, Le TV, Mazier M, Toruneur J, Hau B (1997) Introduction of new traits into cotton through genetic engineering: insect resistance as example. Euphytica 96(1):163–166
Paz-Ares J, The REGIA Consortium (2002) REGIA, an EU Project on Functional Genomics of Transcription Factors from Arabidopsis thaliana. Comput Funct Genomics 3(2):102–108. doi:10.1002/cfg.146
Price HJ, Smith RH (1979) Somatic embryogenesis in suspension cultures of Gossypium klotzsciaanum Andress. Planta 145(3):305–307. doi:10.1007/bf00454456
Rose RJ, Nolan KE (2006) Genetic regulation of somatic embryogenesis with particular reference to Arabidopsis thaliana and Medicago truncatula. In Vitro Cell Dev Biol Plant 42(6):473–481. doi:10.1079/ivp2006806
Sablowski R (2009) Cytokinin and WUSCHEL tie the knot around plant stem cells. Proc Natl Acad Sci USA 106(38):16016–16017. doi:10.1073/pnas.0909300106
Sakhanokho HF, Zipf A, Raiasekaran K, Saha S, Sharma GC (2001) Induction of highly embryogenic calli and plant regeneration in upland (Gossypium hirsutum L.) and pima (Gossypium barbadense L.) cottons. Crop Sci 41(4):1235–1240
Sakhanokho H, Ozias A, May O, Chee P (2004) Induction of somatic embryogenesis and plant regeneration in select Georgia and Pee Dee cotton lines. Crop Sci 44(6):2199–2205
Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T (2000) The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100(6):635-644. (pii:S0092-8674(00)80700-X)
Shani E, Yanai O, Ori N (2006) The role of hormones in shoot apical meristem function. Curr Opin Plant Biol 9(5):484–489. doi:10.1016/j.pbi.2006.07.008
Shoemaker RC, Couche LJ, Galbraith DW (1986) Characterization of somatic embryogenesis and plant regeneration in cotton (Gossypium hirsutum L.). Plant Cell Rep 5(3):178–181
Solis-Ramos LY, Gonzalez-Estrada T, Nahuath-Dzib S, Zapata-Rodriguez LC, Castano E (2009) Overexpression of WUSCHEL in C. chinense causes ectopic morphogenesis. Plant Cell Tissue Organ Cult 96(3):279–287. doi:10.1007/s11240-008-9485-7
Su YH, Zhao XY, Liu YB, Zhang CL, O’Neill SD, Zhang XS (2009) Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. Plant J 59(3):448–460
Sun Y, Zhang X, Huang C, Guo X, Nie Y (2006) Somatic embryogenesis and plant regeneration from different wild diploid cotton (Gossypium) species. Plant Cell Rep 25(4):289–296. doi:10.1007/s00299-005-0085-2
Sunilkumar G, Rathore KS (2001) Transgenic cotton: factors influencing Agrobacterium-mediated transformation and regeneration. Mol Breed 8(1)37–52
Tahir M, Stasolla C (2006) Shoot apical development during in vitro embryogenesis. Can J Bot-Revue Canadienne De Botanique 84(11):1650–1659. doi:10.1139/b06-070
Trolinder N, Goodin J (1987) Somatic embryogenesis and plant regeneration in cotton (Gossypium hirsutum L.). Plant Cell Rep 6(3):231–234
Trolinder N, Goodin J (1988a) Somatic embryogenesis in cotton (Gossypium hirsutum L.) I. Effects of source of explant and hormone regime. Plant Cell Tissue Organ Cult 12(1):31–42
Trolinder N, Goodin J (1988b) Somatic embryogenesis in cotton (Gossypium hirsutum L.) II Requirements for embryo development and regeneration. Plant Cell Tissue Organ Cult 12(1):43–53
Trolinder N, Xhixian C (1989) Genotype specificity of the somatic embryogenesis response in cotton. Plant Cell Rep 8(3):133–136
Umbeck P, Johnson G, Barton K, Swain W (1987) Genetically transformed cotton (Gossypium hirsutum L.) plants. Bio-Technology 5(3):263–266
Verdeil JL, Alemanno L, Niemenak N, Tranbarger TJ (2007) Pluripotent versus totipotent plant stem cells: dependence versus autonomy? Trends Plant Sci 12(6):245–252. doi:10.1016/j.tplants.2007.04.002
Wilkins TA, Mishra R, Trolinder NL (2004) Agrobacterium-mediated transformation and regeneration of cotton. J Food Agric Environ 2(1):179–187
Wu J, Zhang X, Nie Y, Jin S, Liang S (2004) Factors affecting somatic embryogenesis and plant regeneration from a range of recalcitrant genotypes of Chinese cottons (Gossypium hirsutum L.). In Vitro Cell Dev Biol Plant 40(4):371–375. doi:10.1079/ivp2004535
Wu X, Li F, Zhang C, Liu C, Zhang X (2009) Differential gene expression of cotton cultivar CCRI24 during somatic embryogenesis. J Plant Physiol 166(12):1275–1283. doi:10.1016/j.jplph.2009.01.012
Xu YY, Wang XM, Li J, Li JH, Wu JS, Walker JC, Xu ZH, Chong K (2005) Activation of the WUS gene induces ectopic initiation of floral meristems on mature stem surface in Arabidopsis thaliana (vol 58, pg 773, 2005). Plant Mol Biol 58(6):915-915. doi:10.1007/s11103-005-2560-0
Yang X, Zhang X, Yuan D, Jin F, Zhang Y, Xu J (2012) Transcript profiling reveals complex auxin signalling pathway and transcription regulation involved in dedifferentiation and redifferentiation during somatic embryogenesis in cotton. BMC Plant Biol 12(110)
Zeng FC, Zhang XK, Zhu LF, Tu LL, Guo XP, Nie YH (2006) Isolation and characterization of genes associated to cotton somatic embryogenesis by suppression subtractive hybridization and macroarray. Plant Mol Biol 60(2):167–183. doi:10.1007/s11103-005-3381-x
Zhang BH, Wang QL, Liu F, Wang KB, Frazier TP (2009) Highly efficient plant regeneration through somatic embryogenesis in 20 elite commercial cotton (Gossypium hirsutum L.) cultivars. Plant Omics 2(6):259–268
Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, Lohmann JU (2010) Hormonal control of the shoot stem-cell niche. Nature 465(7301):1089–U1154. doi:10.1038/nature09126
Zimmerman J (1993) Somatic embryogenesis—a model for early development in higher plants. Plant Cell 5(10):1411–1423. doi:10.2307/3869792
Zuo J, Niu QW, Frugis G, Chua NH (2002) The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J 30(3):349–359 (pii:1289)
Acknowledgments
We thank P. Hilson, P. Laufs for their critical reading of the manuscript and J. Scarlett for checking English language. We are grateful to JC. Palauqui and F. Bonnot for their generous help in, respectively, confocal Imaging and statistical analysis. Seeds used to initiate our own seed stocks have been provided by CIRAD. This research was supported in part by Agence Nationale de la Recherche (ANR) agreement 05W 37, EUREKA project 3395 F 1203.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Jouanin.
Rights and permissions
About this article
Cite this article
Bouchabké-Coussa, O., Obellianne, M., Linderme, D. et al. Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant Cell Rep 32, 675–686 (2013). https://doi.org/10.1007/s00299-013-1402-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-013-1402-9