Abstract
The GROWTH-REGULATING FACTOR (GRF) and its INTERACTING FACTOR (GIF) have been shown to stimulate regeneration of transgenic plants with studies reporting increased transformation efficiency in multiple species, including wheat, beet, and citrus. The present work evaluated the effects of overexpressing GRF4-GIF1 and GRF5 on the regeneration of transgenic plants in cassava (Manihot esculenta Crantz). Effects of GRF4-GIF1 and GRF5 sequences derived from Vitis vinifera and Arabidopsis thaliana were assessed by cloning expression cassettes under control of strong constitutive promoters. Friable embryogenic callus from cassava varieties 60444 and NASE 13 were transformed with Agrobacterium tumefaciens strains LBA4404 and LBA4404 THY-, and multiple independent transgenic plant lines recovered. Expression of the morphogenic genes did not enhance transformation efficiency, nor efficiency or timing of somatic embryo regeneration or whole plant recovery above the green fluorescent protein (GFP) control. Organogenesis experiments were carried out to observe effects of transgenic expression of these genes on morphogenesis from petiole, leaf-petiole, and stem explants. Results differed between the two genotypes evaluated. Expression of Vitis vinifera GRF4-GIF1 was effective for stimulation of rapid organogenesis and shoot regeneration at 30 to 37% from leaf-petiole explants of cultivar 60444. In contrast, Arabidopsis thaliana GRF5 was superior in stimulating shoot regeneration in cultivar NASE 13 with 40 to 50% of leaf-petiole explants regenerating shoots. In both cultivars, caulogenesis occurred rapidly within 3 to 4 wk culture on medium containing the cytokinin meta-topolin. Effects of overexpression of these morphogenic genes at the whole plant level were accessed by establishing plants in the greenhouse. GRF4-GIF1 overexpression resulted in significantly shorter plants with increased leaf size and total leaf area while AtGRF5 stimulated above average storage root weight.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The root crop cassava (Manihot esculenta Crantz) originated in the southwest Amazon in Brazil (Watling et al. 2018) and since has become an important staple food across the tropical regions of South America, sub-Saharan Africa, and Asia. Cassava is cultivated mostly by smallholder farmers for its large starchy storage roots, providing an important source of dietary calories and industrial starch (Chavarriaga-Aguirre et al. 2016). Transgenic and genome editing technologies have been utilized to address constraints in cassava production, including disease resistance (Wagaba et al. 2017; Veley et al. 2021), modified starch quality (Bull et al. 2018), enhanced nutritional content (Narayanan et al. 2019; 2021), reduced cyanogenic content (Gomez et al. 2023), and herbicide tolerance (Hummel et al. 2018).
Many transgenic and genome editing technologies rely on the production of morphogenic tissues in culture into which transgenes and gene editing tools can be delivered, and whole plants regenerated. Regeneration of plants typically occurs through somatic embryogenesis or organogenesis, the latter via de novo formation of meristems or rearrangement and proliferation of pre-existing meristems. Traditionally, this is achieved in vitro by manipulating auxin/cytokinin ratios in the culture medium. More recently, ectopic overexpression of plant genes that control growth and development has proven effective for stimulating morphogenesis and plant regeneration (Gordon-Kamm et al. 2019). Debernardi et al. (2020) showed how the expression of GROWTH-REGULATING FACTOR 4 (GRF4) and its cofactor GRF-INTERACTING FACTOR 1 (GIF1) dramatically increased the efficiency and speed of plant regeneration in wheat and citrus while the GROWTH-REGULATING FACTOR 5 (GRF5) increased transformation efficiency in beet, canola, soybean, and sunflower (Kong et al. 2020).
The GROWTH-REGULATING FACTOR (GFR) is a family of plant transcription factors defined by the presence of the WRC and QLQ protein domains. These factors have a role in promoting cell proliferation during leaf development and are required for the development and maintenance of the shoot apical meristem (SAM) (Kim et al. 2003; Kim and Lee 2006). GRFs interact with another family of transcription factors, the GROWTH INTERACTING FACTOR (GIF), forming a complex that gives the primordial cells of vegetative and reproductive organs a meristematic specification state, guaranteeing the supply of cells for organogenesis (Lee et al. 2009). GRF expression is post-transcriptionally downregulated by microRNA396 (miR396). In Arabidopsis, the miR396 gene family has two members (ath-mir396a and ath-mir396b). These can induce cleavage of AtGRF mRNA species, except for AtGRF5 and AtGRF6 transcripts, which lack the target site (Kim and Tsukaya 2015). According to Vercruyssen et al. (2015), the transcription factor GRF5 regulates duration of cell proliferation during leaf development in Arabidopsis. They have shown that overexpression of GRF5 also stimulates chloroplast division, resulting in a higher chloroplast number per cell with increased chlorophyll levels in leaves which could maintain higher rates of photosynthesis. Transgenic plants overexpressing GRF5 showed delayed leaf senescence and enhanced tolerance to nitrogen-depleted medium. The authors suggest these changes could potentially improve plant productivity.
The morphogenic and gene transfer systems employed to produce transgenic and genome-edited cassava have improved significantly over recent years. Well-established methods to produce transgenic plants via somatic embryogenesis are in place (Taylor et al. 2012; Segatto et al. 2022), and organogenic systems for regeneration of shoots from petiole and stem tissues have been reported (Chauhan and Taylor 2018). The production of genetically transformed cassava, however, remains a lengthy and skilled process. Four months are required to produce friable embryogenic callus (FEC) target tissues for Agrobacterium or direct gene-mediated transformation, followed by an additional 4 to 6 mo, to regenerate genetically modified or genome-edited plants (Segatto et al. 2022). Plant recovery is also genotype-specific with efficient plant regeneration limited to a relatively small subset of the many 100 s of varieties grown by farmers across the tropics (Utsumi et al. 2022). Continued evaluation of strategies for enhanced plant regeneration therefore remains important and could lead to improvements to the current systems, especially with regard to transformation of recalcitrant cultivars and time required to recover modified plants. This work investigated the effects of overexpression of VviGFR4-GIF1, AtGFR4-GIF1, and AtGFR5 morphogenic gene regulators in cassava, and quantified regeneration rates to observe if these genes had a positive impact on transformation efficiency, organogenesis, and speed of plant recovery.
Material and Methods
Construction of binary plasmids
Plasmid cloning strategies were planned and designed using the SnapGene® version 5.3.2 software (Insightful Science; snapgene.com). The control vector (p8764) was prepared using a modified version of pCAMBIA 2300 in which the nptII selectable marker was driven by the Nos promoter, and the visual marker green fluorescent protein (GFP) was under the control of a 35S promoter and RbcS E-9 terminator.
Plasmid JD631 carrying the Vitis vinifera GRF4-GIF1 gene sequences driven by the 35S promoter was obtained from Jorge Dubcovsky, University of California, Davis (Addgene plasmid #160,399; http://n2t.net/addgene:160399; RRID:Addgene_160399). To produce the plasmid p8765, the donor Vitis GRF4-GIF1 chimera was digested with SbfI and StuI, and the GRF4-GIF1 sequence was purified and ligated into the GFP control plasmid (p8764) at the SmaI site.
A second set of morphogenic genes was prepared using gene sequences from Arabidopsis thaliana. GRF4, GIF1, and GRF5 sequences were searched using The Arabidopsis Information Resource (TAIR) from which two copies of GRF4 and GRF5 plus one GIF1 were identified. The protein coding sequences of GRF4 accession number (AT3G52910), GIF1 (AT5G28640), and GRF5 (AT3G13960) were chosen based on the description of their structure and activity according to published literature (The Arabidopsis Information Resource 2021). These GRF4 and GIF1 sequences were fused with an alanine linker and FASTA sequences sent to Genewiz® for synthesis and fusion to the cassava vein mosaic virus (CsVMV) promoter (Verdaguer et al. 1998) and the NOS terminator. The synthesized AtGRF4-GIF1 in puc57Amp was digested with AhdI, EcoRI, and KpnI, and the top band was gel purified. This fragment was cloned into p8764 in a 3-way ligation where p8764 was cut with KpnI and AscI to separate the plasmid backbone and GFP into two fragments. All three fragments were ligated to form the vector p8788. For AtGRF5, SpeI and PacI were used to remove AtGRF4-GIF1 from p8788 replacing it by inserting AtGRF5 to form the vector p8789. All plasmids were confirmed by restriction analysis and Sanger sequencing before transformation by electroporation into Agrobacterium strains LBA4404 and LBA4404 THY-.
Plant materials and Agrobacterium transformation of cassava
The African cassava varieties 60444 and NASE 13 were used for genetic transformation and plant regeneration studies. Plants were maintained by micropropagation on Murashige and Skoog (MS) basal medium, supplemented with 20.0 g L−1 sucrose (MS2), and solidified with 8.0 g L−1 Noble agar (Segatto et al. 2022).
Production of friable embryogenic callus target tissues
Friable embryogenic callus (FEC) used for transformation was produced from organized embryogenic structures (OES) according to Segatto et al. (2022). Briefly, immature leaf explants approximately 2.5 mm in length were excised from micropropagated mother plants, placed onto MS2 medium supplemented with 50.0 μM picloram and 2.0 μM CuSO4, and cultured in low light (20 µMol m−2 s−1) at 28 °C. After 4 wk, the OES that formed was excised, crushed through a 1-mm mesh, placed onto Gresshoff and Doy (GD) medium supplemented with 20.0 g L−1 sucrose and 50.0 μM picloram, and solidified with 8.0 g L−1 Noble agar (GD2 50P). Tissues were cultured for three, 21-d cycles on GD2 50P medium under low light conditions for cultivar 60444, as above, and in the dark for NASE 13, to generate homogenous FEC tissues for co-culture with Agrobacterium.
Transformation with Agrobacterium and recovery of transgenic plants
All morphogenic gene constructs were transformed into FEC target tissues a minimum of four times with three samples each per transformation experiment. A GFP control and non-transformed control were also performed within each experiment.
A liquid Agrobacterium suspension with OD600 of 0.5 was prepared according to Segatto et al. (2022). When using LBA4404 THY-, 50 mg L−1 thymidine was included in the suspension medium. FEC tissues of 0.5 to 0.7 cc settled cell volume (SCV) per sample were placed in 12-well plates, and each sample was inoculated with 2.0 mL of Agrobacterium suspension for 30 min. FEC tissues were then transferred onto GD2 50P medium supplemented with 200.0 mM acetosyringone and co-cultured for 3 d at 22 °C under light at 90 µMol m−2 s−1 After co-culture, FEC tissues from 60444 were washed with GD liquid medium containing 150.0 mg L−1 carbenicillin and cultured on GD2 50P medium supplemented with 150.0 mg L−1 carbenicillin under low light at 28 °C for 8 d. Tissue was then subcultured onto selection medium consisting of GD2 50P medium containing 150.0 mg L−1 carbenicillin and 27.5 µM paromomycin. For transformation of NASE 13, carbenicillin was replaced with 125.0 mg L−1 cefotaxime at each stage. Tissues were cultured for 21 d, followed by two subsequent regeneration stages on MS2 medium supplemented with 45.0 µM paromomycin, 5.0 µM naphthalene acetic acid (NAA), and 0.5 µM NAA. Cotyledon-stage embryos were germinated on MS2 medium supplemented with 2.0 µM meta-topolin (mT) and solidified with 2.2 g L−1 Gelzan (Segatto et al. 2022).
Tissues and plantlets were monitored for expression of GFP using a Nikon C15304 dissecting microscope equipped with an excitation filter of 460 to 500 nm and barrier filter 510 LP. GPF visual scoring was performed using a 0 to 5 scale according to Chauhan et al. (2015) where 0 = no visible signal; 1 = 1 to 10 GFP-expressing cells; 2 = 11 to 50 cells; 3 = 51 to 100 cells; 4 = 101 to 500 cells; and 5 = greater than 500 GFP-expressing cells visible. The number of growing FEC colonies recovered on callus selection medium, those regenerating somatic embryos, and germinating plantlets expressing GFP were assessed for evaluation of transformation efficiency by dividing the number at each stage by the initial SCV of FEC sample (Segatto et al. 2022). Somatic embryos growing on germination medium were evaluated weekly to observe time required for germination. Regenerated shoots were rooted and multiplied by micropropagation on MS2 medium solidified with 8.0 g L−1 Noble Agar.
Molecular confirmation of transgenic plants-Reverse transcription-polymerase chain reaction (RT-PCR) and RT-qPCR
Regenerated GFP-expressing plant lines were screened for VviGRF4-GIF1, AtGRF4-GIF1, and AtGRF5 gene expression by RT-PCR and RT-qPCR. All primers were designed using the Primer3 program (Primer3web version 4.0.0, https://primer3.org/) and produced by Integrated DNA Technologies (Coralville, IA) with details presented in Supplementary Table 1. Sequences selected for amplification started at the chimeric junction of the GRF4-GIF1 fusion and were therefore not present in the cassava genome.
Young leaf tissues were collected from GFP-expressing in vitro plants, and RNA extraction was performed using the Spectrum™ Plant total RNA kit (Sigma-Aldrich, Saint Louis, MO). Samples were treated with DNAse (DNASE70 Sigma-Aldrich) at room temperature for 15 min and were run in 1.0% agarose gel to check RNA quality. Two micrograms of total RNA was reverse-transcribed using the SuperScript™ III First-Strand Synthesis System (Invitrogen, Waltham, MA). PCR cycling conditions comprised an initial denaturation holding stage at 94 °C for 30 s, followed by 32 cycles of cycling stage at 94 °C for 20 s, 55 °C for 20 s, and 68 °C for 1:30 min, followed by final extension at 68 °C for 5 min.
RT-qPCR was performed with SSO Advanced Universal SYBR® Green Supermix (Bio-Rad laboratories Inc., Hercules, CA) with the endogenous cassava gene PP2A used as an internal control (Moreno et al. 2011). PCR cycling conditions comprised an initial denaturation holding stage at 95 °C for 30 s, followed by 40 cycles of cycling stage at 95 °C for 5 s, 61 °C for 30 s, melt curve stage from 65 to 95 °C with 0.5 increments for 5 s, followed by final extension at 95 °C for 5 min. Reactions were set up in triplicates for each sample. Quantification of the relative transcript levels was performed using the comparative CT (threshold cycle) method (Livak and Schmittgen 2001).
Organogenesis from tissues expressing VviGFR4-GIF1, AtGRF4-GIF, and AtGRF5
Transgenic plant lines regenerated from 60444 and NASE 13, confirmed by RT-PCR to express the morphogenic transgenes, were micropropagated on MS2 medium. Six-week-old plantlets were selected and explants prepared by excision of petioles, petioles with the leaf attached (leaf-petiole), and stem internodes approximately 1 cm in length. Explants were subcultured onto MS2 medium supplemented with 2.0 µM mT solidified with 2.2 g L−1 Gelzan and cultured under light 90 µMol m−2 s−1 at 28 °C for 5 wk.
Tissues were visually scored under a dissection microscope and assessed for presence of callus, compact green nodular tissue, foliose tissues, regenerated shoots, and roots. Callus production was scored using a 0 to 5 scale where 0 was absence of callus and 5 was the most amount of callus observed, reaching approximately 1 cm in diameter. Leaf-petiole regeneration experiments were performed in triplicate and results expressed as mean score and percentage from the replicas.
Assessment of somatic embryo maturation
Transgenic lines of cv 60444 expressing VviGRF4-GIF1 were assessed for regeneration of mature cotyledon-stage embryos from OES. OES was induced from immature leaf explants cultured on MS2 medium supplemented with 50.0 μM picloram and 2.0 μM CuSO4. After 4 wk culture in low light (20 µMol m−2 s−1) at 28 °C, the OES was excised, fragmented with a hypodermic needle, and used to establish 2-mm diameter colonies. Twenty-five OES colonies were established per Petri dish containing MS2 Gelzan medium supplemented with 2.0 µM mT. After 10 d culture under bright light (90 µMol m−2 s−1) at 28 °C, the number of green cotyledon-stage embryos developing from each OES colony was determined.
Plant establishment, growth, and assessment in the greenhouse
In vitro transgenic and control plantlets were planted in Berger BM7 35% Bark HP mixture potting compost (Hummert International, Earth City, MO) in 7.6-cm pots and established in the greenhouse following Segatto et al. (2022). For all experiments, five independent transgenic plant lines were established with five biological replicas along with a GFP control and a non-transgenic control. Plants were grown at 32 °C/26 °C (day/night) with 60 to 70% relative humidity.
Plant height was measured manually, and mean height was calculated. Leaf area was assessed by collecting the fourth expanded leaf below the shoot apical meristem after 14 wk growth in the greenhouse. Leaf images were captured using a Nikon COOLPIX L830 camera mounted on a support set 40 cm above the leaf material. ImageJ (Schneider et al. 2012) web browser (https://ij.imjoy.io/) was used for leaf area calculation where a known distance in cm was set as the scale, and image threshold adjusted to default red, and then analyzed using ROI manager, which selects, sums, and processes mean leaf area in centimeters squared.
After 14 wk in the greenhouse, a destructive harvest was performed. The stem was cut at the base and soil and fibrous roots removed from the storage roots. Stem height, number of nodes, fresh stem weight, number of storage roots, and fresh weight of storage roots were determined for each plant.
Experimental design and statistical analysis
Transformation experiments were performed four times, and organogenesis studies were repeated three times. Data generated was subjected to ANOVA to determine significant differences and where appropriate mean separation was done with the Dunnett test. Both ANOVA and mean separation were done using Minitab 17 statistical software package (Minitab Inc. State College, PA).
Results
Construction of binary plasmids
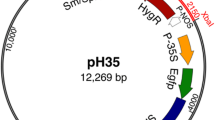
Three binary vectors were produced for integration of GRF morphogenic genes into cassava. Transgenic over expression of genes derived from Vitis vinifera had not been previously attempted in cassava and had unknown functionality in this species. Therefore, a second version of the GRF4-GIF1 and a GRF5 expression vector were generated utilizing sequences from Arabidopsis thaliana. A. thaliana genes are known to function well when overexpressed in cassava (Narayanan et al. 2019). The GRF genes were driven by a strong constitutive promoter, in the case of VviGFR4-GIF1 by the CaMV35S promoter and in AtGRF4-GIF1 and in the case of AtGRF5 by the CsVMV promoter. In all cases, a GFP expression cassette driven by the 35S promoter was included within the T-DNA to enable real-time, non-destructive observation of the transformation process from single cell expression immediately after co-culture to the recovery of fully developed plantlets. Details of all constructs are shown in Table 1 and Fig. 1. Gene constructs were transformed into Agrobacterium tumefaciens strain LBA4404 and the thymidine auxotrophic LBA4404 THY-, a modified version carrying extra virulence genes, previously reported for use with other morphogenic genes studies (Lowe et al. 2016). In this study, LBA4404 THY- was found to enhance efficiency of T-DNA transfer and transient GFP expression in cultivar NASE 13 (Supplementary Table 2).
Schematic representation of gene constructs used to generate transgenic plants in Manihot esculenta Crantz varieties 60444 and NASE 13. GFP-only control plasmid p8764. Vitis vinifera GRF4-GIF1 plasmid p8765. Arabidopsis thaliana GRF4-GIF1 plasmid p8788. A. thaliana GRF5 plasmid p8789. Organization of the T-DNA with arrows representing promoters and coding sequences and rectangles represent terminators. Restriction sites used for the construction are indicated.
Genetic transformation with morphogenic gene constructs
GRF4-GIF1 fusion gene constructs were used for Agrobacterium-mediated transformation of various explants, including cotyledon tissues derived from somatic embryos. However, these resulted in very low efficiency of T-DNA delivery as determined by few, to no, cells expressing GFP and failure to recover transgenic tissues. Friable embryogenic callus was therefore utilized as the target tissue for transgene integration. The varieties 60444 and NASE 13 were selected to observe the effect of GFR4-GIF1 and GRF5 on transformation efficiency and plant regeneration. Variety 60444 has high transformation efficiency and potential of plant recovery (Taylor et al. 2012) while NASE 13, an East African farmer-preferred cultivar, possesses lower capacity for transformation and plant regeneration (Narayanan et al. 2021). FEC tissues were transformed and monitored for GFP expression, recovery of transgenic callus, and regeneration of somatic embryos and plants to assess if morphogenic genes affected transformation and/or plant regeneration efficiency. Data shown in Table 2 is averaged from four experiments.
Recovery of transgenic tissue and plants from cultivar 60444 followed a pattern reported previously (Taylor et al. 2012) with up to 250 callus lines and as many as 29 independent transgenic plants recovered per cm3 SCV of co-cultured FEC for VviGRF4-GIF1. As predicted, efficiencies were lower for NASE 13 with approximately 100 callus lines and 8 to 10 independent transgenic plants recovered per cm3 of starting material for the same construct. A similar response was also seen for the GFP control, with 20 to 30% GFP-expressing callus lines regenerating plants in 60444 compared to 7 to 10% in NASE 13 (Table 2). Time to germination of somatic embryos also differed with shoots of 60444 appearing c. 28 d after being placed on germination medium while cultivar NASE 13 required up to 70 d and 2 to 3 additional cycles on germination medium for plant recovery. Importantly, all morphogenic gene constructs behaved similarly during the stages of transgenic tissue recovery, somatic embryo formation, and plant regeneration. At no stage did presence of the GRF4-GIF1 or GRF5 transgenes, whether derived from Vitis or Arabidopsis, confer measurable advantages for recovery of proliferating callus lines, regeneration of somatic embryos, or germination of plants from transgenic somatic embryos (Table 2). There was also no difference observed in the time required for plant recovery from mature somatic embryos between the morphogenic constructs and the GFP-only control.
Transgene expression confirmed by RT-PCR and RT-qPCR
Zero, or very low, transgene expression could explain why the morphogenic genes failed to simulate recovery of embryos and plants after transformation. Therefore, RT-PCR and RT-qPCR were performed on regenerated plants of 60444 and NASE 13 to confirm transgene expression. GFP-expressing plant lines from each gene construct underwent initial screening by RT-PCR using primers designed to be specific for the transgenes with all found to be expressing the morphogenic transgenes (Supplementary Fig. 2). In order to better determine morphogenic gene expression, RT-qPCR was performed. Figure 2 shows quantitative expression of VviGRF4-GIF1, AtGRF4-GIF1, and AtGRF5 from independent transgenic plant lines and GFP-only control from both cultivars. While no expression was detected from plants of the GFP-only control, expression of morphogenic genes was detected from very low to ten-fold relative gene expression (Fig. 2)., possibly due to position effect of the integrated transgenes.
Quantitative expression of morphogenic genes from in vitro Manihot esculenta Crantz leaf tissues. Expression was compared and normalized to protein phosphatase 2 (pp2A). For cultivar 60444, expression values of 65–3, 88–8, and 89–6 were adjusted to a value of 1 with other values expressed relative to these. For cultivar NASE 13, expression values of 65–1, 88–2, and 89–2 were adjusted to a value of 1 with other values expressed relative to these. Values are means of three technical replicates. Error bars represent SD.
Enhanced organogenesis observed from plants expressing morphogenic transgenes
Experiments were performed to determine if transgenic overexpression of GRF4-GIFI affected morphogenic potential in cassava. Initial evaluation was performed using two plant lines of cultivar 60444 expressing VviGRF4-GIF1 by culturing petiole, leaf-petiole, and stem internode explants on MS2 medium supplemented with 2.0 µM mT. Meta-topolin is a cytokinin produced from Populus × robusta and was previously found to stimulate the production of morphogenic tissues and regeneration of plants in cassava (Chauhan and Taylor 2018). Various tissues and morphogenic structures were produced from the explants over the 5-wk observation period (Supplementary Table 3). These included non-morphogenic callus, which was pale yellow in color with a soft, watery consistency and a compact, green nodular tissue (Figs. 3 and 4). No regeneration was observed from the soft, pale yellow callus. In contrast, green-colored, foliose tissues developed from the compact nodular tissue as soon as 10 d after explanting onto medium supplemented with meta-topolin. These then proliferated to form more foliose tissue or regenerated to produce shoots (Fig. 3; Supplementary Fig. 1). All explant types were capable of generating non-morphogenic yellow callus with petiole-leaf explants responding at the highest frequency of 95 to 100%. Leaf-petiole explants excised from transgenic lines expressing VviGRF4-GIF1 generated green morphogenic tissues at higher frequencies than petioles or internodes and were the only explant type from which shoots and roots were regenerated within the 5-wk observation period (Supplementary Table 3). Subsequent experiments therefore focused on further characterizing the organogenic potential of leaf-petiole explants.
Structures and organs regenerated from Manihot esculenta Crantz leaf-petiole explants of cultivar 60444 after culture on MS2 medium supplemented with 2.0 μM meta-topolin. (a) Initial explant excised from micropropagated mother plant. (b) Response of GFP control: Non-morphogenic callus developing from basal, cut end of the petiole, assessed as a visual quantitative score of 3 (0–5 scale). No further morphogenic response is observed from these tissues. (c–e) Response of GRF-GIF-expressing tissues: (c) compact green nodular tissue developing from basal end of the leaf-petiole explant, (d) development of foliose tissues with putative bud-like structures, and (e) regeneration of a fully formed shoot from the basal end of the leaf-petiole explant after 30-d culture. (f) Root regeneration from the basal end of the leaf-petiole explant after 20-d culture.
Development timeline of morphogenesis from Manihot esculenta Crantz leaf-petiole explants of cultivar NASE 13 after culture on MS2 medium supplemented with 2.0 μM meta-topolin. (a) Beginning of nodular tissue development at 6 d after explanting. (b) Growth of nodular tissue at 12 d after explanting. (c) Shoot-like development 18 d after explanting. (d) Multiple shoot-like structures developing 21 d after explanting. (e) Shoot regeneration 29 d after explanting. (f) Large shoot ready for excision and rooting 35 d after explanting.
Leaf-petiole explants were obtained from five independent transgenic 60444 and NASE 13 plant lines expressing each morphogenic gene construct and cultured on MS2 medium supplemented with meta-topolin. Tables 3 and 4 show data for the production of tissues from explants derived from 60444 and NASE 13, respectively, and Fig. 5 illustrates caulogenesis from these explants in both cultivars. Pale yellow-colored non-morphogenic callus developed from the basal, cut end of the petiole and to a lesser extend from the foliose tissue, starting after approximately 5 d of culture on MS2 medium containing 2.0 µM mT. Production of non-morphogenic callus continued over the 5-wk culture period with 90 to 100% of explants producing such callus in both cultivars across all the constructs and controls (Tables 3 and 4). A more complex response was seen for the production of compact green nodular tissue and foliose tissues (Figs. 3 and 4). In cultivar 60444, transgenic lines expressing VviGRF4-GIF1 formed these tissues at up to 53% and 78%, respectively, versus 12% and 3% for the GFP-only control. In contrast, production of green morphogenic tissues was lower at only 14% and 7% from explants derived from plants transgenic for the Arabidopsis ortholog, AtGRF4-GIF1. Transgenic plants of cultivar 60444 expressing AtGRF5 generated values only slightly elevated from GFP and non-transgenic controls (Table 3). Significantly superior production of green morphogenic tissue was observed from cultivar NASE 13. Transgenic lines expressing VviGRF4-GIF1 in this variety formed foliose tissues at up to 78% versus 4% for the GFP control. Production of green nodular and foliose tissues occurred at 30% and 21% from explants transgenic for AtGRF4-GIF1 and at 70% and 63% for explants expressing AtGRF5 (Table 4).
Caulogenesis in Manihot esculenta Crantz leaf-petioles expressing GRF genes. Average response from leaf-petioles explants after 4 wk culture on MS2 medium supplemented with 2.0 μM meta-topolin. Response is averaged from three replicas with 40 explants each; bars represent mean standard error. *Significantly different from group mean, p < 0.05.
Shoot regeneration only occurred in transgenic lines which produced compact green nodular and foliose tissues, with between one and four shoots produced per responding explant. Shoot regeneration first became visible during the second week of culture on meta-topolin containing medium with clearly defined and recoverable shoots observed within 4 wk. No shoot regeneration occurred from explants derived from GFP controls or from non-transgenic plants of 60444 or NASE 13 over the 5-wk observation period. In cultivar 60444, expression of VviGRF4-GIF1 stimulated caulogenesis with shoot regeneration occurring from all five transgenic lines and a maximum response of 37% seen from event 65–6. No shoot regeneration was observed in tissues transgenic for AtGRF4-GIF1 in 60444 while AtGRF5 stimulated only low shoot regeneration up to 4.5% (Table 3; Fig. 5). NASE 13 displayed a significantly higher shoot regeneration than 60444 with all three morphogenic constructs stimulating caulogenesis (Table 4; Fig. 5). In this cultivar, expression of AtGRF5 resulted in the highest rates of shoot regeneration at up to 46%. Again, VviGRF4-GIF1 was more effective at stimulating caulogenesis than AtGRF4-GIF1 at 33% vs 13%, respectively, with no shoots regenerated from three of the five transgenic AtGRF4-GIF1 lines (Table 4; Fig. 5).
Rhizogenesis was observed in all plant lines, generally taking place within 15 d after explanting. Expression of morphogenic genes did not enhance rhizogenesis with average group means not significantly different from the control. In most cases, explants that produced roots did not regenerate shoots. The lowest rates of rhizogenesis were observed from NASE 13 lines expressing AtGRF5, which also displayed the highest caulogenic potential (Table 4).
Stimulation of somatic embryo maturation
The effect of morphogenic gene expression on somatic embryo maturation was assessed using OES derived from plants expressing VviGRF4-GIF1. OES, induced from immature leaf explants, were placed on MS2 medium supplemented with 2.0 µM mT, and the development of mature, green cotyledon-stage embryos was assessed over a 10-d period. Figure 4 shows the results for three different VviGRF4-GIF1-expressing lines plus controls with an image illustrating the rapid maturity of these embryos compared to the GFP-only control. By the end of the 10-d observation period, VviGRF4-GIF1-expressing lines had developed significantly more cotyledon-stage somatic embryos, and these lines were more mature and approximately 1.5 times larger than those produced by the control (Fig. 6).
Stimulation of somatic embryo maturation from organized embryogenic structures in Manihot esculenta Crantz cultivar 60444 after culture on MS2 medium supplemented with 2.0 μM meta-topolin for 14 d. (a) Average number of embryos that matured to green cotyledon-stage per organized embryogenic structure (OES) fragment. (b) Maturation of somatic embryos from GFP-only control. (c) Development of many mature, green cotyledon-stage embryos from OES expressing VviGRF4-GIF1. Bars represent mean standard error. *Significantly different, p < 0.05.
Growth and development of transgenic plants in the greenhouse
Transgenic plants from cultivar 60444 expressing VviGRF4-GIF1, AtGRF4-GIF1, AtGRF5, and GFP-only were established in soil and grown under greenhouse conditions to assess effects of morphogenic gene overexpression on whole plants. All plant lines were robust and survived transfer to soil at the same rates. After 14 wk of growth in the greenhouse, plants were harvested and plant height, number of nodes, stem fresh weight, leaf area, and storage root number fresh weight were determined from five biological replicates (Table 5). Plants transgenic for GRF4-GIF1 were on average shorter than the GFP control with plants expressing VviGRF4-GIF and AtGRF4-GIF1 and developed broader leaves that resulted in an increased leaf area at levels significantly different in lines 65–1, 65–2, 65–5, 65–6, 88–4, and 88–7 compared to controls (Table 5). AtGRF5-expressing plant lines were similar to the GFP control in height but showed a moderate increase in storage root weight in three out of five transgenic lines. Event 89–5 developed significantly more storage root mass, representing an average gain of 16% compared to the GFP control.
Discussion
The current study reported here that overexpression of GRF4-GIF1 and GRF5 stimulates morphogenesis in cassava. The initial intent was to employ these morphogenic transcription factors to stimulate shoot regeneration from various explants, such as cotyledons from somatic embryos. However, T-DNA transfer to these tissues remained too inefficient to proceed. Agrobacterium-mediated transformation of FEC tissues is well documented in cassava and was therefore utilized to study the effects of these morphogenic transcription factors on the recovery of transgenic tissue and plants in this species. Transformation of the varieties 60444 and NASE 13 clearly showed that expression of these genes had no effect, beneficial or detrimental, on efficiency of the transformation process or on regeneration of somatic embryos and plants. A lack of effect during the recovery of embryogenic tissues is perhaps not surprising and could be related to gene function since meristem formation and proliferation for which these genes are known to function were not occurring. Positive effects were expected, however, during the stages of somatic embryo maturation and germination when meristem formation does occur and foliose cotyledon tissues are being produced. Indeed, this lack of response across several hundred regeneration events in 60444 and NASE 13 contrasts with the significantly accelerated development and maturation, which was subsequently observed from somatic embryos derived from transgenic plants expressing VviGRF4-GIF1 (Fig. 6). This result also contrasts with the positive effects seen when GRF-GIF were overexpressed in beet and citrus. In these species, non-embryogenic tissues were targeted for transgenic overexpression, perhaps explaining the gain in transformation efficiency seen in these species. Regarding regeneration, similarities are seen between organogenesis in citrus epicotyls expressing GRF-GIF (Debernardi, et al. 2020) and cassava petioles with stimulation of de novo meristems in these stem-like explants.
The current authors previously reported regeneration of transgenic plants in cultivar NASE 13 (Narayanan et al. 2021) but did not describe the process or efficiencies for this Ugandan, farmer-preferred variety. As expected, NASE 13 responded at lower efficiencies at all stages of transformation and plant regeneration compared to 60444 but at levels which were still effective for recovery of transgenic plant lines. Data reported here reveals two stages where efforts should be focused to improve the transformation process for this cultivar. T-DNA transfer to the FEC target tissue is relatively efficient, especially when the LBA4404 THY- strain is used (Supplementary Table 2), but initiation of cell division from these transient events occurs at very low frequencies, thus constraining recovery of callus tissue that can then be cultured to regenerate plants. Likewise, germination of the matured cotyledon-stage embryos in NASE 13 is not optimal, occurring at 30% compared to 70% in 60444 and requiring an extra 50 to 70 d for recovery of shoots compared to the latter. Focusing research efforts on stimulating cell division and germination would likely result in enhanced efficiencies for recovery of transgenic and genome-edited events from transgenic FEC and generate knowledge applicable and valuable for improving these processes in additional cassava varieties.
RT-qPCR analysis confirmed expression of GRF4-GIF1 and GRF5 in all transgenic plants tested, although at varying levels (Fig. 2). Three explant types were evaluated for their response to culture on MS2 medium supplemented with 2.0 μM meta-topolin as this cytokinin was previously reported to stimulate organogenesis in cassava cultivars (Chauhan and Taylor 2018). Data generated in the present study confirmed that petioles with leaf tissue attached are the most responsive for production of morphogenic tissues and shoot regeneration (Supplementary Table 3). The superior organogenic potential of leaf-petioles could be due to an interaction between the cytokinin in the growth medium and naturally occurring auxins in the leaf since young leaves are known to be a primary origin of auxin (Taiz and Zeiger 1998).
Investigations using leaf-petiole explants derived from plants transgenic for GRF4-GIF1 and GRF5 indicated a significant difference in morphogenic potential between gene constructs and cultivars. For cultivar 60444, transgenic lines tested expressing Vitis-derived GRF4-GIF1 regenerated shoots at 20 to 30% while no response was observed from plants expressing Arabidopsis-derived GRF4-GIF1 and very low levels of shoot regeneration for AtGRF5 (Fig. 5). This variance could be due to several reasons, such as amino acid sequence similarity, with VviGRF4 protein sequence being 78.26% similar to the Manihot esculenta GRF4 while AtGRF4 is 56.91% similar. Post-transcriptional regulation (Filipowicz et al. 2008; Furlan et al. 2021), ectopic expression (Siefers et al. 2009; Kerr et al. 2018), or functional orthologs (Das et al. 2016) could also play a role. It is worth noting that GRF4 can be post-transcriptionally downregulated by microRNA396 while GRF5 cannot since the latter does not possess the binding site for microRNA396 (Kim and Tsukaya 2015). Interestingly for cultivar NASE 13 AtGRF5 had the best morphogenesis potential with three lines regenerating shoots at 40 to 50%, which outperformed VviGRF4-GIF1 in leaf-petiole organogenesis. In this more recalcitrant cultivar, all morphogenic constructs stimulated fast shoot regeneration.
Chauhan and Taylor (2018) reported the production of compact green nodular tissues produced when leaf-petiole explants were cultured on MS2 containing meta-topolin and that shoots could be regenerated from these tissues after a series of sequential subcultures over a period of 9 to 10 wk. In the present work, shoot regeneration was achieved from the compact green nodular tissue expressing GRF/GIF1 and GFR5 at a higher frequency and much faster with rapid differentiation of foliose tissues and plantlets as soon as 3 wk culture on meta-topolin containing medium (Supplementary Fig. 1). It is postulated therefore that expression of GRF genes is enhancing caulogenesis and proliferation of the shoot meristems from these tissues.
Plants of cultivar 60444, transgenics for GRF4-GIF1 and GRF5, were established in soil and grown in the greenhouse to assess the effects on whole plant development. Cassava plant lines overexpressing GRF4-GIF1 were shorter than the other transgenics and control (Table 5; Fig. 7), and six out of ten lines showed significantly increased leaf area compared to the control (Table 5). This is not an unexpected result as GIF1 is a known positive regulator of cell proliferation in leaves and flowers (Lee et al. 2009; Rodriguez et al. 2010; Debernardi et al. 2014). Increased leaf area did not result in increased productivity in terms of storage root weight, but this may have been due to the relatively small pots used and short growing period of 14 wk in this study. Plants expressing GRF5 did have superior root weight with one line being significantly different from controls with a gain of 16% fresh weight (Table 5). Longer cultivation time in larger pots is therefore recommended to better reveal interesting agronomic characteristics resulting from overexpression of GRF4-GIF1 and GRF5.
This study has shown that overexpression of heterologous GRF4-GIF1 and GRF5 genes can stimulate morphogenesis and shoot regeneration in cassava explants with different rates of regeneration seen between 60444 and NASE 13. It is encouraging that NASE 13 responds with greater caulogenic potential than 60444, indicating that alternative morphogenic pathways could be utilized to enable regeneration across cassava varieties. While use of these genes did not enhance rates of transgenic plant regeneration from FEC tissues, the present authors do consider that the data presented indicates that they have potential to do so in other tissues and could be valuable tools for the recovery of transgenic and gene-edited plants in cassava. Further work should focus on delivery to non-embryogenic tissues and assessing the use of different promoters and arrangements of the GRF and GIF genes to stimulate caulogenesis in non-totipotent tissues.
Data availability
The raw data generated and analyzed during the current study are available from the corresponding author on reasonable request.
References
Bull SE, Seung D, Chanez C, Mehta D, Kuon JE, Truernit E, Hochmuth A, Zurkirchen I, Zeeman SC, Gruissem W, Vanderschuren H (2018) Accelerated ex situ breeding of GBSS- and PTST1-edited cassava for modified starch. Sci Adv 4:eaat6086. https://doi.org/10.1126/sciadv.aat6086
Chauhan RD, Beyene G, Kalyaeva M, Fauquet CM, Taylor NJ (2015) Improvements in Agrobacterium-mediated transformation of cassava (Manihot esculenta Crantz) for large-scale production of transgenic plants. Plant Cell Tiss Org Cult 121:591–603. https://doi.org/10.1007/s11240-015-0729-z
Chauhan RD, Taylor NJ (2018) Meta-topolin stimulates de novo shoot organogenesis and plant regeneration in cassava. Plant Cell Tiss Org Cult 132:219–224. https://doi.org/10.1007/S11240-017-1315-3/FIGURES/3
Chavarriaga-Aguirre P, Brand A, Medina A, Prias M, Escobar R, Martines J, Dias P, Lopes C, Roca WM, Tohme J (2016) The potential of using biotechnology to improve cassava: a review. In Vitro Cell Dev Biol - Plant 52:461–478. https://doi.org/10.1007/s11627-016-9776-3
Das M, Haberer G, Panda A, Laha SD, Ghosh TC, Schäffner AR (2016) Expression pattern similarities support the prediction of orthologs retaining common functions after gene duplication events. Plant Physiol 171:2343–2357. https://doi.org/10.1104/pp.15.01207
Debernardi JM, Mecchia MA, Vercruyssen L, Smaczniak C, Kaufmann K, Inze D, Rodriguez RE, Palatnik JF (2014) Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J 79:413–426. https://doi.org/10.1111/tpj.12567
Debernardi JM, Tricoli DM, Ercoli MF, Hayta S, Ronald P, Palatnik JF, Dubcovsky J (2020) A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat Biotechnol 38:1274–1279. https://doi.org/10.1038/s41587-020-0703-0
Filipowicz W, Bhattacharyya S, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9:102–114. https://doi.org/10.1038/nrg2290
Furlan M, de Pretis S, Pelizzola M (2021) Dynamics of transcriptional and post-transcriptional regulation. Brief Bioinformat 22:389. https://doi.org/10.1093/bib/bbaa389
Gomez M, Berkoff KC, Gill BK, Iavarone AT, Lieberman SE, Ma JM, Schultink A, Karavolias NG, Wyman SK, Chauhan RD, Taylor NJ, Staskawicz BJ, Cho M-J, Rokhsar DS, Lyons JB (2023) CRISPR-Cas9-mediated knockout of CYP79D1 and CYP79D2 in cassava attenuates toxic cyanogen production. Front Plant Sci 17:13. https://doi.org/10.3389/fpls.2022.1079254
Gordon-Kamm B, Sardesai N, Arling M, Lowe K, Hoerster G, Betts S, Jones T (2019) Using morphogenic genes to improve recovery and regeneration of transgenic plants. Plants 8:38. https://doi.org/10.3390/plants8020038
Hummel AW, Chauhan RD, Cermak T, Mutka AM, Vijayaraghavan A, Boyher A, Starker CG, Bart R, Voytas DF, Taylor NJ (2018) Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnol J 16:1275–1282. https://doi.org/10.1111/pbi.12868
Kerr TCC, Abdel-Mageed H, Aleman L, Lee J, Payton P, Cryer D, Allen RD (2018) Ectopic expression of two AREB/ABF orthologs increases drought tolerance in cotton (Gossypium hirsutum). Plant Cell Environ 41:898–907. https://doi.org/10.1111/pce.12906
Kim JH, Choi D, Kende H (2003) The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J 36:94–104. https://doi.org/10.1046/j.1365-313X.2003.01862.x
Kim JH, Lee BH (2006) GROWTH-REGULATING FACTOR4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J Plant Biol 49:463–468. https://doi.org/10.1007/BF03031127
Kim JH, Tsukaya H (2015) Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. J Exp Bot 66:6093–6107. https://doi.org/10.1093/jxb/erv349
Kong J, Martin-Ortigosa S, Finer J, Orchard N, Gunadi A, Batts LA, Thakare D, Rush B, Schmitz O, Stuiver M, Olhoft P, Pacheco-Villalobos D (2020) Overexpression of the transcription factor GROWTH-REGULATING FACTOR5 improves transformation of dicot and monocot species. Front Plant Sci 11:23–57. https://doi.org/10.3389/fpls.2020.572319
Lee BH, Ko JH, Lee S, Lee Y, Pak JH, Kim JH (2009) The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol 151:655–668. https://doi.org/10.1104/pp.109.141838
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho MJ, Scelonge C, Lenderts B, Chamberlin M, Cushatt J, Wang L, Ryan L, Khan T, Chow-Yiu J, Hua W, Yu M, Banh J, Bao Z, Brink K, Igo E, Rudrappa B, Shamseer PM, Bruce W, Newman L, Shen B, Zheng P, Bidney D, Falco C, Register J, Zhao ZY, Xu D, Jones T, Gordon-Kamm W (2016) Morphogenic regulators baby boom and WUSCHEL improve monocot transformation. Plant Cell 28:1998–2015. https://doi.org/10.1105/tpc.16.00124
Moreno I, Gruissem W, Vanderschuren H (2011) Reference genes for reliable Potyvirus quantification in cassava and analysis of cassava brown streak virus load in host varieties. J Virol Methods 177:49–54. https://doi.org/10.1016/j.jviromet.2011.06.013
Narayanan N, Beyene G, Chauhan RD, Gaitan-Solis E, Gehan J, Butts P, Siritunga D, Okwounu I, Woll A, Jimenes-Aguilar DM, Boy E, Grusac MA, Anderson P, Taylor NJ (2019) Biofortification of field-grown cassava by engineering expression of an iron transporter and ferritin. Nat Biotechnol 37:144–151. https://doi.org/10.1038/s41587-018-0002-1
Narayanan N, Beyene G, Chauhan RD, Grusak MA, Taylor NJ (2021) Stacking disease resistance and mineral biofortification in cassava varieties to enhance yields and consumer health. Plant Biotechnol J 19:844–854. https://doi.org/10.1111/pbi.13511
Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF (2010) Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137:103–112. https://doi.org/10.1242/dev.043067
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH image to ImageJ 25 years of image analysis. Nat Methods 9:671–6750. https://doi.org/10.1038/nmeth.2089
Segatto R, Jones T, Stretch D, Albin C, Chauhan RD, Taylor NJ (2022) Agrobacterium-mediated genetic transformation of cassava. Current Protocols 2:620. https://doi.org/10.1002/cpz1.620
Siefers N, Dang KK, Kumimoto RW, Bynun WE, Tayrose G, Holt BF (2009) Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol 149:625–641. https://doi.org/10.1104/pp.108.130591
Taiz L, Zeiger E (1998) Plant physiology. 2nd edn. Sinauer Mass
Taylor NJ, Gaitán-Solís E, Moll T, Trauterman B, Jones T, Pranjal A, Trembley C, Abernathy V, Corbin D, Fauquet C (2012) A high-throughput platform for the production and analysis of transgenic cassava (Manihot esculenta) plants. Trop Plant Biol 5:127–139. https://doi.org/10.1007/s12042-012-9099-4
The Arabidopsis Information Resource (TAIR). https://www.arabidopsis.org/servlets/Search?action=new_search&type=gene. Accessed 19 Mar 2021
Utsumi Y, Utsumi C, Tanaka M, Okamoto Y, Takahashi S, Huong TT, Nguyen AV, Dong NV, Tokunaga H, Taylor NJ, Seki M (2022) Agrobacterium-mediated cassava transformation for the Asian elite variety KU50. Plant Mol Biol 109:271–282. https://doi.org/10.1007/S11103-021-01212-1/TABLES/3
Veley KM, Okwuonu I, Jensen G, Yoder M, Taylor NJ, Meyers BC, Bart RS (2021) Gene tagging via CRISPR-mediated homology-directed repair in cassava. 15:11(4). https://doi.org/10.1093/g3journal/jkab028
Vercruyssen L, Tognetti VB, Gonzalez N, Van Dingenen J, De Milde L, Bielach A, De Rycke R, Van Breusegem F, Inzé D (2015) GROWTH REGULATING FACTOR5 stimulates Arabidopsis chloroplast division, photosynthesis, and leaf longevity. Plant Physiol 167:817–832. https://doi.org/10.1104/pp.114.256180
Verdaguer B, de Kochko A, Fux CI, Beachy RN, Cl F (1998) Functional organization of the cassava vein mosaic virus (CsVMV) promoter. Plant Mol Biol 37:1055–1067. https://doi.org/10.1023/A:1006004819398
Wagaba H, Beyene G, Aleu J, Odipio J, Okao-Okuja G, Chauhan RD, Munga T, Obiero H, Halsey ME, Ilyas M, Raymond P, Bua A, Taylor NJ, Miano D, Alicai T (2017) Field level RNAi-mediated resistance to cassava brown streak disease across multiple cropping cycles and diverse east African agro-ecological locations. Front Plant Sci 7:20–60. https://doi.org/10.3389/FPLS.2016.02060/BIBTEX
Watling J, Shock MP, Mongeló GZ, Almeida FO, Kater T, Oliveira PED, Neves EG (2018) Direct archaeological evidence for southwestern Amazonia as an early plant domestication and food production centre. PLoS ONE 13:e0199868. https://doi.org/10.1371/journal.pone.0199868
Acknowledgements
The methods and systems described here were developed with support from the Donald Danforth Plant Science Center. The authors acknowledge contributions from Getu Beyene, Narayanan Narayanan, Tira Jones, Danielle Stretch, and Claire Albin. This research fulfills part of a MSc thesis in Biotechnology with degree granted to R. Segatto by the Federal University of Mato Grosso do Sul – Brazil.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Rosana Segatto. The first draft of the manuscript was written by Rosana Segatto, and all authors reviewed and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Segatto, R., Paggi, G.M. & Taylor, N.J. Overexpression of GRF-GIF genes enhances plant regeneration in cassava (Manihot esculenta). In Vitro Cell.Dev.Biol.-Plant (2024). https://doi.org/10.1007/s11627-024-10435-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11627-024-10435-y