Abstract
This work aims to discuss the possibility that disordered CUL4B was involved in the pathogenesis of adjuvant-induced arthritis (AIA) in rats. Synovium and FLS from AIA rats both showed increased CUL4B and β-catenin, and up-regulated CUL4B enhanced the canonical Wnt signaling by targeting the GSK3β. Increased CUL4B promoted the FLS abnormal proliferation, activated the secretion of IL-1β and IL-8, and promoted the production of AIA pathology gene MMP3 and fibronectin. Furthermore, miR-101-3p was significantly down-regulated in AIA rats compared with controls, and transfection of AIA FLS with miR-101-3p mimics significantly down-regulated the CUL4B expression, whereas transfection with miR-101-3p inhibitors resulted in an opposite observation. The dual-luciferase reporter assay confirmed that the CUL4B was a direct target of miR-101-3p, and further analysis suggested that lowly expressed miR-101-3p contributed to disordered CUL4B activating the canonical Wnt signaling pathway and further promoting the development of AIA rats. Thus clarification of the CUL4B roles in the pathogenesis of AIA rats and corresponding mechanisms will contribute to the disease diagnosis and treatment for rheumatoid arthritis (RA) patients.
Key messages
-
CUL4B expression is up-regulated in synovium and FLS from AIA rats.
-
Increased CUL4B promotes the canonical Wnt signaling.
-
Increased CUL4B promotes the pathogenesis of AIA rats.
-
Decreased miR-101-3p contributes to disordered CUL4B.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cullin-really interesting new gene (RING) ubiquitin ligase (CRLs) is the largest E3 ubiquitin ligase family in eukaryotes, which is associated with cell cycle, signal transduction, DNA damage response, gene expression, chromatin remodeling, and embryonic development [1, 2]. In the eight cullins (CUL1–7 and PARC) of higher organisms, the CUL4 subfamily of CRLs contains two family members, the CUL4A and CUL4B, which have extensive sequence homology and functional redundancy [3, 4]. In recent years, a large number of studies have confirmed that CUL4A and CUL4B are closely related to cancer pathology [5]. However, the role of CUL4B in cancer pathogenesis is uncertain. Researchers have found that CRL4B interacted with polycomb repressive complex 2 (PRC2), and CRL4B could play a transcriptional inhibitory activity by promoting H2AK119 single ubiquitination or by interacting with PRC2 [6, 7]. Interestingly, removal of Cul4b or depletion of CRL4B, the main component of CUL4B, not only prevented H2AK119 single ubiquitination but also could inhibit H3K27 methylation [8]. In view of in vivo and in vitro experiments, the researchers suggested that CUL4B promoted cell proliferation, invasion, and tumor formation, and its expression was significantly up-regulated in several human cancers [9, 10]. These studies suggested that CUL4B has the important effect of promoting tumor formation. However, the expression of CUL4B in rheumatoid arthritis (RA) and its roles in RA pathological regulation are still unclear.
RA is a chronic and progressive systemic autoimmune disease that primarily affecting the synovial membrane, leading to bone and cartilage destruction, whereas it is frustrating that its pathology is still unknown [11]. RA occurs in 0.5–1.0% of the world population, and the prevalence can be as high as 5% depending on the age group being investigated [12]. Other systems may also be occasionally affected, and the progressive character of RA makes it a potentially disabling disease that affects the life quality and expectancy of RA patients [13]. In recent decades, there have been notable advances toward a deeper understanding of the pathogenesis of RA, and this deeper understanding has led to better diagnostic measures and therapeutic effects for RA patients [14]. For RA treatment, the initial phase of the disease, known as early RA, is a period of time in which the initiation of appropriate treatment results in marked clinical therapeutic effects, so early diagnosis and treatment can change the course of the disease [15]. Therefore, identification of new RA markers is particularly important for early detection and treatment of RA patients.
A large number of studies have shown that CUL4B promoted cell proliferation, invasion, and tumor formation, and its expression was significantly up-regulated in a variety of human cancers, suggesting that CUL4B has the effect of promoting tumor formation [16]. We found that CUL4B expression was significantly up-regulated in adjuvant-induced arthritis (AIA) rats, the RA model rats, compared with control, thus we assumed that CUL4B might have important regulatory effects in AIA pathogenesis.
AIA rats showed multiple peripheral arthritis, local joint swelling, and severe joint deformity, and the pathological changes are mainly proliferative synovitis, articular cartilage destruction, and bone erosion, accompanied by inflammatory cell infiltration. These clinical manifestations are similar to human RA and are the ideal model for screening and studying the treatment of RA drugs [17, 18]. In this work, AIA rats were used as model animals for study of the roles of CUL4B in RA pathogenesis. In view of the key role of FLS in the pathogenesis of RA, the joint synovial tissue was isolated from AIA rats, and tissue culture was used to culture FLS. The expression of CUL4B in synovium and FLS from AIA rats and controls were detected by real-time qPCR and western blotting. The roles of CUL4B in the canonical Wnt signaling and the pathology development of AIA rats, and its targets were investigated by real-time qPCR, western blotting, double-luciferase reporter gene, ELISA, and MTT. Furthermore, bioinformatics predicted that miR-101-3p, miR-144-3p, and miR-146-5p are upstream regulatory factors for CUL4B. In these miRNAs, we found that reduced miR-101-3p was related to the increased CUL4B in AIA rats, thus we investigated the differences in miR-101-3p expression in AIA rats and the regulated roles of this miRNA in AIA pathogenesis.
Materials and methods
Materials and reagents
Anti-Cullin 4B rabbit antibody and Anti-fibronectin mouse antibody were purchased from Abcam (Cambridge, UK). β-Catenin rabbit mAb, phospho-β-catenin rabbit mAb, c-Myc rabbit mAb, cyclin D1 rabbit mAb, and MMP3 rabbit mAb were purchased from Cell Signaling (Beverly, MA, USA). Mouse monoclonal antibody against β-actin was purchased from Santa Cruz Biotechnology (California, USA). MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide) and DMSO (dimethyl sulfoxide) were purchased from Sigma Inc. (St. Louis, MO, USA). MiScript® miRNA PCR Array kit and QuantiFast® SYBR® Green PCR kit were purchased from QIAGEN (Germany). MiR-101-3p mimics, inhibitors, and negative control sequences (NC-miRNA) were purchased from Shanghai GenePharma Co., Ltd., Shanghai, China. CUL4B, β-CATENIN, c-Myc, cyclin D1, MMP3, FIBRONECTIN, and β-ACTIN primers were produced by Shanghai Sangon Biological and Technological Company (Shanghai, China).
AIA rat preparation and FLS culture
Male SD rats were purchased from Experimental Animal Center of Anhui Medical University with a weight range of 160–180 g, and these experimental rats were randomly assigned into each test group. We prepared AIA rats by injecting complete Freund’s adjuvant at the bottom of the right foot of the rats, the rats in the model groups were injected with 0.1 mL complete Freund’s adjuvant, and 0.1 mL PBS was injected in the control group as control. Starting from injection with complete Freund’s adjuvant, the AIA rats were prepared for 28 days. The animal experiments in this study were carried out by the protocols approved by the Anhui Science and Technology University Animal Care and Use committee. After that, the rats in experimental groups were sacrificed, the synovial tissue was isolated, and the FLS were cultured by tissue mass method. FLS were cultured at 37 °C with 5% CO2 in cell culture flasks in high-glucose DMEM medium supplemented with 15% (v/v) heat-inactivated fetal bovine serum (FBS) (Hyclone, USA) and the penicillin–streptomycin solution (Beyotime, China). The experimental cells we used were the second generation to the fifth generation of primary FLS.
Transient transfection of miR-101-3p mimics and inhibitors
MiR-101-3p mimics, inhibitors, and negative control sequences (NC-miRNA) were purchased from Shanghai GenePharma Co., Ltd., China, and these small RNAs were transfected into cultured FLS by the Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. FLS used for cell transfection were cultured in DMEM with 15% FBS at a density of 0.5–1 × 105 cells/mL. After 6 h of transfection, the transfection reagent was discarded and cell culture medium was added, then transfected FLS were cultured for another 24 h for further analysis. MiR-101a-3p mimics sense 5′ UACAGUACUGUGAUAACUGAA 3′, antisense 5′ CAGUUAUCACAGU ACUGUAUU 3′. Mimics NC sense 5′ UUCUCCGAACGUGUCACGUTT 3′, antisense 5′ ACGUGACACG UUCGGAGAATT 3′.
Cell proliferation assay
The treated FLS of each group with a cell density of approximately 0.5–1 × 105/mL were seeded in 96-well plates for 24 h. Then the FLS were cultured with 20 μL MTT (5 mg/mL) (Sigma, USA) for another 4 h in a cell incubator. After incubation, the FLS with MTT reagent were resuspended in 150 μL DMSO (Sigma, USA) and the bed shaken at low-speed oscillation for 10 min to fully dissolve the formazan transformed from MTT. The absorbance was measured at the wavelength of 490 nm using a Thermomax microplate reader (bio-tek EL, USA); the detection value indirectly reflected the number of viable cells.
Real-time qPCR
For real-time qCPR, we prepared the total RNA using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions and carried out the reverse transcription using 1 μg of total RNA by the instruction provided by the RevertAid First Strand cDNA Synthesis Kit (Fermentas, USA). After that, real-time qPCR analyses for target genes were performed using the QuantiFast® SYBR® Green PCR kit (QIAGEN, Germany) with the following primers: CUL4B (forward, 5′-GCCCCTGGAATAGAGGATGGA-3′; reverse, 5′-TCGGTCTGTAGTGCTTGCTTGT-3′); β-CATENIN (forward, 5′-CTTAC GGCAATCAGGAAAGC-3′; reverse, 5′-ACAGAC AGCACCTTCAGCACT-3′); c-Myc (forward, 5′-ATTTCTATCACCAGCAACAGCA-3′; reverse, 5′-ATTTCTATCACCAGCAACAGCA-3′); cyclin D1 (forward, 5′-GCCCTCCGTTTCTTACTTCAA-3′; reverse, 5′-CTCTTCGC ACTTCT GCTCCTC-3′); MMP3 (forward, 5′-TGAT GAACG ATGGACAGATGA-3′; reverse, 5′-AGCATTGGCTGAGTGAAAGAG-3′); fibronectin (forward, 5′-GACACTATGCGGGTCA CTTG-3′; reverse, 5′-CCCAGG CAGGAGATTTGTTA-3′); β-ACTIN (forward, 5′-CCCATCTATGAGGGTTACGC-3′; reverse, 5′-TTTAATGTCACGCACGATTTC-3′). The amplification steps were based on the method provided by the kit, and the specific amplification parameters include 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s. The real-time qPCR for miR-101-3p expression analysis was followed by denaturation at 95 °C for 3 min, 95 °C for 12 s, and 62 °C for 40s for 40 cycles. The relative expression level of miR-101-3p was calculated using 2−ΔΔCt method and U6 as internal control. U6 F primer CAGCACATATACTAAAATTGGAACG, R primer ACGAATTTGCGTGTCATCC.
Immunoblot analysis
Synovium and FLS were isolated from AIA rats and control individuals and were lysed by the cell lysis buffer for Western and IP kit which is purchased from Beyotime Biological Technology Co., Ltd., China. Then the protein concentration was determined by the Enhanced BCA Protein Assay Kit (Beyotime, China) according to instructions provided by the manufacturer. After that, 20 mg total protein was separated by SDS-PAGE and blotted onto PVDF membranes (Millipore Corp., Billerica, MA, USA) for immunoblot analysis. Nitrocellulose blots were blocked and incubated for 6 h in the primary antibody dilution buffer (Beyotime, China) including primary antibodies, such as CUL4B, β-catenin, c-Myc, cyclin D1, and MMP3, and these primary antibodies were used at 1:500 or 1:800, respectively. Mouse monoclonal antibody β-actin was used at 1:800. The protein blots were detected using the ECL-chemiluminescent kit (ECL-plus; Thermo Scientific).
Dual-luciferase reporter assay
The dual-luciferase reporter plasmid used in the experiment was the GP-miRGLO plasmid. The CUL4B-3′UTR WT, MUT gene fragments were obtained by annealing, and the fragments were cloned into the plasmid GP-miRGLO. The primers were synthesized by Shanghai GenePharma Co., Ltd., and the upstream and downstream primers of the target gene were respectively added with the homologous sequences of both sides of SacI and XhoI on GP-miRGLO plasmid for subcloning. After the PCR reaction is completed, the target gene fragments were recovered by agarose electrophoresis. The plasmid GP-miRGLO was digested with SacI and XhoI at 37 °C for 2 h. The amplified fragments were cloned into the linearized GP-miRGLO plasmid using the ClonExpress® Entry One Step Cloning Kit. The experiment was set up in four groups: CUL4B 3′UTR WT + rno-miR-101a-3p, CUL4B 3′UTR WT + mimics NC, CUL4B 3′UTR MUT + miR-101a-3p, and CUL4B 3′UTR MUT + mimics NC. Then the culture medium in the cell culture plate was decanted and the cells were washed twice with PBS. PLB (1×) was added to the culture wells, the plate was gently shaken at room temperature for 15 min, and the lysis solution was transferred to a test plate. The Tecan M1000 microplate reader was opened, preheated, the dual-luciferase detection system selected, the LARII reagent added to each well, and the firefly luciferase activity detected on the microplate reader. The test plate was removed and the Stop & Glo® Reagent was added to each well, and the Renilla luciferase activity was detected on the microplate reader.
CUL4B expression vector construction
The CUL4B gene encoding CDS sequence was constructed on PEX-3 plasmid to obtain CUL4B overexpression vector. The upstream and downstream primers of the target gene were respectively added with the homologous sequences on both sides of SalI and BamHI of the pEX-3 plasmid for subcloning. Primers were synthesized by Shanghai GenePharma Co., Ltd. After the PCR reaction is completed, the CUL4B gene fragments were recovered by agarose electrophoresis. PEX-3 was digested with SalI and BamHI at 37 °C for 2 h. After electrophoresis, double-digested pEX-3 plasmids were recovered using a DNA gel recovery kit. The amplified fragments were recombinantly cloned into the linearized pEX-3 plasmids using the ClonExpress® Entry One Step Cloning Kit. Then the competent cells were prepared and the ligation products were transformed into competent cells. The colonies were picked from the plate, vectors pumped, and the positive clones identified. On a six-well cell culture plate, 250 μL of serum-free DMEM medium was added to a 1.5-mL EP tube and 2.5 μg of vector was added. Another 1.5-mL EP tube was taken, and 250 μL serum-free DMEM medium and 5 μL lipofectamine 2000 were added. After 5 min at room temperature, it was mixed and allowed to stand at room temperature for 20 min. The medium from the six-well plate was removed and the transfection mixture added to the six-well plate dropwise. After that, the mixture was incubated for 5 h in the incubator. The transfection solution was removed and 1 mL of DMEM containing 15% FBS was added, further incubated for 24 or 48 h at 37 °C, 5% CO2, and samples were used for further studies. PEX-3 empty vector served as a negative control for overexpression.
ELISA
We used the ELISA to determine the roles of CUL4B in the production of IL-1β and IL-8 in FLS from AIA rats. The quantification ELISA kit was purchased from Shanghai YuanYe Bio-Technology Co., Ltd., the detection procedure referred to the method provided by the kit, and the optical density values were read at 450 nm. Three replicate wells were quantified for every sample, and all experiments were performed in triplicate.
Statistical analysis
Statistical significance was determined by either the Student’s t test for comparison between means or one-way analysis of variance with a post hoc Dunnett’s test. Data are represented as mean ± SE. Significance was defined as p < 0.05 versus controls.
Results
CUL4B and β-catenin are significantly up-regulated in synovium and FLS of AIA rats
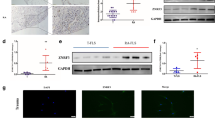
We firstly detected the levels of CUL4B and β-catenin in synovium and FLS from AIA rats and controls. SD rats were randomly assigned to each group: day 0, day 7, day 14, day 21, and day 28. After these rats were treated with complete Freund’s adjuvant, rats in each group were sacrificed and their synovium were isolated on day 0, day 7, day 14, day 21, and day 28, respectively. The expression of CUL4B in each group of AIA rats and control rats were detected by real-time qPCR, and results showed that the expression of CUL4B mRNA was significantly up-regulated starting from the 14th day of model preparation (Fig. 1a). The cells we used are the primary cultured FLS of the second generation to fifth generation, and we found that CUL4B mRNA was significantly up-regulated in all fourth-generation FLS (Fig. 1a). We determined the levels of β-catenin, the key member of canonical Wnt signaling, in synovium and FLS from AIA rats and controls, and found that the levels of β-catenin in synovium and FLS from AIA rats were significantly higher than that in normal rats (Fig. 1b). Furthermore, we used western blotting to determine the protein expression of CUL4B and β-catenin in AIA rats and controls, and found that the protein levels of CUL4B and β-catenin were significantly higher in synovium (Fig. 1c) and FLS (Fig. 1d) of AIA rats than that in normal rats. These observations indicate that the abnormal up-regulated CUL4B and β-catenin may contribute to the pathogenesis of AIA rats.
CUL4B and β-catenin are both significantly up-regulated during the development of AIA rats. The expression of CUL4B and β-catenin in AIA rats and normal rats was detected by real-time qPCR, western blotting. Data showed that the mRNA expression of the CUL4B (a) and β-catenin (b) in synovium and FLS from AIA rats was significantly up-regulated compared with normal control. Western blotting assays confirmed that the CUL4B and β-catenin protein both up-regulated in synovium (c) and FLS (d) from AIA rats compared with normal control. *p < 0.05 and #p < 0.05 versus respective normal group. Values are the mean ± SD of three different experiments
CUL4B enhances the canonical Wnt signaling in AIA rats
In view of the above findings, we next investigated the role of CUL4B in the β-catenin mRNA and the total β-catenin protein in AIA FLS by real-time qPCR and western blotting. Data suggested that CUL4B knockdown used in the CUL4B siRNA significantly suppressed the expression of β-catenin mRNA and total β-catenin protein (Fig. 1a) and that overexpression of CUL4B could promote the expression of the β-catenin mRNA and total β-catenin protein (Fig. 2a). Moreover, the role of CUL4B in the nuclear β-catenin protein in AIA FLS was detected by western blotting, and the results showed that down-regulated CUL4B significantly reduced the nuclear β-catenin protein production and overexpression of CUL4B significantly promoted the expression of nuclear β-catenin protein (Fig. 2b). c-Myc and cyclin D1 are important downstream genes of the canonical Wnt signaling, and the roles of CUL4B in the expression of c-Myc and cyclin D1 were detected by real-time qPCR and western blotting. Data suggested that CUL4B knockdown significantly suppressed the mRNA expression of c-Myc and cyclin D1 and CUL4B overexpression up-regulated their production (Fig. 2c). Western blotting showed a similar regulatory role of CUL4B in protein expression of c-Myc and cyclin D1 (Fig. 2d). Altogether, these observations indicate that CUL4B may display as a positive regulator of the canonical Wnt signaling activity.
CUL4B enhances the canonical Wnt signaling in AIA FLS. The roles of CUL4B in the canonical Wnt signaling in AIA FLS were determined by real-time qPCR and western blotting, respectively. The results suggested that overexpression of CUL4B significantly promoted the expression of β-catenin mRNA and CUL4B knockdown inhibited the expression of β-catenin mRNA and total protein in AIA FLS (a). Western blotting assays confirmed the regulatory role of CUL4B in β-catenin protein in the nucleus of AIA FLS (b). Real-time qPCR (c) and western blotting (d) results showed that CUL4B knockdown inhibited and overexpression of CUL4B promoted the expression of c-Myc and cyclin D1, two members of the canonical Wnt signaling pathway. *p < 0.05 versus SiControl, #p < 0.05 versus vector control. Values are the mean ± SD of three different experiments
CUL4B activates the canonical Wnt signaling via the GSK3β inhibition in AIA rats
GSK3β is an important regulator for β-catenin ubiquitination degradation in the canonical Wnt signaling, and the GSK3β expression in AIA FLS and the effect of GSK3β siRNA on the GSK3β expression were determined by real-time qPCR. The results of real-time qPCR showed that GSK3β was significantly up-regulated in AIA FLS compared with control and SiGSK3β interference significantly down-regulated the production of GSK3β (Fig. 3a). Western blotting assays suggested that SiGSK3β transfection for 12, 24, and 36 h all could down-regulated the GSK3β production (Fig. 3b). The roles of SiGSK3β in the regulation β-catenin and the phospho-β-catenin in RA model FLS were detected by real-time qPCR and western blotting. Consistent with the classical Wnt signaling mechanism, data suggested that treatment with SiGSK3β up-regulated the β-catenin expression but reduced the phospho-β-catenin expression. The GSK3β inhibitors LiCl, CHIR99021, and SB216763 were used to investigate the role of GSK3β in the regulation of β-catenin in AIA FLS, and data indicated that GSK3β inhibition significantly promoted the expression of β-catenin (Fig. 3c). Furthermore, the GSK3β inhibitors were used to investigate the role of GSK3β inhibition in the process that CUL4B knockdown will suppress the β-catenin expression by targeting the up-regulated GSK3β in SiCUL4B-treated AIA FLS, and data suggested that the GSK3β inhibition significantly restored the inhibited β-catenin in SiCUL4B-treated AIA FLS (Fig. 3d, e). In SiCUL4B-treated AIA FLS, we also found the effect of GSK3β knockdown on the c-Myc and cyclin D1 expression (Fig. 3f). These findings confirm that the CUL4B activates the canonical Wnt signaling via the GSK3β inhibition in AIA rats.
CUL4B activates the canonical Wnt signaling via the GSK inhibition in RA model rats. Real-time qPCR results showed that the GSK3β expression in AIA FLS was significantly higher than that in untreated FLS, and that GSK3β knockdown reduced its expression in AIA FLS (a). Western blotting assays confirmed the effect of SiGSK3β on the GSK3β expression (b). Real-time qPCR and western blotting results suggested that GSK3β knockdown significantly promoted the β-catenin expression and inhibited the β-catenin phosphorylation degradation in AIA FLS (c). The GSK3β inhibitors LiCl, SB216763, or CHIR99021 up-regulated the β-catenin expression in AIA FLS (c). Real-time qPCR (d) and western blotting (e) results found that the GSK3β inhibitors LiCl, SB216763, or CHIR99021 significantly up-regulated the β-catenin expression in SiCUL4B-treated AIA FLS. The effect of SiGSK3β on the c-Myc and cyclin D1 expression in SiCUL4B-treated AIA FLS was detected by real-time qPCR, and data suggested that SiGSK3β significantly promoted the expression of c-Myc and cyclin D1, indicating that GSK3β mediated the role of CUL4B in the canonical Wnt signaling in AIA FLS (f). *p < 0.05 versus normal FLS or SiControl or NaCl group or DMSO group or the group of SiCUL4B + SiControl in AIA FLS. Values are the mean ± SD of three different experiments
CUL4B promotes the pathology development of AIA rats
Compared with the control, CUL4B expression was significantly up-regulated in the synovium and FLS from AIA rats, and increased CUL4B was related to the disordered β-catenin, suggesting that CUL4B may participate in the pathology of AIA rats. We firstly investigated the effect of CUL4B on the FLS growth using the cell count method. Data suggested that the FLS proliferation multiplication of SiCUL4B group is lower than that in SiControl group in AIA FLS, while the CUL4B overexpression accelerated the FLS proliferation compared with control (Fig. 4a). MTT assays of CUL4B-knockdown or CUL4B overexpression in AIA FLS confirmed these findings (Fig. 4b). IL-1β and IL-8 are involved in the pathogenesis of RA, resulting in inflamed synovium, increased angiogenesis, and decreased lymphangiogensis during the disease development [19]. We next determined the effect of CUL4B on the production of IL-1β and IL-8 in AIA FLS using ELISA. After transfection of AIA FLS with SiCUL4B for 12 h, the IL-1β and IL-8 were both significantly down-regulated. However, transfection with CUL4B vectors for 24 h enhanced the expression of IL-1β and IL-8 (Fig. 4c). As far as we know, MMP3 and fibronectin are RA pathology-related genes, and the abnormal increased expression of two genes is accompanied by the development of RA pathology. The test results show that CUL4B knockdown by SiCUL4B significantly suppressed the expression of MMP3 and fibronectin (Fig. 4d), and that CUL4B overexpression promoted their production in AIA FLS (Fig. 4e). These results suggest that CUL4B may display a positive regulator during development of AIA rats and may participate in its pathogenesis.
CUL4B promotes the pathology development of AIA rats. The effect of CUL4B on the FLS growth was evaluated using the cell count method in AIA FLS, and data showed that CUL4B knockdown inhibited and CUL4B overexpression promoted the FLS growth (a). MTT assays confirmed the effect of CUL4B on the FLS proliferation in AIA FLS (b). ELISA assays suggested that CUL4B knockdown inhibited and CUL4B overexpression promoted the expression of IL-1β and IL-8 (c). Real-time qPCR and western blotting results confirmed that CUL4B knockdown significantly inhibited the expression of MMP3 and fibronectin (d). However, CUL4B overexpression promoted their expression in AIA FLS (e). *p < 0.05 versus SiControl, #p < 0.05 versus vector control. Values are the mean ± SD of three different experiments
Reduced miR-101-3p is related to the increased CUL4B in AIA rats
Bioinformatics predicted that miR-101-3p may be a regulatory factor for disordered CUL4B in rats, thus we investigated the possible role of miR-101-3p in CUL4B-induced AIA pathology. After analysis of real-time qPCR, we found that miR-101-3p expression was significantly down-regulated both in synovium and FLS of AIA rats compared with normal individuals (Fig. 5a). Importantly, transfection of AIA FLS with miR-101-3p mimics significantly down-regulated the CUL4B expression, and transfection with miR-101-3p inhibitors significantly up-regulated the production of CUL4B in AIA FLS (Fig. 5b, c). The influence of miR-101-3p on CUL4B activity was also determined by a well-established dual-luciferase reporter assay, the TOP/FOP Flash assay. The TOP Flash reporter contains CUL4B 3′UTR, whereas the FOP Flash reporter was used as a negative control. As shown in Fig. 5d, increased miR-101-3p significantly down-regulated the TOP Flash luciferase activity (Fig. 5d).
Reduced miR-101-3p is related to the increased CUL4B in AIA rats. Real-time qPCR assays showed that the miR-101-3p expression was significantly down-regulated in both synovium and FLS from AIA rats compared with control (a). The miR-101-3p overexpression inhibited and miR-101-3p knockdown promoted the expression of CUL4B mRNA (b) and protein (c) in AIA FLS. d The dual-luciferase reporter assay was used to observe the influence of miR-101-3p on CUL4B, and data suggested that miR-101-3p directly regulated the expression of CUL4B, and CUL4B may be a direct target of miR-101-3p. *p < 0.05, #p < 0.05 versus normal group, or control FLS, or miR-101-3p mimic/inhibitor control, or the NC miRNA group. Values are the mean ± SD of three different experiments
Compared with control, the expression of miR-144-3p and miR-146-5p in AIA rats was not significantly different. Taken together, these results indicate that miR-101-3p is a negative regulator for increased CUL4B in AIA rats, and miR-101-3p may participate in the pathology of AIA through the disordered CUL4B.
The CUL4B influences the regulation of β-catenin by miR-101-3p
According to the previous study, the highly expressed CUL4B promoted the expression of β-catenin through GSK3β, while overexpression of miR-101-3p inhibited CUL4B expression, so we hypothesized that miR-101-3p might affect the canonical Wnt signaling in AIA rats. In this part of the study, the effect of miR-101-3p on β-catenin was investigated by real-time qPCR, and results showed that overexpression of miR-101-3p significantly inhibited the β-catenin expression, and decreased miR-101-3p resulted in a significant up-regulation of β-catenin (Fig. 6a). Western blotting assay confirmed this control relationship (Fig. 6b). Furthermore, the effect of miR-101-3p on c-Myc and cyclin D1 was investigated by real-time qPCR, and results suggested that miR-101-3p negative regulated the expression of c-Myc and cyclin D1 in AIA FLS (Fig. 6c). These findings indicated that miR-101-3p might be a regulator of the canonical Wnt signaling in AIA pathogenesis. In addition, the role of CUL4B in the regulation of β-catenin by miR-101-3p was detected by real-time qPCR and western blotting. In model FLS transfected with miR-101-3p mimics, CUL4B overexpression strongly promoted the expression of β-catenin, whereas SiCUL4B significantly suppressed the β-catenin production in AIA FLS transfected with miR-101-3p inhibitors (Fig. 6d, e). These findings suggest that miR-101-3p influences the canonical Wnt signaling through the CUL4B.
MiR-101-3p affects the canonical Wnt signaling by targeting the CUL4B in AIA rats. Real-time qPCR showed that miR-101-3p mimics significantly inhibited and miR-101-3p inhibitors promoted the expression β-catenin in AIA FLS (a), and the western blotting assays confirmed these regulatory effects (b). Similar to the effect on the β-catenin, further analysis showed that miR-101-3p has negative regulatory roles in the expression of c-Myc and cyclin D1 in AIA FLS (c). In AIA FLS transfected with miR-101-3p mimics or inhibitors, the CUL4B vectors or CUL4B siRNAs were added to investigate the role of CUL4B in the expression of β-catenin regulated by miR-101-3p. Real-time qPCR (d) and western blotting (e) assays showed that CUL4B mediated the β-catenin expression regulated by miR-101-3p, indicating that miR-101-3p affected the canonical Wnt signaling by targeting the CUL4B. *p < 0.05 versus their respective control groups, #p < 0.05 versus miR-101-3p mimic or inhibitor transfection, ##p < 0.05 versus miR-101-3p mimics + vector control or miR-101-3p inhibitors + SiCUL4B control. Values are the mean ± SD of three different experiments
Discussion
RA is a chronic, systemic, autoimmune disease with unknown etiology, and the disease is mainly characterized by multi-joint, symmetry, and invasive arthritis, often accompanied by extra-articular organ involvement and positive serum rheumatoid factors. The incidence of RA may be related to genetics, infection, sex hormones, and so on, and a variety of pathogenic factors cause the synovial lining cell proliferation, interstitial inflammatory cell infiltration, microvascular neonatal, pannus formation, and cartilage and bone destruction [20]. The pathogenesis of this disease is very complex and is still not fully elucidated [21]. Although much knowledge has been gained with respect to the regulation of RA pathogenesis and prognosis, the precise molecular mechanisms orchestrating and guiding persistent synovial hyperplasia, cartilage erosion, and bone damage during the RA development have not been fully elucidated. However, much attention has been focused on the RA FLS in this regard. For example, the abnormal proliferation of FLS produces more chemokines and cytokines such as stromal cell derived factor 1 (SDF-1), IL-1β, IL-8, and IL-15, which can promote infiltration and activation of lymphocytes in the synovial tissue [22]. In addition, FLS foster the differentiation of B lymphocytes to plasma cells, and also make matrix metalloproteinases such as pro-MMP3, which in its mature form enhances cartilage degradation [23]. In view of the key role of FLS in RA pathology, our group used FLS as the target cells to study the important role of CUL4B in the pathogenesis of AIA in this work.
CUL4B is a member of the cullin4 family, and the CUL4B is highly sequence homologous with CUL4A with about sharing 83% sequence identity, whereas more and more evidence indicates that there are almost completely different roles and mechanisms between the two molecules [24]. For example, CUL4A and CUL4B both can interact with the substrate adaptor DDB21, and they may target the same substrate and function redundantly in genome integrity maintenance, DNA replication, and cell cycle regulation. However, CUL4B can target the WD repeat containing protein5 (WDR5) and peroxiredoxin III (PrxIII), but these are not targeted by CUL4A [25]. In addition, CUL4A is highly expressed in the pancreas, testes, and T cells, while CUL4B is most highly expressed in endocrine glands, pancreatic tissue, the cerebellum, bone marrow, and the testes [26].
Evidence has shown that the expression of CUL4B is abnormal in a wide variety of human diseases and physiological process [27]. The function of CUL4B in solid tumors has been gradually uncovered and attracts a lot of interest, and these studies have demonstrated clearly that CUL4B acts pivotal roles in cell abnormal proliferation, invasion, DNA damage and repair, DNA methylation, and histone acetylation modification, also including the regulation of related signaling pathways [28].
In view of the important roles of CUL4B in human diseases, we hypothesize that CUL4B may play an important regulatory role in RA pathogenesis. In order to reveal the fact that CUL4B is involved in the RA pathogenesis, we used the AA rats as the RA model rats and firstly detected the expression levels of the CUL4B in synovium and FLS which were isolated from AA rats and normal rats. It was encouraging that the expression of CUL4B mRNA was significantly up-regulated starting from the 14th day after the model was prepared. Many literatures reported that CUL4B could regulate the β-catenin expression, a key member of the canonical Wnt signaling pathway, affecting the canonical Wnt signaling, thus we determined the levels of β-catenin in synovium and FLS from AA rats and controls, and found that the β-catenin expression in AIA model rats were significantly higher than that in normal rats. These data indicate that there may be a certain regulated relationship between CUL4B and β-catenin in the AIA pathogenesis.
In this work, data showed that abnormal CUL4B positive regulated the production of β-catenin in AIA FLS, leading to activation of the canonical Wnt signaling. GSK3β is an important regulator for β-catenin ubiquitination degradation in the canonical Wnt signaling pathway, and disordered GSK3β affects the activation of the Wnt signaling [29]. In this work, we suggested that GSK3β mediated the positive regulated role of CUL4B in β-catenin since the GSK3β siRNA and the GSK3β inhibitors LiCl, CHIR99021, or SB216763 all significantly restored the inhibited β-catenin in SiCUL4B-treated AIA FLS. These findings revealed the mechanism that the CUL4B activated the canonical Wnt signaling in the pathogenesis of AIA rats. Results also suggested that the FLS proliferation multiplication of SiCUL4B group is lower than that in SiControl group in AIA FLS, whereas the CUL4B overexpression accelerated the FLS proliferation compared with control. Increased CUL4B enhanced the expression of IL-1β, IL-8, and RA pathology-related gene MMP3 and fibronectin, suggesting that CUL4B might display a positive regulator for the pathogenesis of AIA.
MicroRNAs are a class of endogenous, small RNAs of about 20–24 nucleotides in length that have a number of important regulatory effects in pathophysiological mechanisms [30, 31]. It is speculated that miRNA regulates one third of the human gene [32]. Our group’s interest is to investigate the relationship between miRNAs and the pathogenesis of AIA, thus we particularly want to know whether the abnormal expression of CUL4B is related to specific miRNA in AIA rats. Bioinformatics predicted that miR-101-3p, miR-144-3p, and miR-146-5p are upstream regulatory factors for CUL4B, thus we investigated the expression of these miRNA in AIA rats and control, as well as their regulated roles in AIA rats. In these miRNAs, miR-101-3p expression was significantly down-regulated in both synovium and FLS from AIA rats compared with normal rats. Transfection of miR-101-3p mimics significantly down-regulated the CUL4B expression, miR-101-3p inhibitors significantly up-regulated the expression of CUL4B in AIA FLS, and miR-101-3p was confirmed as a direct negative regulator of CUL4B in the pathogenesis of AIA.
Furthermore, data analysis showed that overexpression of miR-101-3p significantly inhibited the β-catenin expression, and decreased miR-101-3p resulted in a significant up-regulation of β-catenin, leading to an activation of β-catenin downstream signaling. In AIA FLS transfected with miR-101-3p mimics, CUL4B overexpression strongly promoted the expression of β-catenin, whereas SiCUL4B significantly suppressed the β-catenin production in AIA FLS transfected with miR-101-3p inhibitors. These findings suggest that miR-101-3p influences the canonical Wnt signaling through the CUL4B and that miR-101-3p may be a potential diagnostic and therapeutic strategy targeting the CUL4B/GSK3β/β-catenin axis.
As far as we know, miRNAs play an important role in the process of cell differentiation, biological development and disease development, and more and more researchers concern this research area [33]. With further research into the mechanisms of RA and miRNAs, the combination of miRNA and RA will lead to a new level of understanding of the pathology of RA regulation. This will also make miRNAs as new biological markers for the diagnosis of RA disease and may also make these molecules as drug targets, or to mimic these molecules for new drug development, which may provide a new treatment for human RA.
Acknowledgements
This project was supported by the National Science Foundation of China (no. 81302783).
Abbreviations
- RA:
-
rheumatoid arthritis
- CUL4B:
-
Cullin 4B
- AIA:
-
adjuvant-induced arthritis
- RING:
-
really interesting new gene
- CRLs:
-
Cullin-RING ubiquitin ligase
- PRC2:
-
polycomb repressive complex 2
- MTT:
-
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide
- DMSO:
-
dimethyl sulfoxide
- FBS:
-
fetal bovine serum
- WDR5:
-
WD repeat containing protein5
- PrxIII:
-
peroxiredoxin III
- SDF-1:
-
stromal cell derived factor 1
References
Vriend J, Reiter RJ (2016) Elatonin, bone regulation and the ubiquitin–proteasome connection: a review. Life Sci 145:152–160
Vittal V, Stewart MD, Brzovic PS, Klevit RE (2015) Regulating the regulators: recent revelations in the control of E3 ubiquitin ligases. J Biol Chem 290:21244–21251
Hadchouel J, Ellison DH, Gamba G (2016) Regulation of renal electrolyte transport by WNK and SPAK-OSR1 kinases. Annu Rev Physiol 78:367–389
Lampert F, Brodersen MM, Peter M (2017) Guard the guardian: a CRL4 ligase stands watch over histone production. Nucleus 8:134–143
Antonioli M, Di Rienzo M, Piacentini M, Fimia GM (2017) Emerging mechanisms in initiating and terminating autophagy. Trends Biochem Sci 42:28–41
Wang X, Chen Z (2016) Knockdown of CUL4B suppresses the proliferation and invasion in non-small cell lung cancer cells. Oncol Res 24:271–277
Hu H, Yang Y, Ji Q, Zhao W, Jiang B, Liu R, Yuan J, Liu Q, Li X, Zou Y, Shao C, Shang Y, Wang Y, Gong Y (2012) CRL4B catalyzes H2AK119 monoubiquitination and coordinates with PRC2 to promote tumorigenesis. Cancer Cell 22:781–795
Mi J, Zou Y, Lin X, Lu J, Liu X, Zhao H, Ye X, Hu H, Jiang B, Han B, Shao C, Gong Y (2017) Dysregulation of the miR-194-CUL4B negative feedback loop drives tumorigenesis in non-small-cell lung carcinoma. Mol Oncol 11:305–319
Qu Z, Li D, Xu H, Zhang R, Li B, Sun C, Dong W, Zhang Y (2016) CUL4B, NEDD4, and UGT1As involve in the TGF-β signalling in hepatocellular carcinoma. Ann Hepatol 15:568–576
Chen P, Yao GD (2016) The role of cullin proteins in gastric cancer. Tumour Biol 37:29–37
Bluett J, Barton A (2017) Precision medicine in rheumatoid arthritis. Rheum Dis Clin N Am 43:377–387
van Drongelen V, Holoshitz J (2017) Human leukocyte antigen–disease associations in rheumatoid arthritis. Rheum Dis Clin N Am 43:363–376
Knevel R, Huizinga TWJ, Kurreeman F (2017) Genomic influences on susceptibility and severity of rheumatoid arthritis. Rheum Dis Clin N Am 43:347–361
Sapir-Koren R, Livshits G (2017) Postmenopausal osteoporosis in rheumatoid arthritis: the estrogen deficiency-immune mechanisms link. Bone 103:102–115
Laufer VA, Chen JY, Langefeld CD, Bridges SL Jr (2017) Integrative approaches to understanding the pathogenic role of genetic variation in rheumatic diseases. Rheum Dis Clin N Am 43:449–466
Hannah J, Zhou P (2015) Distinct and overlapping functions of the cullin E3 ligase scaffolding proteins CUL4A and CUL4B. Gene 573:33–45
Abd El-Rahman RS, Suddek GM, Gameil NM, El-Kashef HA (2011) Protective potential of MMR vaccine against complete Freund’s adjuvant-induced inflammation in rats. Inflammopharmacology 19:343–348
Mbiantcha M, Almas J, Shabana SU, Nida D, Aisha F (2017) Anti-arthritic property of crude extracts of Piptadeniastrum africanum (Mimosaceae) in complete Freund’s adjuvant-induced arthritis in rats. BMC Complement Altern Med 17:111–127
Alam J, Jantan I, Bukhari SNA (2017) Rheumatoid arthritis: recent advances on its etiology, role of cytokines and pharmacotherapy. Biomed Pharmacother 92:615–633
Yuan FL, Li X, Xu RS, Jiang DL, Zhou XG (2014) DNA methylation: roles in rheumatoid arthritis. Cell Biochem Biophys 70:77–82
Angelotti F, Parma A, Cafaro G, Capecchi R, Alunno A, Puxeddu I (2017) One year in review 2017: pathogenesis of rheumatoid arthritis. Clin Exp Rheumatol 35:368–378
Kim KW, Cho ML, Kim HR, Ju JH, Park MK, Oh HJ, Kim JS, Park SH, Lee SH, Kim HY (2007) Up-regulation of stromal cell-derived factor 1 (CXCL12) production in rheumatoid synovial fibroblasts through interactions with T lymphocytes: role of interleukin-17 and CD40L–CD40 interaction. Arthritis Rheum 56:1076–1086
Li Y, Wang LM, Xu JZ, Tian K, Gu CX, Li ZF (2017) Gastrodia elata attenuates inflammatory response by inhibiting the NF-κB pathway in rheumatoid arthritis fibroblast-like synoviocytes. Biomed Pharmacother 85:177–181
Sertic S, Evolvi C, Tumini E, Plevani P, Muzi-Falconi M, Rotondo G (2013) Non-canonical CRL4A/4B(CDT2) interacts with RAD18 to modulate post replication repair and cell survival. PLoS One 8:e60000
Li X, Lu D, He F, Zhou H, Liu Q, Wang Y, Shao C, Gong Y (2011) Cullin 4B protein ubiquitin ligase targets peroxiredoxin III for degradation. J Biol Chem 286:32344–32354
Sharifi HJ, Furuya AK, Jellinger RM, Nekorchuk MD, de Noronha CM (2014) Cullin4A and cullin4B are interchangeable for HIV Vpr and Vpx action through the CRL4 ubiquitin ligase complex. J Virol 88:6944–6958
Sarikas A, Hartmann T, Pan ZQ (2011) The cullin protein family. Genome Biol 12:220–228
Shen E, Shulha H, Weng Z, Akbarian S (2014) Regulation of histone H3K4 methylation in brain development and disease. Philos Trans R Soc Lond Ser B Biol Sci 369:1652
Daud M, Rana MA, Husnain T, Ijaz B (2017) Modulation of Wnt signaling pathway by hepatitis B virus. Arch Virol doi 162:2937–2947
Kaneko H, Terasaki H (2017) Biological involvement of MicroRNAs in proliferative vitreoretinopathy. Transl Vis Sci Technol 6:5–13
Rajman M, Schratt G (2017) MicroRNAs in neural development: from master regulators to fine-tuners. Development 144:2310–2322
Awan HM, Shah A, Rashid F, Shan G (2017) Primate-specific long non-coding RNAs and microRNAs. Genomics Proteomics Bioinformatics 15:187–195
Yan Y, Wang R, Guan W, Qiao M, Wang L (2017) Roles of microRNAs in cancer associated fibroblasts of gastric cancer. Pathol Res Pract 213:730–736
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest statement
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Miao, C., Chang, J., Zhang, G. et al. CUL4B promotes the pathology of adjuvant-induced arthritis in rats through the canonical Wnt signaling. J Mol Med 96, 495–511 (2018). https://doi.org/10.1007/s00109-018-1635-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-018-1635-8