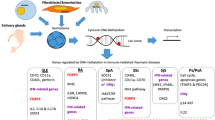
Abstract
Rheumatoid arthritis (RA) is an immune-mediated disease of unknown cause that primarily affects the joints and ultimately leads to joint destruction. In recent years, the potential role of DNA methylation in the development of RA is raising great expectations among clinicians and researchers. DNA methylation influences diverse aspects of the disease and regulates epigenetic silencing of genes and behavior of several cell types, especially fibroblast-like synoviocytes (FLS), the most resident cells in joints. The activation of FLS is generally regarded as a key process in the development of RA that actively results in the promotion of ongoing inflammation and joint damage. It has also been shown that aberrant DNA methylation occurs in the pathogenesis of RA and contributes to the development of the disease. Recently, there has been an impressive increase in studies involving DNA methylation in RA. In this paper, we consider the role of DNA methylation in the development of RA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rheumatoid arthritis (RA) is a chronic systemic inflammatory disease that is characterized by the inflammatory proliferation of synovial tissues, leading to damage of cartilage and bone, and eventual disability [1, 2]. Both genetic and environmental factors, as well as an aberrant immune response, play a key role in the pathogenesis of RA [3]. The main pathologic feature of RA is the large amount of pro-inflammatory cytokines secreted by activated B cells and T cells, as well as synovial hyperplasia [4]. Other cell populations, including T helper 17 cell (Th17) [5], monocytes [6], mast cells [7], macrophages [6], and epithelial cells [8], result in destruction of cartilage and bone. Characteristically, peripheral joints are involved in a symmetric distribution. Disease is two-to-three times more common in women than in men, with a peak symptom onset between 30 and 55 years of age [9]. RA affects between 0.5 and 1 % of the population worldwide [10], representing a crucial cause of musculoskeletal dysfunction.
In recent years, knowledge regarding the pathogenic mechanisms underlying RA has significantly improved. It has been appreciated that fibroblast-like synoviocytes (FLS) play a key role in the pathogenesis of RA [4, 11, 12]. Activation of FLS in RA leads to the production of a wide range of inflammatory mediators that promote the recruitment, retention, and activation of cells in the affected joints and the immune system, thus, causing the promotion of an ongoing inflammatory response and tissue damage. Interestingly, RA FLS exhibit a unique aggressive phenotype that can increase invasiveness into the extracellular matrix (ECM) and further exacerbates joint damage [4, 13]. Recently, Karouzakis et al. [14] have revealed that epigenetic determinants, such as DNA methylation, might contribute to these phenotypic changes in RA FLS.
DNA methylation is a novel area of research in RA pathogenesis with the potential to answer unsolved problems. Analysis of DNA methylation in T cells derived from patients with RA has shown global hypomethylation [14]. DNA hypomethylation has also been shown in RA FLS and RA tissues, as well as in peripheral blood mononuclear cells (PBMCs) derived from RA patients [15–17]. Demethylation of the IL-6 and IL-10 promoter in PBMCs leads to their mRNAs overexpression, suggesting that changes in RA DNA methylation potentially contribute to disease pathogenesis [17–20]. Different DNA methyltransferases (DNMTs) linked to the DNA methylation process in RA have been described [21, 22]. Here, we attempt to piece together data based on aberrant DNA methylations in FLS and PBMCs from RA patients and highlight the epigenetic etiology and molecular basis of RA.
DNA Methylation
Epigenetics is defined as heritable given patterns of gene expression that occurs without changes in DNA sequences. Thus, the epigenetic process is crucial for controlling gene expression patterns during the cell cycle, development or differentiation, and in response to biological or environmental changes [23]. The predominant mechanisms of epigenetic changes include DNA methylation and histone modifications [24].
In mammalian genomes, DNA methylation, occurring almost exclusively at cytosine residues within CpG dinucleotides, involves the migration of a methyl group at the 5th carbon in the pyrimidine ring and the formation of 5-methylcytosine (5-mC) [25]. The majority of CpG dinucleotides in the human genome are methylated, while a variety of the non-methylated CpG dinucleotides is found at the higher concentration of CpG sites (CpG islands) and associated with the promoter regions or 5′-end of genes (Fig. 1). Methylation of CpG islands in the promoter region correlates with gene inactivation, whereas unmethylation of this region can induce transcriptional activation [26].
Schematic diagram of DNA methylation mediating gene silencing. a Transcriptional silencing of gene promoters via DNA methylation. If a promoter region is methylated, the gene cannot be expressed. b In genomic DNA, a methyl group may be added to cytosine to form 5-methylcytosine. Cytosine residues in DNA at CpG sites are added to cytosine to form 5-methylcytosine in the cytosine pyridine ring. This reaction is catalyzed by the enzymatic activity of DNMTs. The reversible DNA methylation reaction is catalyzed by DNMT inhibitors
DNA methylation in mammalian cells is catalyzed by several DNMTs, which transfer a methyl group from S-adenosylmethionine to cytosine [27, 28]. There are at least four independent methyltransferases, including DNMT1, DNMT3a, DNMT3b, and DNMT3L, which are involved in the process of DNA methylation related to inhibition of gene transcription [25, 29]. DNMT1 is a critical enzyme which is mainly responsible for the maintenance of existing methylation patterns during DNA replication. DNMT3a and DNMT3b are involved in de novo methylation and also participate in the maintenance of methylation activity. DNMT3L modulates the DNA methylation activity of DNMT3a and DNMT3b. Moreover, the methyl-CpG-binding domain (MBD) proteins, including MBD1, MeCP2, MBD3, and MBD4, play an important role in the mechanism of DNA methylation and interact with histone deacetylases leading to local changes in chromatin structure [30].
A change in the methylation pattern has been reported to influence the development of autoimmune diseases [31], aging [32], and cancers [33]. For example, hypomethylation of proto-oncogenes has been confirmed, particularly in liver tumors, colorectal cancer, and leukemia, and this may increase malignant transformation through promotion of proto-oncogene expression [34]. In addition to the tumor findings, a possible important role of abnormal methylation changes has been shown in the development of autoimmune diseases, including RA [35].
Aberrant DNA Methylation in RA
Methylation of CpG dinucleotide clusters, which is mediated by DNMTs, is the best-characterized mechanism of epigenetic modification. DNA methylation modulates gene transcription through two basic models: DNA methylation represses transcription by direct disruption of interactions between transcription factors and cognate DNA sequences [36]; and MBP proteins specifically recognize methylated DNA and recruit co-repressors to silence gene expression [37]. In cancer cells, hypomethylation results in chromosomal instability, and the expression of proto-oncogenes or hypermethylation affects silencing of tumor suppressor genes [38]. Interestingly, many chronic diseases, including RA, are now known to exhibit similar changes in the methylation status of promoter regions of genes that modulate survival and activation of cells that contribute to the pathogenesis of RA [39].
Global DNA Methylation in RA
Recent studies have highlighted the possible important role of DNA methylation in the pathogenesis of RA. Hypomethylation of DNA in T cells from RA patients was first introduced by Richardson et al. [40]. By studying DNA methylation in T cells, Richardson et al. [40] identified that the global genomic hypomethylation in T cells from patients with RA was less than that in T cells from healthy controls. Supporting these findings, Corvetta et al. [41] have reported low DNA methylcytosine levels in PBMCs, synovial mononuclear cells, and synovial in RA. The same phenomenon of DNA hypomethylation in the pathogenesis of RA was also discussed by Neidhart et al. [15]. The authors found the presence of the retrotransposable element long interspersed nuclear element-1 (LINE-1) in RA synovial fluid pellets and in RA synovial tissue at the site of inflammation, suggesting a decreased methylation level in RA. Retrotransposons not only can integrate into host genomes but also affect gene expression, resulting in an impaired cellular phenotype. In addition, the expression of LINE1 is significantly increased in cultured synovial fibroblasts derived from RA synovial tissue, constituting clear evidence that DNA hypomethylation is associated with gene expression in RA. Furthermore, enforced expression of LINE-1 sequences to RA synovial fibroblasts (RA FLS) has been shown to induce the transcription of p38δ mitogen-activated protein kinase and therefore thought to contribute to the phenotype of RA FLS. A recent study by Karouzakis et al. [14] reported that the decreased global DNA methylation in RA FLS is due to an increased recycling of polyamines associated with a decreased level of S-adenosylmethionine. More recently, a genome-wide evaluation of DNA methylation with 1,859 differentially methylated loci in FLS has resulted in the identification of a number of hypomethylated genes relevant to RA, such as CHI3L1, CASP1, STAT3, MAP3K5, MEFV, and WISP3. Grouped analysis identified 207 hypermethylated or hypomethylated genes with multiple differentially methylated loci, including COL1A1, MEFV, and TNF [42].
Although, it has been suggested that DNA hypomethylation is also associated with the pathogenesis of RA. It has recently been demonstrated that hypermethylation of EBF3 and IRX1 genes interacts with transforming growth factor-β (TGF-β) pathway components and down-modulates mRNA expression in RA FLS, which may result in the activated phenotype of FLS in RA pathogenesis [43].
Aberrant Gene Promoter Methylation in RA
In addition to global DNA methylation in RA, different CpG island promoters located in regulatory regions of genes can become aberrantly methylated, giving rise to an aberrant transcription of the associated genes [44]. Some related studies have demonstrated that even demethylation of a single CpG motif in the promoters of the IL-6 in PBMCs from RA leads to IL-6 mRNA overexpression and increased levels of serum IL-6 and therefore play a role in the pathogenesis of RA [18]. Fu et al. [17] showed that PBMCs derived from patients with RA contain characteristic hypomethylation of CpG sites in the IL-10 promoter associated with elevated IL-10 expression, which may regulate IL-10 transcription and be responsible for the pathogenesis of RA. Moreover, hypomethylation of promoter regions of single genes, such as chemokine (C-X-C motif) ligand 12 (CXCL-12) in RASFs and (Interleukin 1 receptor, type II) IL1R2, has also been reported [45]. In contrast, some promoter regions can also be associated with hypermethylation in RA. The promoter region of the death receptor 3 gene (DR3), a member of the apoptosis-inducing Fas gene family, was shown to have significant hypermethylation of CpG dinucleotides in RA patients, which may enhance resistance to apoptosis in RA synovial fibroblasts [46].
X-Chromosome Inactivation Defects in RA
There is also evidence that DNA methylation may be involved in the pathogenesis of RA, and a potential explanation for the increased risk of RA in women came from studies exploring the pattern of X-chromosome inactivation. The gender-bias (3:1 female-to-male ratio) of RA suggests that the X-chromosome may play a role in the development of RA. Chabchoub et al. [47] showed an increased presence of a skewed X-chromosome inactivation pattern in the peripheral blood from RA patients. Instead of a random X-chromosome inactivation, 80 % or more of the cells exhibited inactivation of the same X-chromosome. CD40 ligand (CD40L) is primarily expressed at the surface of activated CD4+ T helper cells, which binds the CD40 receptor on B cells [48]. The CD40–CD40L pathway plays a crucial role in the immune response. Increased expression of CD40L has been detected in CD4+ T helper cells of female RA patients. In addition, the promoter region of the X-chromosome-encoded gene CD40L was demethylated in CD4+ T cells from female RA patients. CD40L demethylation in female RA patients raises the possibility that other X-chromosome genes may also demethylate in RA. Demethylation of genes on the inactive X chromosome may contribute to female-biased RA patients.
Possible Mechanisms of DNA Hypomethylation in RA
It is possible that DNA hypomethylation in T cells from patients with autoimmune disease could be due to diminished activity of DNMTs. However, the present study evaluating the regulation of DNMTs gene expression and activity in RA are limited. Richardson et al. [40] demonstrated that patients with RA showed a lower level of DNMTs activity but not reaching a statistically significant difference compared with controls. Similarly, new evidence demonstrated that DNMT1 protein in RA was significantly lower than in osteoarthritis (OA) synovial tissue [22]. DNMT expression and function are inhibited in FLS after IL-1 stimulation. However, this effect is transient. An expanded data has been shown the stability of the methylome signature in RA FLS [49]. Moreover, the DNMT1 protein levels in RA FLS also remained low but were not suppressed further by cytokines and growth factors. Recent work has demonstrated that exposure to IL-1 decreases the mRNA levels of DNMT1 and DNMT3a in RA FLS and also inhibits global methylation of RA FLS, suggesting that the cytokine milieu can contribute to differential DNA methylation in RA FLS through altered expression of DNMTs. However, the available data seem to be controversial. In a previous study, the expression of DNMT1mRNA was markedly increased in RA, which may be an indirect response to global DNA hypomethylation through a feedback mechanism [21].
DNA methylation has been shown to regulate the level of expression of microRNAs (miRNAs) in the pathogenesis of RA [50–52]. It is well known that miRNAs are a class of short, single-stranded, and non-coding RNAs which function as post-translational repressors of gene expression, which result in degradation and/or inhibition of translation [53]. There is evidence that DNA demethylation with 5-azacytidine (5-azaC) increases the expression of miR-203 in R [50]. Importantly, the increased levels of miR-203 result in elevated secretion of MMP-1 and IL-6 via the NF-κB pathway, and thereby, contribute to the activated phenotype of RASFs. MeCP2 is a chromatin-binding protein that preferentially binds to methylated CpGs and is highly abundant in the synovium of RA rats. It has also been shown that increasing global methylation levels are regulated by overexpression of MeCP2 in a RA rat model [54].
Pharmacologic Inhibition of DNA Methylation Causes an Enhanced Development of RA
Drugs targeting DNA methylation is being introduced in cancer therapy. The inhibitors of DNMT anti-tumorigenic properties restore the expression of pathologically silenced genes [55]. A number of study results also show that pharmacologic inhibition of DNA methylation creates a hypomethylating milieu and leads to the activated phenotype of FLS of RA. 5-azacytidine (5-aza-CR) and 5-Aza-2′-deoxycytidine (5-Aza-CdR), the inhibitors of DNMT, are known to induce demethylation and reactivation of silenced genes. In fact, exciting discoveries on the effects of DNMT inhibitor in RA have recently been made. Long-term administration of DNA methylation inhibitor 5-azaC in normal synovial fibroblasts induced an activated phenotype similar to that observed in RASFs, and treatments were associated with increase in expression adhesion molecules, cytokines, and receptors for growth factors, as well as matrix-degrading enzymes (MMPs) [14]. In addition, Karouzakis et al. [45] reported that 5-azaC increases CXCL-12 protein secretion from the OA synovial fibroblasts. Furthermore, stimulation of RASFs with CXCL-12 induced the expression of MMPs, including MMP-1, MMP-13, and MMP-3. Another study showed that In vitro treatment of FLS with DNA methylation inhibitor 5-azadC blocked FLS proliferation and increased the secreted frizzled-related protein 4 (SFRP4) expression, a physiological inhibitor can attenuate Wnt signaling in RA. Treatment of FLS with DNA methylation inhibitor 5-azadC or silence of MeCP2 increases SFRP4 the SFRP4 expression, a physiological inhibitor can attenuate Wnt signaling in RA. Enforced SFRP4 inhibits the activation of Wnt pathway which promotes the FLS abnormal proliferation in RA [56]. Moreover, RA FLS were stimulated with 5-azaC increased the expression of miR-203. Increased expression of miR-203 led to significantly enforced levels of MMP-1 and IL-6 in RA [50].
Conclusion and Prospective
Collectively, there is now increasing evidence that activation of genes due to hypomethylation causes the progression of RA, and FLS activation can contribute to epigenetic changes, resulting in abnormal cellular gene function and an alteration in expression. Advances in understanding of the epigenetic mechanisms of DNA methylation in the pathogenesis of RA could provide important clues to develop targeted therapeutic strategies for RA, as well as a deeper understanding of the etiology of this disease.
References
Li, X., Yuan, F. L., Lu, W. G., Zhao, Y. Q., Li, C. W., Li, J. P., et al. (2010). The role of interleukin-17 in mediating joint destruction in rheumatoid arthritis. Biochemical and Biophysical Research Communications, 397, 131–135.
Yuan, F. L., Li, X., Lu, W. G., Li, C. W., Xu, R. S., & Dong, J. (2011). IL-33: A promising therapeutic target for rheumatoid arthritis? Expert Opinion on Therapeutic Targets, 15, 529–534.
Klein, K., Ospelt, C., & Gay, S. (2012). Epigenetic contributions in the development of rheumatoid arthritis. Arthritis Research and Therapy, 14, 227.
Bartok, B., & Firestein, G. S. (2010). Fibroblast-like synoviocytes: Key effector cells in rheumatoid arthritis. Immunological Reviews, 233, 233–255.
Peck, A., & Mellins, E. D. (2009). Breaking old paradigms: Th17 cells in autoimmune arthritis. Clinical Immunology, 132, 295–304.
Davignon, J. L., Hayder, M., Baron, M., Boyer, J. F., Constantin, A., Apparailly, F., et al. (2013). Targeting monocytes/macrophages in the treatment of rheumatoid arthritis. Rheumatology (Oxford), 52, 590–598.
Maruotti, N., Crivellato, E., Cantatore, F. P., Vacca, A., & Ribatti, D. (2007). Mast cells in rheumatoid arthritis. Clinical Rheumatology, 26, 1–4.
Zhao, Y., Zhang, A., Du, H., Guo, S., Ning, B., & Yang, S. (2012). Tolerogenic dendritic cells and rheumatoid arthritis: Current status and perspectives. Rheumatology International, 32, 837–844.
Noss, E. H., & Brenner, M. B. (2008). The role and therapeutic implications of fibroblast-like synoviocytes in inflammation and cartilage erosion in rheumatoid arthritis. Immunological Reviews, 223, 252–270.
Firestein, G. S. (2003). Evolving concepts of rheumatoid arthritis. Nature, 423, 356–361.
Miao, C. G., Yang, Y. Y., He, X., Li, X. F., Huang, C., Huang, Y., et al. (2013). Wnt signaling pathway in rheumatoid arthritis, with special emphasis on the different roles in synovial inflammation and bone remodeling. Cellular Signalling, 25(10), 2069–2078.
Bottini, N., & Firestein, G. S. (2013). Duality of fibroblast-like synoviocytes in RA: Passive responders and imprinted aggressors. Nature Reviews Rheumatology, 9, 24–33.
Zhang, Y., Dong, J., He, P., Li, W., Zhang, Q., Li, N., et al. (2012). Genistein inhibit cytokines or growth factor-induced proliferation and transformation phenotype in fibroblast-like synoviocytes of rheumatoid arthritis. Inflammation, 35, 377–387.
Karouzakis, E., Gay, R. E., Michel, B. A., Gay, S., & Neidhart, M. (2009). DNA hypomethylation in rheumatoid arthritis synovial fibroblasts. Arthritis and Rheumatism, 60, 3613–3622.
Neidhart, M., Rethage, J., Kuchen, S., Kunzler, P., Crowl, R. M., Billingham, M. E., et al. (2000). Retrotransposable L1 elements expressed in rheumatoid arthritis synovial tissue: association with genomic DNA hypomethylation and influence on gene expression. Arthritis and Rheumatism, 43, 2634–2647.
Karouzakis, E., Gay, R. E., Gay, S., & Neidhart, M. (2012). Increased recycling of polyamines is associated with global DNA hypomethylation in rheumatoid arthritis synovial fibroblasts. Arthritis and Rheumatism, 64, 1809–1817.
Fu, L. H., Ma, C. L., Cong, B., Li, S. J., Chen, H. Y., & Zhang, J. G. (2011). Hypomethylation of proximal CpG motif of interleukin-10 promoter regulates its expression in human rheumatoid arthritis. Acta Pharmacologica Sinica, 32, 1373–1380.
Nile, C. J., Read, R. C., Akil, M., Duff, G. W., & Wilson, A. G. (2008). Methylation status of a single CpG site in the IL6 promoter is related to IL6 messenger RNA levels and rheumatoid arthritis. Arthritis and Rheumatism, 58, 2686–2693.
Ishida, K., Kobayashi, T., Ito, S., Komatsu, Y., Yokoyama, T., Okada, M., et al. (2012). Interleukin-6 gene promoter methylation in rheumatoid arthritis and chronic periodontitis. Journal of Periodontology, 83, 917–925.
Lin, S. Y., Hsieh, S. C., Lin, Y. C., Lee, C. N., Tsai, M. H., Lai, L. C., et al. (2012). A whole genome methylation analysis of systemic lupus erythematosus: Hypomethylation of the IL10 and IL1R2 promoters is associated with disease activity. Genes and Immunity, 13, 214–220.
Liu, C. C., Fang, T. J., Ou, T. T., Wu, C. C., Li, R. N., Lin, Y. C., et al. (2011). Global DNA methylation, DNMT1, and MBD2 in patients with rheumatoid arthritis. Immunology Letters, 135, 96–99.
Nakano, K., Boyle, D. L., & Firestein, G. S. (2013). Regulation of DNA methylation in rheumatoid arthritis synoviocytes. Journal of Immunology, 190, 1297–1303.
Delcuve, G. P., Rastegar, M., & Davie, J. R. (2009). Epigenetic control. Journal of Cellular Physiology, 219, 243–250.
Li, E. (2002). Chromatin modification and epigenetic reprogramming in mammalian development. Nature Reviews Genetics, 3, 662–673.
Turek-Plewa, J., & Jagodzinski, P. P. (2005). The role of mammalian DNA methyltransferases in the regulation of gene expression. Cellular and Molecular Biology Letters, 10, 631–647.
Weber, M., & Schubeler, D. (2007). Genomic patterns of DNA methylation: Targets and function of an epigenetic mark. Current Opinion in Cell Biology, 19, 273–280.
Malygin, E. G., & Hattman, S. (2012). DNA methyltransferases: Mechanistic models derived from kinetic analysis. Critical Reviews in Biochemistry and Molecular Biology, 47, 97–193.
Zhou, Y., & Lu, Q. (2008). DNA methylation in T cells from idiopathic lupus and drug-induced lupus patients. Autoimmunity Reviews, 7, 376–383.
Pradhan, S., & Esteve, P. O. (2003). Mammalian DNA (cytosine-5) methyltransferases and their expression. Clinical Immunology, 109, 6–16.
Hendrich, B., & Tweedie, S. (2003). The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends in Genetics, 19, 269–277.
Baranzini, S. E., Mudge, J., van Velkinburgh, J. C., Khankhanian, P., Khrebtukova, I., Miller, N. A., et al. (2010). Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature, 464, 1351–1356.
Boyd-Kirkup, J. D., Green, C. D., Wu, G., Wang, D., & Han, J. D. (2013). Epigenomics and the regulation of aging. Epigenomics, 5, 205–227.
Gros, C., Fahy, J., Halby, L., Dufau, I., Erdmann, A., Gregoire, J. M., et al. (2012). DNA methylation inhibitors in cancer: recent and future approaches. Biochimie, 94, 2280–2296.
Kulis, M., & Esteller, M. (2010). DNA methylation and cancer. Advances in Genetics, 70, 27–56.
Viatte, S., Plant, D., & Raychaudhuri, S. (2013). Genetics and epigenetics of rheumatoid arthritis. Nature Reviews Rheumatology, 9, 141–153.
Watt, F., & Molloy, P. L. (1988). Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes and Development, 2, 1136–1143.
Nan, X., Ng, H. H., Johnson, C. A., Laherty, C. D., Turner, B. M., Eisenman, R. N., et al. (1998). Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature, 393, 386–389.
Majumdar, S., Buckles, E., Estrada, J., & Koochekpour, S. (2011). Aberrant DNA methylation and prostate cancer. Current Genomics, 12, 486–505.
Backdahl, L., Bushell, A., & Beck, S. (2009). Inflammatory signalling as mediator of epigenetic modulation in tissue-specific chronic inflammation. International Journal of Biochemistry and Cell Biology, 41, 176–184.
Richardson, B., Scheinbart, L., Strahler, J., Gross, L., Hanash, S., & Johnson, M. (1990). Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis and Rheumatism, 33, 1665–1673.
Corvetta, A., Della Bitta, R., Luchetti, M. M., & Pomponio, G. (1991). 5-Methylcytosine content of DNA in blood, synovial mononuclear cells and synovial tissue from patients affected by autoimmune rheumatic diseases. The Journal of Chromatography, 566, 481–491.
Nakano, K., Whitaker, J. W., Boyle, D. L., Wang, W., & Firestein, G. S. (2013). DNA methylome signature in rheumatoid arthritis. Annals of the Rheumatic Diseases, 72, 110–117.
Park, S. H., Kim, S. K., Choe, J. Y., Moon, Y., An, S., Park, M. J., et al. (2013). Hypermethylation of EBF3 and IRX1 genes in synovial fibroblasts of patients with rheumatoid arthritis. Molecules and Cells, 35, 298–304.
Bian, E. B., Huang, C., Wang, H., Wu, B. M., Zhang, L., Lv, X. W., et al. (2013). DNA methylation: New therapeutic implications for hepatic fibrosis. Cellular Signalling, 25, 355–358.
Karouzakis, E., Rengel, Y., Jungel, A., Kolling, C., Gay, R. E., Michel, B. A., et al. (2011). DNA methylation regulates the expression of CXCL12 in rheumatoid arthritis synovial fibroblasts. Genes and Immunity, 12, 643–652.
Takami, N., Osawa, K., Miura, Y., Komai, K., Taniguchi, M., Shiraishi, M., et al. (2006). Hypermethylated promoter region of DR3, the death receptor 3 gene, in rheumatoid arthritis synovial cells. Arthritis and Rheumatism, 54, 779–787.
Chabchoub, G., Uz, E., Maalej, A., Mustafa, C. A., Rebai, A., Mnif, M., et al. (2009). Analysis of skewed X-chromosome inactivation in females with rheumatoid arthritis and autoimmune thyroid diseases. Arthritis Research and Therapy, 11, R106.
Liao, J., Liang, G., Xie, S., Zhao, H., Zuo, X., Li, F., et al. (2012). CD40L demethylation in CD4(+) T cells from women with rheumatoid arthritis. The Journal of Clinical Immunology, 145, 13–18.
Whitaker, J. W., Shoemaker, R., Boyle, D. L., Hillman, J., Anderson, D., Wang, W., et al. (2013). An imprinted rheumatoid arthritis methylome signature reflects pathogenic phenotype. Genome Medicine, 5, 40.
Stanczyk, J., Ospelt, C., Karouzakis, E., Filer, A., Raza, K., Kolling, C., et al. (2011). Altered expression of microRNA-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthritis and Rheumatism, 63, 373–381.
Niederer, F., Trenkmann, M., Ospelt, C., Karouzakis, E., Neidhart, M., Stanczyk, J., et al. (2012). Down-regulation of microRNA-34a* in rheumatoid arthritis synovial fibroblasts promotes apoptosis resistance. Arthritis and Rheumatism, 64, 1771–1779.
de la Rica, L., Urquiza, J. M., Gomez-Cabrero, D., Islam, A. B., Lopez-Bigas, N., Tegner, J., et al. (2013). Identification of novel markers in rheumatoid arthritis through integrated analysis of DNA methylation and microRNA expression. Journal of Autoimmunity, 41, 6–16.
Miao, C. G., Yang, Y. Y., He, X., Xu, T., Huang, C., Huang, Y., et al. (2013). New advances of microRNAs in the pathogenesis of rheumatoid arthritis, with a focus on the crosstalk between DNA methylation and the microRNA machinery. Cellular Signalling, 25, 1118–1125.
Miao, C. G., Yang, Y. Y., He, X., & Li, J. (2013). New advances of DNA methylation and histone modifications in rheumatoid arthritis, with special emphasis on MeCP2. Cellular Signalling, 25, 875–882.
Catalano, M. G., Fortunati, N., & Boccuzzi, G. (2012). Epigenetics modifications and therapeutic prospects in human thyroid cancer. Frontiers in Endocrinology (Lausanne), 3, 40.
Miao, C. G., Huang, C., Huang, Y., Yang, Y. Y., He, X., Zhang, L., et al. (2013). MeCP2 modulates the canonical Wnt pathway activation by targeting SFRP4 in rheumatoid arthritis fibroblast-like synoviocytes in rats. Cellular Signalling, 25, 598–608.
Acknowledgments
This work was supported by the China National Science Foundation grants (30901526, 81101372, and 81270011).
Conflict of interest
The authors declare that there are no conflicts of interest associated with this study.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Yuan, FL., Li, X., Xu, RS. et al. DNA Methylation: Roles in Rheumatoid Arthritis. Cell Biochem Biophys 70, 77–82 (2014). https://doi.org/10.1007/s12013-014-9913-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-014-9913-8