Abstract
Various carbohydrate-active enzymes in Aspergillus are produced in response to physiological inducers, which is regulated at the transcriptional level. To elucidate the induction mechanisms in Aspergillus, we screened for new regulators involved in cellulose-responsive induction from approximately 10,000 Aspergillus aculeatus T-DNA-inserted mutants. We constructed the T-DNA-inserted mutant library using the host strain harboring the orotidine 5′-monophosphate decarboxylase gene (pyrG) under the control of the FIII-avicelase gene (cbhI) promoter. Thus, candidate mutants deficient in cellulose-responsive induction were positively screened via counter selection against 5-fluoroorotic acid (5-FOA). Among less than two hundred 5-FOA-resistant mutants, one mutant that the T-DNA inserted into the AasepM locus reduced the cbhI expression in response to cellulose. Since AaSepM is similar to Schizosaccharomyces pombe Cdc14p (E-value, 2e-20; identities, 33%), which is a component of the septation initiation network (SIN)-complex, we constructed an AasepM deletion mutant (ΔAasepM). We analyzed the expression of cellulase and xylanase genes in response to cellulose, septation, and conidiation in ΔAasepM. The AasepM deletion leads to delayed septation and decreased formation of the conidium chain in A. aculeatus but does not affect hyphal growth on minimal media. We also confirmed AaSepM’s involvement in multiple cellulose-responsive signaling pathways of cellulase and xylanase genes under the control of the ManR-dependent, XlnR-dependent, and ManR- and XlnR-independent signaling pathways.
Key points
• A new regulator for cellulolytic gene expression has been identified.
• AaSepM is involved in septation and conidiation in A. aculeatus.
• AasepM is involved in multiple cellulose-responsive signaling pathways.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Filamentous fungi are extensively used to produce enzymes that degrade lignocellulose into compounds (Payne et al. 2015). Among filamentous fungi, Trichoderma reesei strains are important hosts involved in cellulase production (Bischof et al. 2016). However, the secreted cellulase mix of T. reesei lacks sufficient β-glucosidase activity (Saloheimo et al. 2002). Murao and Sakamoto (1979) screened and identified Aspergillus aculeatus no. F-50 [NBRC 108789] produces enzymes that synergistically hydrolyze pulp along with a cellulase enzyme mix of T. reesei. β-Glucosidase is a key enzyme of A. aculeatus that synergistically accelerates cellulose hydrolysis with the T. reesei system (Baba et al. 2015; Nakazawa et al. 2012). Although A. aculeatus no. F-50 has useful enzyme components for lignocellulose degradation, there is little information about the regulatory mechanisms of the associated gene expressions.
Multiple pathways regulate cellulase and hemicellulase genes’ expression in Aspergillus (Kunitake and Kobayashi 2017; Tani et al. 2014). CreA in Aspergillus nidulans plays a central role in carbon catabolite repression (CCR) (Dowzer and Kelly 1989). However, Kunitake et al. (2019) recently reported that protein kinase A (pkaA) and GanB, which is one of the trimeric G-protein α subunits, are involved in CCR in a CreA-independent manner in A. nidulans. The contribution of CreA, PkaA, and GanB in CCR differs depending on the inducers, repressing carbon sources, and culture conditions.
In contrast to CCR, various transcriptional activators have been identified over the past few decades. The first identified transcriptional activator of cellulolytic and xylanolytic genes was XlnR, a Zn(II)2Cys6 type transcriptional activator that coordinates xylanolytic expression in Aspergillus niger (van Peij et al. 1998b). Further analysis revealed that cellulolytic gene expression is also controlled by XlnR in Aspergillus (Marui et al. 2002; Noguchi et al. 2009; van Peij et al. 1998a). In Neurospora, the expression of a transcriptional activator gene, clr-2, is triggered by a transcriptional activator, CLR-1, which further triggers the expression of many cellulase genes in response to cellulosic carbon sources (Coradetti et al. 2012). CLR-2 is conserved in the genomes of most filamentous ascomycete fungi that produce cellulolytic enzymes (Coradetti et al. 2012). ClrB, a CLR-2 ortholog in A. nidulans, regulates cellulolytic gene expression (Coradetti et al. 2012). ClrB forms a homodimer or heterocomplex with McmA to regulate cellulosic expression in A. nidulans (Li et al. 2016). Cellulolytic and mannolytic genes in response to carbon sources liberated from cellulose and mannan are also regulated by ManR, which is a ClrB ortholog in Aspergillus oryzae (Ogawa et al. 2013). The regulation mechanisms of cellulolytic enzymes in Aspergillus are complex (Tani et al. 2014). Therefore, elucidating cellulase genes’ regulatory mechanisms in each Aspergillus species is necessary to improve enzyme production by transcription factor modification.
The function of XlnR in A. aculeatus seemed to be conserved among Aspergillus. Therefore, we first screened genes that were not regulated by XlnR (Tani et al. 2012). We found that cellulosic carbon sources induced the FIII-avicelase gene (cbhI) via an XlnR-independent signaling pathway. To identify new factors involved in the XlnR-independent signaling pathway in A. aculeatus, we established a positive screening system using the cbhI promoter fused to the orotidine 5′-phosphate decarboxylase gene. From a random insertional mutagenesis library generated by Agrobacterium tumefaciens–mediated transformation of A. aculeatus, the ClbR transcription factor and dipeptidyl peptidase IV were identified (DppIV) as regulators for cellulolytic gene expression in A. aculeatus (Kunitake et al. 2011, 2013; Tani et al. 2017).
In this study, further screening revealed the involvement of AaSepM in septation and conidiation, as well as in multiple cellulose-responsive signaling pathways under the control of the ManR-dependent, XlnR-dependent, and ManR- and XlnR-independent signaling pathways.
Experimental procedures
Strains, transformation, marker recycling, and T-DNA insertion
All A. aculeatus strains used in this study were derived from wild-type A. aculeatus no. F-50 [NBRC 108796]. Unless otherwise stated, all strains were propagated at 30°C in an appropriately supplemented minimal medium (MM) or complete medium (Adachi et al. 2009; Kunitake et al. 2015). A. aculeatus NCP2 (niaD1::niaD::PCHB1-pyrG, pyrG1) was used as a host to construct A. aculeatus strains for T-DNA insertion by A. tumefaciens–mediated transformation. Counter selection on 5-FOA and marker recycling were done as described previously (Kunitake et al. 2011, 2013). We used A. aculeatus MR12 (pyrG1, Δku80) as a host for the disruption and complementation of the AasepM gene (Tani et al. 2013). Moreover, we used Escherichia coli DH5αF′ (Takara, Kyoto, Japan) for plasmid construction.
Disruption and complementation of AasepM
The primer pair Fno12/Rno12 was used for the amplification of an AasepM fragment that possesses a SmaI site at the 5′ end of each primer to include 1119 nt before the translation start site and 267 nt after the stop codon. The amplified DNA fragment was digested with SmaI and subcloned into the pPTRII SmaI site (Takara) to yield pPTRII-sepM. The T-DNA-inserted mutant, namely, the A. aculeatus no. 12 mutant, was subsequently transformed by pPTRII and pPTRII-sepM (Kubodera et al. 2000). Pyrithiamine was added at a final concentration of 0.1 μg/ml to the selection media.
We generated the A. aculeatus sepM-deficient mutant (pyrG1, Δku80, ΔAasepM) by replacing the AasepM gene by the A. nidulans orotidine 5′-phosphate decarboxylase gene (AnpyrG), followed by marker recycling (Adachi et al. 2009; Tani et al. 2013). To construct the AasepM deletion cassette, the 5′ and 3′ regions of the AasepM gene, which play a key role in homologous recombination to replace AasepM with AnpyrG, were amplified from A. aculeatus genomic DNA using the primer pair no12_1F/no12_1R and no12_2F/no12_2R, respectively. The AnpyrG gene was amplified from the A. nidulans genomic DNA using primer pair no12pyrGF/no12pyrGR. Moreover, the flanking region on the 3′ side of AasepM was amplified using the primer pair no12_3F/no12_3R to eliminate AnpyrG by intramolecular homologous recombination at the AasepM locus. The 5′ region, which is responsible for marker recycling, AnpyrG, and the 3′ region were fused by PCR using the primer pair no12_1F/no12_2R. The amplified fragment was introduced into MR12 (pyrG1, Δku80) by the protoplast-PEG method to yield the A. aculeatus ΔsepM plus pyrG strain (pyrG1, Δku80, ΔAasepM::AnpyrG). Then, 1 × 104 spores of transformants were spread onto the MM, supplemented with 0.01% (w/v) uridine and 1 mM 5-FOA, to perform marker recycling. After monospore isolation, A. aculeatus ΔsepM (pyrG1, Δku80, ΔAasepM) was obtained (Supplementary Fig. S1). The primers used in this study are summarized in Supplementary Table S1.
To complement A. aculeatus ΔsepM, we first amplified the AasepM promoter, open reading frame, and 3′ untranslated region using the primer pair CsepM_1F/CsepM_1R. Subsequently, we amplified AnpyrG and the 3′ region required for homologous recombination at the AasepM locus with primer pairs CsepM-pF/CsepM-pR and CsepM_2F/CsepM_2R, respectively. The three DNA fragments were fused by PCR using the primer pair CsepM_1F/CsepM_2R. Finally, we used the amplified DNA fragments to transform A. aculeatus ΔsepM into the AasepM-complemented strain (pyrG1, Δku80, ΔAasepM::AasepM::AnpyrG) (Supplementary Fig. S1).
Disruption and complementation of manR
The A. aculeatus manR-deficient mutant (pyrG1, Δku80, ΔmanR) was created with the replacement of the manR gene by AnpyrG followed by marker recycling. To construct the manR deletion cassette as described above, we amplified the 5′ region of manR, which is the fragment for marker recycling; the selection marker gene; and the 3′ regions of manR, using primer pair DmanR5F/DmanR5R, DmanRrecF/DmanRrecR, Dm-AnpyrGF/Dm-AnpyrGR, and DmanR3F/DmanR3R, respectively (Supplementary Table S1). We simultaneously subcloned the 5′ region, digested with BglII and NotI, and the fragment for marker recycling, digested with NotI and EcoRV, into pBluescript II KS (+) (Takara), which was digested with BglII and EcoRV. This plasmid was digested with EcoRV and HindIII and subsequently ligated with AnpyrG, which was digested with EcoRV and PacI, and the 3′ region, which was digested with PacI and HindIII. This process yielded pDmanR. The ligated fragment was amplified from pDmanR using the primer pair DmanR5F/DmanR3R. We used the protoplast-PEG method to introduce the amplified fragment into MR12 (pyrG1, Δku80) to yield the A. aculeatus ΔmanR plus pyrG strain (pyrG1, Δku80, ΔmanR::AnpyrG). Marker recycling was performed as described above. After monospore isolation, A. aculeatus ΔmanR (pyrG1, Δku80, ΔmanR) was obtained (Supplementary Fig. S2).
To complement A. aculeatus ΔmanR, we first amplified the manR promoter, open reading frame, and 3′ untranslated region using the primer pair CmanR5F/CmanR5R. Subsequently, we amplified AnpyrG and the 3′ region required for homologous recombination at the manR locus with the primer pairs CmanR3F/CmanR3R and Cm-AnpyrGF/Cm-AnpyrGR, respectively. The three DNA fragments were fused by PCR using the primer pair CmanR5F/CmanR3R. Finally, A. aculeatus ΔmanR was transformed with the amplified DNA fragments to yield the manR-complemented strain (pyrG1, Δku80, ΔmanR::manR::AnpyrG) (Supplementary Fig. S2).
Gene expression analysis by real-time quantitative reverse transcription PCR
We used real-time quantitative reverse transcription PCR (qRT-PCR) to quantify cellulase expression and hemicellulase gene expression based on a previously described method (Tani et al. 2017). In this study, 0.1% Bacto Tryptone (Thermo Fisher Scientific, Tokyo, Japan) was used as a neutral carbon source (noninducing condition). We added the indicated carbon sources to media supplemented with 0.1% Bacto Tryptone to investigate test genes’ expression. Total RNA (500 ng) was used to amplify cDNA with ReverTra Ace qPCR RT-Master Mix (Toyobo, Tokyo, Japan). qRT-PCR was performed in a Thermal Cycler Dice Real-Time System (Takara). As for the amplification reactions, we performed a SYBR Green I assay using THUNDERBIRD™ SYBR qPCR Mix (Toyobo) in a reaction volume of 20 μl. Supplementary Table S1 lists the primers used for qRT-PCR. The internal control was set to the expression of the glyceraldehyde-3-phosphate dehydrogenase A gene (gpdA). The specificity of the PCR amplification was confirmed by melting curve analysis. We analyzed each gene’s expression profile using the delta-delta CT method (Livak and Schmittgen 2001). More than three biological replicates were performed for each experiment, and each was tested in a triplicate.
Yeast two-hybrid assay
We performed a yeast two-hybrid assay (Y2H) using the Matchmaker GAL4-based Two-Hybrid System 3 (Clonetech, Tokyo, Japan) according to the manufacturer’s instructions. We amplified the coding sequences of the AasepM cDNA and the A. aculeatus sepL (AasepL) cDNA with the primer sets Y2sepMF/Y2sepMR2 and sepL-cDNAFa/sepLcDNARb, respectively. The amplified DNA fragments of AasepM and AasepL were digested with NdeI and PstI or NdeI and BamHI, respectively. We fused the digested fragments in-frame with the GAL4 DNA-binding domain in the pGBKT7 to yield pGBKT7-sepM and, subsequently, the GAL4-activation domain in the pGADT7 to yield pGADT7-sepL.
Counting of septa
Conidia (1 × 103) were grown for 16 to 24 h on coverslips at 30°C in MM supplemented with 0.1% yeast extract. We monitored septation based on a previous method that was lightly modified (Harris et al. 1994). We stained germlings with a solution containing 0.5 μg/ml calcofluor white (Sigma-Aldrich, Tokyo, Japan), monitored them using Zeiss LSM-700 (ZEISS, Tokyo, Japan), and counted the number of septa that formed in 100 μm of a hypha. We counted more than 60 hyphae in different areas of the three biological replicates and then calculated the average and standard deviations.
Additional methods
We performed genomic DNA preparation and Southern blot analysis, as previously described (Kunitake et al. 2013). We used an in-house A. aculeatus [NBRC 108789] draft genome database to obtain the genomic sequence of the AasepM gene and analyzed two independently amplified cDNA fragments to determine the AasepM cDNA sequence. Conidia in the A. aculeatus strains were collected with 0.1% Tween 80/0.8% NaCl solution and counted with a hemocytometer. We normalized the number of conidia by the colony area. Supplementary Table S2 lists the genes studied in this manuscript. A scanning electron microscope (SEM) SU-1510 (Hitachi, Tokyo, Japan) was used to observe the conidium formation.
Nucleotide sequence data
The nucleotide sequence data studied in this manuscript have been deposited in Japan’s DNA Data Bank (DDBJ) Nucleotide Sequence Data Libraries. Supplementary Table S2 lists the genes and their accession numbers.
Results
A. aculeatus SepM is a new factor controlling the cellulose-responsive induction of cbhI
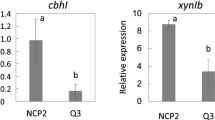
To isolate a new factor controlling the cellulose-responsive expression of cellulase genes, we screened the T-DNA-inserted mutant library in A. aculeatus to identify cellulose-responsive expression-deficient mutants. The T-DNA-inserted mutant library was constructed by A. aculeatus NCP2 harboring the orotidine 5′-phosphate decarboxylase gene (pyrG) under the control of the promoter of cbhI, which is induced in response to cellulose. Therefore, we assumed that only cellulose-utilization-deficient mutants could grow on 5-FOA media supplemented with wheat bran as a cellulosic carbon source (Kunitake et al. 2011, 2013). After counter selection on the 5-FOA, fewer than two hundred mutants out of an estimated number of 10,000 T-DNA-inserted mutants were observed to grow on the media. Afterward, we selected five isolated mutants with poor growth on cellulose media but normal growth on D-glucose, D-xylose, and beechwood xylan media from the total pool. We assessed the cbhI expression in response to Avicel in the isolated five mutants for the last screening step. The cbhI transcripts were quantified using RNA extracted from mycelia grown for 9 h under Avicel-inducing or noninducing conditions. Only one mutant, namely, no. 12, significantly reduced the cbhI expression under the Avicel-inducing condition to approximately 50% in A. aculeatus NCP2 (Fig. 1a). At the same time, there was no difference under the noninducing condition. These data suggest that the T-DNA disrupted a gene required for the cellulose-responsive expression of cbhI in the no. 12 strain.
Identification of cellulose-responsive induction-deficient mutants. a qRT-PCR analysis of the cbhI expression at 9-h post-induction by 1% (w/v) Avicel was conducted in the host (NCP2) and the no. 12 strains (gray bars). The filled bars indicate the cbhI expression level under an uninduced condition. The relative expression corresponds to the mean expression levels of cbhI divided by those of the gpdA, a reference gene. The relative expression levels are the means of three independent experiments, and the error bars indicate standard deviations. b A schematic representation of the AasepM locus and the deduced T-DNA integration pattern. c qRT-PCR analysis of the cbhI in the no. 12 strains harboring pPTRII (-) and pPTRII-sepM (AasepM) was performed as described above. Letters indicate significant differences between groups (p < 0.05, Student’s t test)
To identify the gene disrupted by T-DNA insertion, we recovered the T-DNA flanking sequences by an inverse PCR. Since the T-DNA was inserted into the genome of the no. 12 strain as a single copy (Supplementary Fig. S3), we used the genomic DNA of the no. 12 strain, digested with only EcoRI, to amplify the T-DNA-flanking sequences, which recovered the right border sequence of the T-DNA. The genomic DNA digested with XbaI and SpeI was used to recover the left border sequence (Fig. 1b). Sequence analyses of the amplified DNA segments revealed that the right border of the T-DNA was inserted into 510 bp upstream of the gene orthologous to the septation initiation network (SIN) component (sepM in A. nidulans, AN0655) in the recipient genome, namely, A. aculeatus sepM (AasepM). The flanking sequence of the left border was not determined, which was likely due to the vector backbone. Based on our previous analyses, T-DNA tends to be inserted with an average 1.4-kb deletion of the recipient genome at the T-DNA-inserted locus (Kunitake et al. 2011). Therefore, we selected AasepM as the first candidate gene for a factor involved in the cellulose-responsive expression of cbhI.
The no. 12 strain was transformed with pPTRII and pPTRII-sepM. The insertion of the AasepM gene complemented the cellulose-responsive induction-deficient phenotype of the no. 12 strain, but not pPTRII. This finding suggests that AaSepM is involved in the cellulose-responsive expression of cbhI in A. aculeatus (Fig. 1c). Therefore, the deletion mutant of AasepM (ΔAasepM) and its complement strain (AasepM+) were newly constructed (Supplementary Fig. S1). We quantified the cbhI transcripts in MR12, ΔAasepM, and AasepM+ strains, respectively. The Avicel-induced expression of cbhI in ΔAasepM was reduced to 16% compared to that of the control strain. The reduced expression of cbhI in ΔAasepM recovered in AasepM+, which confirms that AaSepM is involved in the cellulose-responsive induction of cbhI in A. aculeatus (Fig. 2).
The effect of AasepM deletion on cbhI expression. Results of the qRT-PCR analysis of the cbhI expression at 9-h post-induction with (gray bars) or without (filled bars) 1% (w/v) Avicel in MR12, ΔAasepM, and AasepM+. The relative expression corresponds to the ratio of the mean expression levels of cbhI divided by those of gpdA. The relative expression levels are the means of three independent experiments, and the error bars indicate standard deviations. Letters indicate significant differences between groups (p < 0.05, Student’s t test)
AasepM comprises 965 bp, with two exons divided by one intron, which encode a 295 amino acid protein. This protein has homology with AN0655, A. nidulans SepM, (E-value, 3e-166; identities, 84%), and Schizosaccharomyces pombe Cdc14p (E-value, 2e-20; identities, 33%), which was a component of the SIN complex (Kim et al. 2009).
AaSepM is involved in septation and conidiation in A. aculeatus
Cdc14p is required for septum formation since it interacts with the carboxyl terminus of Sid1p kinase, which is a group II p21-activated kinase/germinal center kinase family member, and is necessary for the Sid1p catalytic activity and intracellular localization to control the cytokinesis in S. pombe (Guertin et al. 2000; Guertin and McCollum 2001). It has been reported that the SepL, a Sid1p ortholog in A. nidulans, is required for septation and conidiation in A. nidulans (de Souza et al. 2013). In contrast, there are no experimental studies of SepM’s function in filamentous fungi.
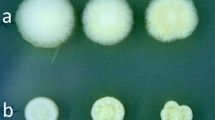
To test whether AaSepM is involved in septum formation in A. aculeatus, germlings of MR12, ΔAasepM, and AasepM+ were fixed and stained with calcofluor white for chitin visualization. Septum formation in ΔAasepM reduced to 4% and 37% of that in the control strain in a MM at 16-h and 24-h post-inoculation, respectively (Fig. 3a–c). Several putative immature septa were randomly observed in the hyphae in ΔAasepM (Fig. 3a, arrowhead). This septation-deficient phenotype was restored in AasepM+. In contrast, there was no detectable septum-deficient phenotype in ΔAasepM at 16-h post-inoculation in the complete medium, probably due to the rapid hyphal growth and septum formation in complete media (data not shown). These data confirmed that AaSepM is involved in septum formation but is not strictly necessary. Subsequently, we investigated the effect of the AasepM deletion on hyphal growth and conidiation on MM. Conidia in ΔAasepM (average 3.2 × 106 conidia/cm2) was significantly reduced (n = 3, p < 0.05) to 25% of conidia in the control strain (average 1.3 × 107 conidia/cm2), which was restored in AasepM+ (average 1.1 × 107 conidia/cm2), although there was no significant difference in the hyphal growth among test strains (Fig. 4a). SEM analysis revealed conidium chain in MR12 and AasepM+ but not in ΔAasepM (Fig. 4b). These data indicated that AaSepM is involved in septation and formation of the conidium chain. These phenotypes were similar to those of the A. nidulans sepL-deletion mutant, suggesting that AaSepM functions cooperatively with AaSepL. Afterward, we assessed whether AaSepM interacts with AaSepL by yeast two-hybrid analysis. The only strain expressing both AaSepM and AaSepL was grown on quadruple dropout media (Fig. 5). These data demonstrated that AaSepM interacts with AaSepL, demonstrating that AaSepM and AaSepL cooperatively regulate septum formation and conidiation.
The effect of the AasepM deletion on septum formation. a MR12, ΔAasepM, and AasepM+ were grown for 16 or 24 h at 30°C. Hyphae were fixed, stained with calcofluor white, and analyzed with a confocal laser scanning microscope. Representative images are shown. Arrowheads indicate immature septa. We counted the number of septa formed in 100 μm of a hypha. More than 60 hyphae in different areas were counted in three biological replicates, and the average and standard deviations were calculated. The number of septa after 16-h and 24-h incubation is shown in b and c, respectively. Letters indicate significant differences between groups (p < 0.05, Student’s t test)
The effect of AasepM deletion on hyphal growth and conidiation on the MM. a Representative images of MR12, ΔAasepM, and AasepM+, which were grown for 4 days at 30°C. b Representative images of MR12, ΔAasepM, and AasepM+ observed by SEM, showing a deficiency in the formation of the conidium chain in ΔAasepM. Bar is 50 μm
AaSepM and AaSepL interaction by the Y2H system. Growth tests of yeasts co-transformed with pGBKT7 and pGADT7, pGBKT7 and pGBKT7-AaSepL, pGBKT7-AaSepM and pGADT7, and pGBKT7-AaSepM and pGADT7-AaSepL were conducted on double-dropout media (SD-Leu-Trp) and quadruple-dropout media (SD-Leu-Trp-His-Ade), respectively. Representative data from three independent experiments are shown
The expression profile of AasepM
To investigate the expression of AasepM under the inducing, noninducing, or repressing conditions of the cellulase and xylanase genes, we quantified transcripts of AasepM under 1% (w/v) d-glucose, 1% (w/v) d-xylose, 1% (w/v) beechwood xylan, 1% (w/v) cellobiose with 50 μg/l deoxy-nojirimycin (DNJ), 1% (w/v) Avicel, and no-carbon conditions. We added DNJ to investigate the direct effect of cellobiose on the gene expression since A. aculeatus secretes β-glucosidase effectively hydrolyze cello-oligosaccharides (Baba et al. 2015). The wild-type A. aculeatus was grown for the best duration to repress or induce the expression of cellulase and xylanase genes; e.g., d-glucose (3 h), d-xylose (3 h), beechwood xylan (6 h), cellobiose with DNJ (3 h), and Avicel (6 h), respectively (Fig. 6). Although the expression of AasepM was significantly lower in the presence of d-glucose and beechwood xylan in comparison with that under a carbon-free condition (3 h), AasepM seemed to be expressed on nearly the same level as gpdA, which is known to be a highly expressed gene. Therefore, we concluded that AasepM is abundantly expressed under all test conditions.
qRT-PCR analysis of the AasepM expression in A. aculeatus wild-type at 9-h incubation under the following conditions: NC noninducing condition, G 1% glucose, X 1% xylose, BX 1% beechwood xylan, C 0.1% cellobiose with 50 μg/l DNJ, A 1% Avicel. The relative expression corresponds to the ratio of the mean expression levels of AasepM divided by those of gpdA. The relative expression levels are the means of three independent experiments, and the error bars indicate the standard deviations. Letters indicate significant differences between groups (p < 0.05, one-way ANOVA)
AaSepM is involved in the cellulose-responsive expression of the cellulase and hemicellulase genes
To elucidate the role of AaSepM on the cellulose-responsive expressions of cellulase and xylanase genes, we quantified the expressions of the cellobiohydrolase II gene (cbhII), hydrocellulase gene (cel7b), FI-carboxymethyl cellulase gene (cmc1), FII-carboxymethyl cellulase gene (cmc2), FIa-xynalase gene (xynIa), and FIb-xylanase gene (xynIb) in response to 1% (w/v) Avicel at 9-h post-induction. Since all test genes’ expression levels in response to Avicel were the same in MR12 and AasepM+, AasepM+ was used as a control. The expression level of each test gene in ΔAasepM was reduced to 4% (cbhII), 29% (cmc1), 28% (cmc2), 53% (cel7b), 49% (xynIa), and 20% (xynIb) of those in AasepM+ under the Avicel condition, respectively. These reductions were observed only under the Avicel-inducing condition, but not under the noninducing condition. Therefore, the fold induction of each test gene (the gene expression level under the inducing condition divided by that under the noninducing condition) is reflected by the reduction of the expression level under the inducing condition (Fig. 7a).
The effect of AasepM deletion on the expression of the cellulase and hemicellulase genes. a qRT-PCR was conducted on each gene in ΔAasepM and AasepM+ incubated for 9 h under a noninduced condition (filled bars) and 1% Avicel-inducing condition (gray bars), respectively. b qRT-PCR of each gene in the strains incubated for 9 h under a noninduced condition (filled bars) and the 1% beechwood xylan-inducing condition (gray bars) was conducted. The relative expression corresponds to the ratio of the mean expression levels of each gene divided by that of gpdA. The relative expression levels are the means of three independent experiments, and the error bars indicate the standard deviations. An asterisk indicates a significant difference between test genes’ expression under the inducing condition in ΔAasepM and in AasepM+ (p < 0.05, Student’s t test)
Furthermore, we investigated whether the deletion of AasepM affected cellulase and xylanase gene expressions in response to beechwood xylan. Transcripts of cmc1 and xynIb were quantified in the presence of 1% beechwood xylan at 9-h post-induction, since these genes’ expressions, but not the other test genes, were induced in response to d-xylose. The expressions of cmc1 and xynIb in response to beechwood xylan at 9-h post-induction were not affected by the deletion of AasepM (Fig. 7b). These data demonstrated that AaSepM was involved in the cellulose-responsive expression in A. aculeatus.
AaSepM is involved in multiple signaling pathways in response to the cellulosic substrate
We previously reported that a transcription factor, XlnR, controls the induction of cmc1 and xynIb in response to both Avicel and beechwood xylan in A. aculeatus, while the other test genes were not under the control of XlnR (Tani et al. 2012). One candidate regulator for the XlnR-independent signaling pathway is ClrB/ManR, which controls cellulase gene expression in response to cellulose in A. nidulans and A. oryzae (Coradetti et al. 2012; Ogawa et al. 2012, 2013). Therefore, the potential genes under the control of ManR in the cellulose-responsive expression in A. aculeatus were assessed. The cellulose-responsive expression levels of cbhI, cbhII, cel7b, and cmc2 were abolished in ΔmanR, which was restored in the manR-complemented strain (manR+) (Fig. 8). Furthermore, the cellulose-responsive induction of xynIa, cmc1, and xynIb was reduced to 50%, 10%, and 22% compared to that in manR+ (Fig. 8).
qRT-PCR analysis of the cellulase and hemicellulase genes in the manR deletion mutant (ΔmanR) and the manR-complemented strain (manR+) grown at 9-h post-induction by 1% Avicel. We normalized each gene’s expression level by that of gpdA. The relative expression corresponds to the ratio of the mean expression levels of each gene in ΔmanR divided by those in manR+. The relative expressions are the means of three independent experiments, and error bars indicate standard deviations. Letters indicate significant differences between groups (p < 0.05, one-way ANOVA)
The involvement of AaSepM in the gene expression of known transcription factors such as ManR, ClbR, and XlnR, which are involved in the cellulose-responsive expression in A. aculeatus, was investigated (Fig. 9). We quantified transcripts of the manR, clbR, and xlnR genes in ΔAasepM and AasepM+. The expression of manR under the Avicel condition in ΔAasepM was reduced to 24% compared to that in AasepM+. In contrast, the deletion of AasepM resulted in a 1.7-fold (clbR) and 1.8-fold (xlnR) increase, which indicates that AaSepM is also involved in the cellulose-responsive induction of manR. These data demonstrated that AaSepM participates broadly in cellulose-responsive induction in A. aculeatus.
The effect of AasepM deletion on the expression of transcription factor genes. The expression of selected transcription factor genes in ΔAasepM and AasepM+ following a 9-h shift to Avicel via qRT-PCR. The relative expression corresponds to the ratio of the mean expression levels of each gene in ΔAasepM divided by those in AasepM+. The relative expressions are the means of three independent experiments, and the error bars indicate standard deviations. Student’s t test assessed statistical significance (*p < 0.05)
Discussion
Expression of the cellulase and xylanase genes is regulated in response to physiological conditions in filamentous fungi. Here, we described a novel factor, AaSepM, which is involved in septation and conidiation and regulates the cellulose-responsive induction of the cellulase genes, xylanase genes, and manR gene in A. aculeatus.
Cytokinesis completes the cell cycle and marks the birth of newly formed daughter cells. In S. pombe, several proteins intricately control septum formation. These proteins form the SIN and regulate cytokinesis initiation at the end of the anaphase. The SIN consists of nine proteins, including Sid1p and Cdc14p (Krapp and Simanis 2008). Cdc14p is required for full catalytic activity and localization of Sid1p. Cdc14p regulates the kinase activity of Sid1p by interacting with the C-terminus of Sid1p in S. pombe (Guertin et al. 2000; Guertin and McCollum 2001). SIN mutants proceed through additional nuclear division cycles without septation, thus becoming long and multinucleate before they eventually lyse (Balasubramanian et al. 1998). In contrast to these ascomycetous yeasts, the sepL-deletion mutant in A. nidulans and its Sid1p ortholog, showed extremely poor conidiation and a complete absence of the septa in A. nidulans (de Souza et al. 2013). However, the sepL-deletion mutant and ΔAasepM were not lethal, which is probably due to the general nature of the multi-nuclear Aspergillus cells. A yeast two-hybrid assay showed that AaSepM interacted with AaSepL (Fig. 5), which showed good correlation between the septation-deficient and conidiation-deficient phenotypes in ΔAasepM, while septa formed gradually in ΔAasepM, which suggested that AaSepM is not necessary for septum formation stimulated by AaSepL in A. aculeatus.
To induce cellulase gene expression, soluble saccharides such as cellobiose, a physiological inducer, must first be liberated from the cellulose in A. aculeatus (Tani et al. 2012). Therefore, reduced production of cellulases, i.e., cellobiohydrolase and endoglucanase, could result in an insufficient supply of physiological inducers and reduced expression of their genes. In filamentous fungi, exocytosis occurs mainly at growing hyphal apexes where the Golgi apparatus and endoplasmic reticulum compartments accumulate abundantly (Shoji et al. 2014; Wösten et al. 1991). However, exocytosis of α-amylase at the septa has been reported in Aspergillus oryzae (Hayakawa et al. 2011). If the enzymes’ exocytosis is reduced in ΔAasepM due to a deficiency in septation, their genes’ reduced expression is necessary. However, the accumulation of endoglucanase and β-1,4-xylanase in submerged MM and complete medium supplemented with wheat bran as a carbon source was not affected by the deletion of AasepM (data not shown). Furthermore, our results showed that AasepM deletion affected only gene induction in response to cellulosic carbon sources, but not beechwood xylan. Therefore, the reduced expression of the cellulase genes is probably caused by the specific function of AaSepM in the cellulose-responsive signaling pathway.
Factors controlling the expression of the cellulase and hemicellulase genes are required to adapt to environmental changes or alterations in physiological conditions, such as pH, light, and cell wall integrity in filamentous fungi. VEL1, the T. reesei ortholog of A. nidulans VeA, forms the VELVET protein complex consisting of VeA, LaeA, and VelB, regulating secondary metabolism sexual and asexual reproduction. VEL1 is also involved in the expression of cellulase and xylanase genes in response to lactose and sophorose, governed by the main regulator, Xyr1, in T. reesei (Karimi Aghcheh et al. 2014). Interestingly, different signaling pathways are crossed via pathway-specific transcription factors, suggesting that a comprehensive analysis of both ManR and AaSepM might reveal molecular mechanisms that cooperatively regulate morphological development and inducible enzyme production.
Of particular interest in the present study was the observation that AaSepM was involved in three different cellulose-responsive induction pathways: (i) the gene expressions of cbhI, cbhII, cmc2, and cel7b, which were regulated via the ManR-dependent signaling pathway; (ii) the gene expressions of cmc1 and xynIb, which were regulated via the XlnR-dependent signaling pathway; and (iii) the expression of xynIa, which was regulated via the ManR- and XlnR-independent signaling pathway. Furthermore, among the genes under the control of the ManR-dependent signaling pathway, it should be noted that the induced expression of cbhII was abolished by the AasepM deletion but not the others (Fig. 7a). We first considered the possibility that AaSepM directly or indirectly affects the ManR activity in gene expression, which in response to cellulose is regulated by the homologs, ClrB in A. nidulans, ManR in A. oryzae (Coradetti et al. 2013; Ogawa et al. 2013), and ManR in A. aculeatus. Their function, ClrB/ManR, could be conserved in cellulase-responsive gene expression. ClrB in A. nidulans positively regulates the expression of cellulase genes in two different forms. The first is that ClrB forms a homodimer and binds to CGGN8CCG. The second is that ClrB forms a heterocomplex with McmA, which is an SRF-MADS box protein in A. nidulans that binds to CCGN2CCN6GG, namely, the cellulose-responsive element (CeRE) (Endo et al. 2008; Li et al. 2016; Yamakawa et al. 2013). To investigate the possibility of the involvement of AaSepM in switching the complex formation of ClrB/ManR, we mined the ClrB recognition sites in the promoter region of test genes in A. aculeatus. Among the genes under the control of ManR, three and one homodimer recognition sites were discovered within the 1-kb upstream region of the translation start site of cmc2 and cel7b in A. aculeatus, respectively. One CeRE site was found in cbhI, while no conserved sequence was found within the 1-kb promoter region of cbhII in A. aculeatus. There was no correlation between the dependency of AaSepM on the cellulose-responsive induction of the test genes and the DNA-binding mode of ClrB/ManR. This lack of correlation suggests that AaSepM might not be involved in switching the dimerization form of ManR, although further analyses are needed to confirm this.
Our data also indicated that the ManR- and XlnR-independent signaling pathway regulated the expression of xynIa in response to cellulose, which also was under the control of AaSepM. ClbR, a Zn2(II)Cys6 binuclear cluster transcription factor, helped induce cellulase and hemicellulase genes in response to cellulose in A. aculeatus (Kunitake et al. 2013). Our previous study revealed that ClbR is a limiting factor for xynIa expression in response to cellulosic carbon sources. clbR overexpression resulted in xynIa overexpression, increasing XynIa protein production (Kunitake et al. 2015). No coactivators were known to bind to Zn2(II)Cys6 binuclear cluster transcription factors and control gene regulation. Therefore, AaSepM should interact with another protein, such as protein kinase(s), responsible for governing the cellulose-responsive expression upstream of the transcription factors. Perhaps, the new roles of AaSepM in A. aculeatus reflect the evolutionary divergence between fungi. It will be an important focus of future studies to identify counterparts of AaSepM on the cellulose-responsive signaling pathway in A. aculeatus and determine their function.
Data availability
All data generated or analyzed during this study are included in this published article and its Supplementary Information files. DNA sequences will be available at the DDBJ database when this article is released for publication.
References
Adachi H, Tani S, Kanamasa S, Sumitani J, Kawaguchi T (2009) Development of a homologous transformation system for Aspergillus aculeatus based on the sC gene encoding ATP-sulfurylase. Biosci Biotechnol Biochem 73(5):1197–1199. https://doi.org/10.1271/bbb.80772
Baba Y, Sumitani J, Tani S, Kawaguchi T (2015) Characterization of Aspergillus aculeatus beta-glucosidase 1 accelerating cellulose hydrolysis with Trichoderma cellulase system. AMB Express 5:3. https://doi.org/10.1186/s13568-014-0090-3
Balasubramanian MK, McCollum D, Chang L, Wong KC, Naqvi NI, He X, Sazer S, Gould KL (1998) Isolation and characterization of new fission yeast cytokinesis mutants. Genetics 149(3):1265–1275
Bischof RH, Ramoni J, Seiboth B (2016) Cellulases and beyond: the first 70 years of the enzyme producer Trichoderma reesei. Microb Cell Factories 15(1):106. https://doi.org/10.1186/s12934-016-0507-6
Coradetti S, Craig J, Xiong Y, Shock T, Tian C, Glass N (2012) Conserved and essential transcription factors for cellulase gene expression in Ascomycete fungi. Proc Natl Acad Sci U S A 109(19):7397–7402. https://doi.org/10.1073/pnas.1200785109
Coradetti ST, Xiong Y, Glass NL (2013) Analysis of a conserved cellulase transcriptional regulator reveals inducer-independent production of cellulolytic enzymes in Neurospora crassa. Microbiol Open 2(4):595–609. https://doi.org/10.1002/mbo3.94
de Souza CP, Hashmi SB, Osmani AH, Andrews P, Ringelberg CS, Dunlap JC, Osmani SA (2013) Functional analysis of the Aspergillus nidulans kinome. PLoS One 8(3):e58008. https://doi.org/10.1371/journal.pone.0058008
Dowzer CE, Kelly JM (1989) Cloning of the creA gene from Aspergillus nidulans: a gene involved in carbon catabolite repression. Curr Genet 15(6):457–459. https://doi.org/10.1007/BF00376804
Endo Y, Yokoyama M, Morimoto M, Shirai K, Chikamatsu G, Kato N, Tsukagoshi N, Kato M, Kobayashi T (2008) Novel promoter sequence required for inductive expression of the Aspergillus nidulans endoglucanase gene eglA. Biosci Biotechnol Biochem 72(2):312–320. https://doi.org/10.1271/bbb.70278
Guertin DA, Chang L, Irshad F, Gould KL, McCollum D (2000) The role of the Sid1p kinase and Cdc14p in regulating the onset of cytokinesis in fission yeast. EMBO J 19(8):1803–1815. https://doi.org/10.1093/emboj/19.8.1803
Guertin DA, McCollum D (2001) Interaction between the noncatalytic region of Sid1p kinase and Cdc14p is required for full catalytic activity and localization of Sid1p. J Biol Chem 276(30):28185–28189. https://doi.org/10.1074/jbc.M103802200
Harris SD, Morrell JL, Hamer JE (1994) Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics 136:517–532
Hayakawa Y, Ishikawa E, Shoji J, Nakano H, Kitamoto K (2011) Septum-directed secretion in the filamentous fungus Aspergillus oryzae. Mol Microbiol 81(1):40–55. https://doi.org/10.1111/j.1365-2958.2011.07700.x
Karimi Aghcheh R, Nemeth Z, Atanasova L, Fekete E, Paholcsek M, Sandor E, Aquino B, Druzhinina IS, Karaffa L, Kubicek CP (2014) The VELVET A orthologue VEL1 of Trichoderma reesei regulates fungal development and is essential for cellulase gene expression. PLoS One 9(11):e112799. https://doi.org/10.1371/journal.pone.0112799
Kim JM, Zeng CJ, Nayak T, Shao R, Huang AC, Oakley BR, Liu B (2009) Timely septation requires SNAD-dependent spindle pole body localization of the septation initiation network components in the filamentous fungus Aspergillus nidulans. Mol Biol Cell 20(12):2874–2884. https://doi.org/10.1091/mbc.E08
Krapp A, Simanis V (2008) An overview of the fission yeast septation initiation network (SIN). Biochem Soc Trans 36(3):411–415. https://doi.org/10.1042/bst0360411
Kubodera T, Yamashita N, Nishimura A (2000) Pyrithiamine resistance gene (ptrA) of Aspergillus oryzae: cloning, characterization and application as a dominant selectable marker for transformation. Biosci Biotechnol Biochem 64(7):1416–1421. https://doi.org/10.1271/bbb.64.1416
Kunitake E, Kawamura A, Tani S, Takenaka S, Ogasawara W, Sumitani J, Kawaguchi T (2015) Effects of clbR overexpression on enzyme production in Aspergillus aculeatus vary depending on the cellulosic biomass-degrading enzyme species. Biosci Biotechnol Biochem 79(3):488–495. https://doi.org/10.1080/09168451.2014.982501
Kunitake E, Kobayashi T (2017) Conservation and diversity of the regulators of cellulolytic enzyme genes in Ascomycete fungi. Curr Genet. https://doi.org/10.1007/s00294-017-0695-6
Kunitake E, Li Y, Uchida R, Nohara T, Asano K, Hattori A, Kimura T, Kanamaru K, Kimura M, Kobayashi T (2019) CreA-independent carbon catabolite repression of cellulase genes by trimeric G-protein and protein kinase A in Aspergillus nidulans. Curr Genet 65(4):941–952. https://doi.org/10.1007/s00294-019-00944-4
Kunitake E, Tani S, Sumitani J, Kawaguchi T (2011) Agrobacterium tumefaciens-mediated transformation of Aspergillus aculeatus for insertional mutagenesis. AMB Express 1(1):46. https://doi.org/10.1186/2191-0855-1-46
Kunitake E, Tani S, Sumitani J, Kawaguchi T (2013) A novel transcriptional regulator, ClbR, controls the cellobiose- and cellulose-responsive induction of cellulase and xylanase genes regulated by two distinct signaling pathways in Aspergillus aculeatus. Appl Microbiol Biotechnol 97(5):2017–2028. https://doi.org/10.1007/s00253-012-4305-8
Li N, Kunitake E, Aoyama M, Ogawa M, Kanamaru K, Kimura M, Koyama Y, Kobayashi T (2016) McmA-dependent and -independent regulatory systems governing expression of ClrB-regulated cellulase and hemicellulase genes in Aspergillus nidulans. Mol Microbiol 102(5):810–826. https://doi.org/10.1111/mmi.13493
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Marui J, Kitamoto N, Kato M, Kobayashi T, Tsukagoshi N (2002) Transcriptional activator, AoXlnR, mediates cellulose-inductive expression of the xylanolytic and cellulolytic genes in Aspergillus oryzae. FEBS Lett 528(1-3):279–282. https://doi.org/10.1016/s0014-5793(02)03328-8
Murao S, Sakamoto R (1979) β-Glucosidase of Aspergillus aculeatus. Agric Biol Chem 43(8):1791–1792
Nakazawa H, Kawai T, Ida N, Shida Y, Kobayashi Y, Okada H, Tani S, Sumitani J-i, Kawaguchi T, Morikawa Y, Ogasawara W (2012) Construction of a recombinant Trichoderma reesei strain expressing Aspergillus aculeatus beta-glucosidase 1 for efficient biomass conversion. Biotechnol Bioeng 109(1):92–99. https://doi.org/10.1002/bit.23296
Noguchi Y, Sano M, Kanamaru K, Ko T, Takeuchi M, Kato M, Kobayashi T (2009) Genes regulated by AoXlnR, the xylanolytic and cellulolytic transcriptional regulator, in Aspergillus oryzae. Appl Microbiol Biotechnol 85(1):141–154. https://doi.org/10.1007/s00253-009-2236-9
Ogawa M, Kobayashi T, Koyama Y (2012) ManR, a novel Zn(II)2Cys6 transcriptional activator, controls the beta-mannan utilization system in Aspergillus oryzae. Fungal Genet Biol 49(12):987–995. https://doi.org/10.1016/j.fgb.2012.09.006
Ogawa M, Kobayashi T, Koyama Y (2013) ManR, a transcriptional regulator of the beta-mannan utilization system, controls the cellulose utilization system in Aspergillus oryzae. Biosci Biotechnol Biochem 77(2):426–429. https://doi.org/10.1271/bbb.120795
Payne CM, Knott BC, Mayes HB, Hansson H, Himmel ME, Sandgren M, Ståhlberg J, Beckham GT (2015) Fungal cellulases. Chem Rev 115:1308–1448. https://doi.org/10.1021/cr500351c
Saloheimo M, Paloheimo M, Hakola S, Pere J, Swanson B, Nyyssönen E, Bhatia A, Ward M, Penttilä M (2002) Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur J Biochem 269(17):4202–4211. https://doi.org/10.1046/j.1432-1033.2002.03095.x
Shoji J, Kikuma T, Kitamoto K (2014) Vesicle trafficking, organelle functions, and unconventional secretion in fungal physiology and pathogenicity. Curr Opin Microbiol 20:1–9. https://doi.org/10.1016/j.mib.2014.03.002
Tani S, Kanamasa S, Sumitani J, Arai M, Kawaguchi T (2012) XlnR-independent signaling pathway regulates both cellulase and xylanase genes in response to cellobiose in Aspergillus aculeatus. Curr Genet 58(2):93–104. https://doi.org/10.1007/s00294-012-0367-5
Tani S, Kawaguchi T, Kobayashi T (2014) Complex regulation of hydrolytic enzyme genes for cellulosic biomass degradation in filamentous fungi. Appl Microbiol Biotechnol 98(11):4829–4837. https://doi.org/10.1007/s00253-014-5707-6
Tani S, Tsuji A, Kunitake E, Sumitani J, Kawaguchi T (2013) Reversible impairment of the ku80 gene by a recyclable marker in Aspergillus aculeatus. AMB Express 3:4. https://doi.org/10.1186/2191-0855-3-4
Tani S, Yuki S, Kunitake E, Sumitani J, Kawaguchi T (2017) Dipeptidyl peptidase IV is involved in the cellulose-responsive induction of cellulose biomass-degrading enzyme genes in Aspergillus aculeatus. Biosci Biotechnol Biochem 81(6):1227–1234. https://doi.org/10.1080/09168451.2017.1295800
van Peij N, Gielkens M, de Vries R, Visser J, de Graaff L (1998a) The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl Environ Microbiol 64(10):3615–3619. https://doi.org/10.1128/AEM.64.10.3615-3619.1998
van Peij N, Visser J, de Graaff L (1998b) Isolation and analysis of XlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol Microbiol 27(1):131–142. https://doi.org/10.1046/j.1365-2958.1998.00666.x
Wösten HA, Moukha SM, Sietsma JH, Wessels JGH (1991) Localization of growth and secretion of proteins in Aspergillus niger. J Gen Microbiol 137:2017–2023
Yamakawa Y, Endo Y, Li N, Yoshizawa M, Aoyama M, Watanabe A, Kanamaru K, Kato M, Kobayashi T (2013) Regulation of cellulolytic genes by McmA, the SRF-MADS box protein in Aspergillus nidulans. Biochem Biophys Res Commun 431(4):777–782. https://doi.org/10.1016/j.bbrc.2013.01.031
Funding
JSPS KAKENHI supported this work under Grant 19K05777. This work was also partly supported by the New Energy and Industrial Technology Development Organization (NEDO) Project under Grant P07015.
Author information
Authors and Affiliations
Contributions
S.T. and T.K. conceived the project. R.T., K.S., E.K., J. S., and S.T. designed the project. R.T., K.S., and E.K. conducted the experiments. The paper was written by S.T. and E.K. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies that involve human participants or animals.
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(PDF 2743 kb)
Rights and permissions
About this article
Cite this article
Tsumura, R., Sawada, K., Kunitake, E. et al. A component of the septation initiation network complex, AaSepM, is involved in multiple cellulose-responsive signaling pathways in Aspergillus aculeatus. Appl Microbiol Biotechnol 105, 1535–1546 (2021). https://doi.org/10.1007/s00253-021-11110-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11110-7