Abstract
Soil salinity is a widespread issue that reduces the productivity of major cereal crops including rice. Plant growth-promoting endophytic bacteria are known to alleviate salt stress via wide range of mechanisms, such as production of aminocyclopropane-1-carboxylate deaminase (ACCD). In this study, salt-tolerant endophytic bacterium identified as Bacillus altitudinis NKA32 was isolated from the roots of rice. Multiple plant growth-promoting (PGP) traits of NKA32 were analyzed under increasing salt concentrations, among which ACCD activity was confirmed by enzyme assay and Fourier transform infrared spectroscopy. Proline, trehalose, antioxidant production and sodium ion accumulation were also estimated at different levels of salt stress. The PGP characteristics (indole acetic acid, siderophore, zinc and phosphate solubilization) and ACCD were significantly enhanced in case of NKA32 at concentrations of 300 and 600 mM NaCl, while osmolyte production increased at 900 mM NaCl. Pot study showed that rice plants inoculated with NKA32 exhibited maximum fresh and dry weight (188.16 and 76.68 g/plant), total chlorophyll (4.49 mg/g FW), total phenols and flavonoids (4.43 and 2.23 mg/g FW), along with grain yield, protein, carbohydrate and relative water content, in non-saline soil. Additionally, inoculated rice plants showed 53.67% and 71.53% lower malondialdehyde and ethylene content respectively, as compared to control plants under saline conditions. Elevated level of antioxidant enzymes- superoxide dismutase (44.82%) and ascorbate peroxidase (67.61%); hydrogen peroxide scavenging activity (49.30%), 2,2-diphenyl-1-picrylhydrazyl scavenging activity (11.23%) and proline (39.38%) were reported in inoculated plants under saline conditions. The current findings show that the application of plant growth-promoting endophytic strain is a suitable option for increasing crop growth and productivity in salt-stressed farming systems.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Soil salinity is one of the major abiotic stress that affects plant growth and causes significant loss of crop productivity worldwide (Arora et al. 2020). Globally, 20% of irrigated land has been severely damaged by the accumulation of salts and it is expected to reach up to 50% by 2050 (Nawaz et al. 2020). In India, about 6.74 million hectares (m ha) of land is affected by salinity corresponding to a loss of 16.8 million tonnes of agricultural output annually (Sheoran et al. 2021). High salt concentrations have negative consequences such as increased root ethylene synthesis, hyper-osmotic shock, and plant ionic imbalance (Saghafi et al. 2019). Salinity stress produces reactive oxygen species (ROS) like superoxide radical (O2−), hydrogen peroxide (H2O2), and hydroxyl radicals (OH−), which cause oxidative damage, lipid peroxidation, membrane deterioration and a significant reduction in plant growth and development (Fatima et al. 2020). Plants have evolved several mechanisms to safeguard themselves from stressful conditions, including enzymatic (superoxide dismutase, peroxidase, and catalase) and non-enzymatic (accumulation of compatible solutes such as soluble sugars and proline, glycine betaines) mechanisms (Litalien and Zeeb 2020; Mishra et al. 2021). These enzymes and osmolytes help plants scavenge harmful ROS, resulting in less oxidative damage and improved plant growth (Mahmud et al. 2019).
Rice is one of the most widely grown crops and a staple food for half of the world’s population (Ji et al. 2020). It is considered as one of the most salt-sensitive crops with a threshold of 3–18 dSm−1 for most cultivated varieties (Minh et al. 2016). Soil salinity has drastically reduced rice’s biomass production and yield worldwide (Kakar et al. 2016). Over the next 30 years, a significant increase in global rice production will be required to meet the demands of the growing population (Sarker et al. 2023). Several physical and chemical methods have been employed for remediation of saline soil impacting microbial biodiversity and soil quality. To overcome these limitations and rehabilitate saline-degraded areas, sustainable ways are necessary that are both less expensive and environment friendly (Sunita et al. 2020; Arora et al. 2024).
Use of salt-tolerant plant growth-promoting endophytic bacteria (ST-PGPEB) for seed priming or bio-augmentation can be a sustainable approach to mitigate salinity stress (Choudhury et al. 2023). Aminocyclopropane-1-carboxylate deaminase (ACCD) (EC. 3.5.99.7), an enzyme produced by bacteria alleviates salt-induced damage in crops by catalysing the conversion of aminocyclopropane-1-carboxylate (ACC) into α-ketobutyrate and ammonia, resulting in lowered ethylene stress (Glick et al. 2014). The endophytic bacteria reside within different parts of plants, remaining protected from competitive environments and usually promote plant growth by producing phytohormones, siderophores and by solubilizing minerals (Patel et al. 2023). Additionally, ST-PGPEB can synthesize antioxidants such as superoxide dismutase (SOD), ascorbate peroxidase (APX), and catalase (CAT), that detoxify excess ROS produced when plants are under stress conditions (Afridi et al. 2019). Production of ACCD, antioxidant enzymes and osmolytes by endophytic bacteria can induce an array of chemical variations in plants, including total protein, carbohydrate, flavonoids and phenolic content, which may improve their ability to withstand salinity stress (Choudhury et al. 2021). ACCD-producing salt tolerant plant growth promoting rhizobacteria (PGPR) including Achromobacter sp., Enterobacter sp. PR14, Methylobacterium oryzae, Enterobacter, Achromobacter, Bacillus, and Stenotrophomonas have been previously reported (Shahid et al. 2020; Sagar et al. 2020; Jhuma et al. 2021) to protect rice plants from salinity stress. Among plant growth promoting bacteria (PGPB), the genus Bacillus is extensively researched because of its widespread distribution and compelling characteristics. The genus is at present known to have 379 species (including sub-species) and is found in varieties of habitats, including plants, soils, water, and marine sediments (source: list of prokaryotic names in nomenclature, http://www.bacterio.net/bacillus.html, accessed on 2019.07.30) (Yue et al. 2019). Many of them have been shown to protect plants from biotic and abiotic stresses. For example, Bacillus pumilus, Bacillus koreensis, and Bacillus megaterium alleviate salinity and cadmium stress in tomato and rice plants (Kumar et al 2021; Zhou et al. 2021; Romero-Munar and Aroca 2023). Additionally, some PGPB known as endophytes have been isolated from the interior tissues of plants. Strain Bacillus altitudinis WR10 isolated from roots of wheat improved seed germination and root dry weight under low phosphorus and saline conditions. The strain WR10 produced indole acetic acid (IAA), ACCD, formed biofilm, solubilized both inorganic and organic phosphates and resisted salt toxicity (12% NaCl). Furthermore, B. altitudinis SRI-178 was found to enhance the uptake of iron (Fe) and zinc (Zn) in chickpea (Kushwaha et al. 2021). Draft genome of B. altitudinis showed that they possess genes responsible for plant growth promotion, osmolytes (proline and glycine betaine) production and tolerance against heat stress (Chang et al. 2024). However, only a few studies (Siddikee et al. 2021; Choudhury et al. 2023) are known to be done on salt-tolerant bacterial endophytes with ACCD activity that alleviate the negative effects of salt stress in rice. The current study also details about the bacterial mechanisms involved in mitigation of salt and ethylene stress (under saline conditions) through ACCD activity along with other salt tolerance traits including osmoregulation, antioxidants production, and ion accumulation. The present study explains the mechanisms of selected ST-PGPE strain along with the development of bioformulation for stressed agro-ecosystems specifically for a global staple crop, rice. The objective of this work was to find the beneficiary effect of ST-PGPEB on the growth, biochemical traits, salt tolerance mechanisms and yield of rice under non-saline and saline conditions. The study also focused on the mechanism of ST-PGPE bacterial survival and adaptation under salinity stress.
Materials and methods
Isolation of salt-tolerant endophytic ACCD producing bacteria
Rice plant samples were taken from village Usri, Kanpur Dehat, Uttar Pradesh (26.7108° N, 79.7540° E) in July 2021. At flowering stage, healthy plants (randomly chosen) of Oryza sativa (L.) and soil samples were collected and transferred to the laboratory for analysis. Electrical conductivity (EC) and pH of the collected soil were 8.6 dS m−1 and 8.92 respectively. For the isolation of root endophytic bacteria, rice root samples were removed from each plant, washed under running water and surface sterilized according to Mahgoub et al. (2021). The isolation was done on nutrient agar (NA) medium as per Dif et al. (2022). A total of forty-four distinct endophytic bacterial colonies were found. Pure cultures were stored at − 20 °C in 20% glycerol stock for further study.
To examine the ACCD activity of endophytic bacteria, fresh cultures grown in nutrient broth (NB) up to late log phase (OD600 = 0.1) were spot inoculated on DF (Dworkin and Foster) minimal agar medium containing 3 mM ACC instead of (NH4)2SO4 as nitrogen source (Glick et al. 1995). In addition, 300–1200 mM NaCl (w/v) and a non-saline control was used in the culture media to evaluate the effect of NaCl on ACCD activity of isolates. Isolates capable of using ACC as sole N-source were designated as salt-tolerant ACCD producing bacteria. Furthermore, NKA32, a bacterial isolate with high salt-tolerance (up to 1200 mM) and positive for ACCD activity, was selected and further characterized.
Salt tolerance assay of selected bacterium
The salt tolerance of ACCD producing NKA32 was checked using NB supplemented with 300–1200 mM NaCl (w/v) and a control without additional NaCl in 150 ml flasks. Log phase culture of NKA32 (108 CFU/ml) was inoculated (1 ml) into the medium and incubated for 48 h in shaking incubator (150 rpm) at 28 ± 2 °C. The growth was measured every four hours upto stationary phase by taking optical density at 600 nm using Thermo Scientific™ Evolution 201 UV–Vis spectrophotometer and growth curve was prepared (Fatima and Arora 2021). The sample were taken in triplicates.
Biochemical and molecular characterization of endophytic bacterium
The selected isolate was biochemically characterized according to Bergey’s Manual of Determinative Bacteriology (1994). The molecular identification of ~ 1200 bp 16S rRNA gene sequencing was done using universal primers 16F27 [5′-CCA GAG TTT GAT CMT GGC TCA G-3′] and 16R1492 [5′-TAC GGY TAC CTT GTT ACG ACT T-3′] (El-Shakh et al. 2015). The obtained sequence of bacterial endophyte was compared for homology with type strain sequences found in NCBI database (http://www.ncbi.nlm.nih.gov/BLAST/) (Yoon et al. 2017). The isolate was then submitted to the NCBI GenBank. The phylogenetic tree was constructed by using the neighbour-joining method in MEGA X software (Kimura 1980; Kumar et al. 2016). The isolate (after identification) was submitted to National Agriculturally Important Microbial Culture Collection (ICAR-NAIMCC), Mau, India.
ACCD enzyme activity under salinity stress and confirmation through Fourier transform infrared (FTIR)
ACCD activity of the selected isolate was checked by calculating the amount of α-ketobutyrate produced by the enzymatic hydrolysis of ACC. Briefly, isolate NKA32 was grown overnight in DF medium supplemented with ACC (5 mM) under salt stress (up to 1200 mM NaCl). After incubation, the cells were collected and washed with Tris–HCl buffer (pH = 7.6). Further, the cell suspension was mixed with toluene and 20 µl ACC (5 mM), followed by incubation at 30 °C for 15 min. For detailed quantitative analysis, the procedure of El-Tarabily (2008) was followed. ACCD activity was determined as the amount of α-ketobutyrate generated per nmol mg−1 protein h−1. The amount of α -ketobutyrate was estimated by comparing the absorbance obtained at 540 nm to a standard curve of α-ketobutyrate ranging from 0.02 to 0.2 mol. Protein content in toluenized cell suspension was determined according to Lowry et al. (1951) using bovine serum albumin (BSA) as standard. FTIR (Model: NicoletTM 6700, ThermoFisher Scientific, USA) spectral analysis was used to confirm the formation of α -ketobutyrate under salt stress (uo to 1200 mM NaCl) via the breakdown of ACC (Honma and Shimomura 1978; Penrose and Glick 2003). For comparison, FTIR analysis of pure α-ketobutyrate solution was also done. The wavelength was recorded from 1000 to 400 cm−1.
Determination of PGP traits of the identified endophytic bacterium under salinity stress
Plant growth-promoting activities such as phosphate solubilization, IAA production and siderophore production were checked for the selected endophyte under various salt concentrations (upto 1200 mM NaCl). The phosphate solubilizing activity was assayed on Pikovskaya agar medium. For quantitative analysis, a 100 µl bacterial culture (108 CFU/ml) was inoculated in freshly prepared NBRIP broth and incubated for 10 days at 28 °C. Stannous chloride was used to quantify soluble phosphate at optical density of 600 nm (Yue et al. 2019). IAA production was measured in LB broth (amended with 200 μg/ml l-tryptophan) using Salkowski reagent according to Etesami et al. (2015). Siderophore production was evaluated by following the method of Schwyn and Neilands (1987). Briefly, fresh culture supernatant (centrifuged at 6000 rpm for 15 min) was spot inoculated on chrome azurol S (CAS) agar plates and incubated for 5 days at 28 ± 2 °C to observe for a halo orange zone around colonies. Quantitative analysis for siderophore production was checked using CAS shuttle solution. The Zn solubilizing property of the bacterial isolate was tested in vitro by inoculating aseptically on Tris-minimal medium and the solubilization index was calculated (Fasim et al. 2002).
Bacterial physiology under salinity stress
To check the effect of salt stress on ion accumulation, osmolite accumulation, and antioxidants production in selected bacteria, pre-grown culture of NKA32 (with O.D600 = 0.1) was inoculated (1 ml) in NB medium containing various concentrations of NaCl (upto 1200 mM) and incubated at 28 °C, 150 rpm for 48 h. The cells were harvested by centrifugation (at 6000 rpm for 15 min) in the late log phase (OD600 = 0.1; adjusted using sterilized distilled water), filtrate and biomass were collected. For checking cellular accumulation of Na+ ions in bacteria, the cell pellets were washed three times with isotonic MgCl2 solution (10 mM) and lysed with 30% HNO3. The intracellular Na+ ions were measured with a flame photometer (Fatima et al. 2020). Proline accumulation was determined by the method of Bates et al. (1973). The accumulation of disaccharide trehalose accumulation in the cell-free extract was quantified according to Orozco-Mosqueda et al. (2019). The antioxidant activity was calculated based on the reduction of DPPH (2,2-diphenyl-1-picrylhydrazyl) (Xing et al. 2015).
Pot trial and experimental design
The pot experiment was conducted for two consecutive years (2021 and 2022) from July to October. The rice seeds (var. CSR10) were obtained from ICAR-Central Soil Salinity Research Institute (CSSRI) Lucknow, U.P., India. The seeds were surface sterilized for five minutes using 95% (v/v) ethanol, followed by ten minute treatment with 3% (v/v) sodium hypochlorite. Further, the seeds were rinsed 5–7 times and soaked for 6 h using sterile distilled water. The seeds were germinated at room temperature on sterilized wet germination paper for five days. The germinated seedlings were then inoculated with a cell suspension of B. altitudinis NKA32 (108 CFU ml−1) by root dip method in a beaker containing liquid bioformulation and soaked for 2 h at room temperature (Musson et al. 1995). The liquid bioformulation was prepared as per method of Chompa et al. (2024). Briefly, Luria Bertani (LB) medium was used as a base to develop liquid bioinoculants. The LB broth was augmented with glycerol (5 mM) as carrier additive to improve the survival of B. altitudinis NKA32. After sterilizing the medium, log-phase culture of B. altitudinis NKA32 was inoculated to the broth and incubated at 28 °C for 24 h. A control treatment with uninoculated plants was also taken. Treated and untreated seedlings (3 seedlings from each beaker) were subsequently transferred and grown in pots (35 × 21 × 21 cm) containing 7.0 kg of un-sterilized soil. For pot study, two different sets (saline soil and non-saline soil) of earthen pots were prepared in three replicates. The saline soil was collected from isolation site from where rice plants were collected (i.e. village Usri, Kanpur Dehat, Uttar Pradesh) and non-saline soil was taken from Environmental Science Research Field, BBA University Lucknow. The physiochemical characteristics of both soil samples were estimated according to Odlare et al. (2008) (results mentioned in Table S1). The pot experiment was set up in a randomised block design with four treatments: (T1) seedlings grown in non-saline soil, (T2) seedlings treated with NKA32 grown in non-saline soil (T3) seedlings grown in saline soil (T4) seedlings treated with NKA32 grown in saline soil. The study was conducted in open greenhouse with maximum temperature range from 37 °C and 26 °C in July to October 2021, and 38 °C and 23 °C in July to October 2022, respectively, with an average humidity of about 75% (Inostroza et al. 2017).
The ability of selected endophytic bacteria to colonize roots
To check endophytic colonization under saline and non-saline conditions, rice seeds were inoculated with developed bioformulation as mentioned above. Scanning electron microscopy (SEM) and re-isolation was done to check interior colonization (Li et al. 2016). Treated and untreated seeds (ten seeds) were sown in culture tube amended with water agar medium containing 10 and 90 mM NaCl (w/v) under controlled conditions and maintained for 10 days under a 12 h dark/light cycle at 26–30 °C. In the experiment, salt concentrations of 10 and 90 mM NaCl were utilized, corresponding to EC of 0.97 and 8.92 dS m−1, respectively. These concentrations were chosen to closely match the EC levels of non-saline and saline soils used in the pot experiment, as indicated in Table S2. Ten days later, fresh root samples from both untreated and treated plants were collected, surface-sterilized, crushed, and plated on NA medium using the same procedure as used for isolation of endophytic bacteria. The bacterial colonies (CFU/g of fresh root weight) were counted after 3 days of incubation at 28 °C ± 2 °C. The obtained colonies were identified by conducting 16S rRNA sequencing. The experiment was conducted in triplicates.
For SEM analysis, fresh root samples from both control and treated plants grown under 10 mM and 90 mM NaCl were removed at 10 days after sowing (DAS). The plant root samples were then processed as per method of Nowell and Parules (1980). The root samples were uprooted, rinsed with distilled deionized water, cut into thin transverse sections and fixed with 2.5% (v/v) glutaraldehyde in 0.1 M phosphate buffer, pH 7.4, for 2 h at 4 °C. The fixed roots were aseptically air-dried under laminar flow and mounted on an aluminium cylinder with silver paste for observation at various magnifications. Finally, the presence of bacterial cells in plant roots was observed using SEM (JSM 6490 LV, JOEL Japan).
Growth parameters and yield
The plants were uprooted from each treatment, rinsed with water to remove adhering soil and various growth parameters including shoot length, root length, fresh weight, dry weight, and tiller number were checked at 120 days after sowing (DAS). In addition, extracts of the leaves were prepared in variety of solvents, including methanol, ethanol and acetone as described by Das et al. (2014). The supernatant was obtained after centrifugation and stored at − 20 °C for later use. The quantitative analysis of ethylene was also measured in the leaves of rice at 120 DAS. The biochemical traits such as protein, carbohydrate, relative water content (RWC), total phenolic content (TPC), and total flavonoid content (TFC) of leaf extracts was determined at 120 DAS, while chlorophyll content was checked at 90 DAS. Further salt tolerance properties (SOD, APX, DPPH and malondialdehyde (MDA)) of inoculated and uninoculated plants was estimated at 120 DAS.
Determination of biochemical and salt tolerance properties of plants
Chlorophyll a, b and carotenoid in rice leaves was determined according to Arnon (1974). The absorbance of leaf extracts (prepared in 80% ethanol) were taken at 645, 663 and 470 nm using UV–vis spectrophotometer (Thermoscientific, Evolution201) (Mukhtar et al. 2020). Protein estimation in fresh leaves was done according to Lowry et al. (1951). Carbohydrate concentration was determined according to Irigoyen et al. (1992), using the anthrone reagent. RWC of plant leaves was checked according to the method of Weatherley (1950). TPC of the methanolic extract solution was estimated by using Folin-Ciocalteu (Hi-media, HPLC grade) reagent (Kiani et al. 2021). TFC of the methanolic extract solution was determined using the aluminium chloride assay (Kalita et al. 2013).
SOD activity (EC 1.15.1.1) in fresh leaves of rice was evaluated by measuring photochemical reduction capacity of nitro blue toluene (Nawaz et al. 2020). APX (EC 1.11.1.11) activity was assessed by monitoring the decline of ascorbate and measuring the change in absorbance at 290 nm for 1 min (Fatnassi et al. 2015). Proline accumulation in leaf extracts was done according to Bates et al. (1973). H2O2 scavenging activity and DPPH free radical scavenging activity of leaf extracts was determined according to Parejo et al. (2003). The amount of MDA produced in the samples was analyzed using the method of Gowtham et al. (2020).
Ethylene evolution analysis
The amount of ethylene production was determined according to Rasheed et al. (2022). Briefly, leaf samples (0.5 g) were taken from each treatment, cut into small pieces and placed in a 25 ml vial capped with an air-sealed rubber stopper. The vials were kept in the dark at 30 ± 2 °C for 4 h. To measure the ethylene, 250 μl of gas was withdrawn from the vials and injected into a gas chromatograph (GC-2010 SHIMADZU, 130,000, USA) equipped with a flame ionization detector (FID) and RXi-5 Sil MS column (30.0 m × 0.25 mm) with nitrogen as the carrier gas. Nitrogen, hydrogen, and oxygen flow rates were 30, 40, and 400 ml min−1, respectively. The detector was set to 200 °C. Based on the retention time, ethylene was identified and quantified by comparing it with peaks from standard ethylene concentration. Samples were analyzed at Advanced Instrumentation Research Facility (AIRF), Jawaharlal Nehru University, India.
Statistical analysis
All the results are average of three replicates. Satistical analysis was performed using MS Excel 2019 (version 16.65) and IBM SPSS software version 25, One-way analysis of variance (ANOVA) with Duncan’s multiples range test (DMRT) was used to find the level of significance (P < 0.05). Correlation and cluster heat map were created by Past4 and Metaboanalyst 5.0 software.
Results
Isolation of salt-tolerant endophytic ACCD producing bacteria
In this study, forty four endophytic bacteria with diverse morphology were isolated from the roots of rice. Out of these isolates, NKA32 showed growth up to 1200 mM NaCl on DF-ACC agar medium. On the other hand, other isolates were able to grow only up to 600 mM NaCl concentration on ACC-DF agar medium. Isolate NKA32 demonstrating salt tolerance and ACC utilizing capabilities was selected for further study and experiments.
Salt tolerance assay of selected bacteria
The growth and survival of isolate NKA32 at different salt concentrations was assessed (Fig. S2). NKA32 was able to grow on 300 mM NaCl with minimal impact on growth pattern; however, growth rate decreased sharply above 900 mM NaCl concentration. The generation time was found to be 119 min under non-saline conditions and it rose to 210, 338 and 463 min at 300, 600 and 900 mM NaCl stress, respectively. The doubling time increased with additional salt levels, reaching maximum at 1200 mM NaCl (662 min).
Biochemical and molecular characterization of endophytic bacteria
Isolate NKA32 was found to be Gram-positive, rod-shaped, motile, aerobic, flagellated, endospore-forming, showing positive results for the enzymes catalase, citrate, amylase, lipase, protease and nitrate, but was negative for indole, urease and methyl red. It also showed positive results for Voges-Proskauer tests, triple sugar iron test and ammonia production. The 16S rRNA (∼1200 bp) sequencing showed 99.77% similarity with Bacillus altitudinis 41KF2b(T) (accession number ASJC01000029) through BLASTn and the sequence was submitted to NCBI Genbank under the accession number OQ918302 (Fig. S1). Isolate NKA32 has been submitted to the international culture collection centre (NAIMCC, India) as B. altitudinis with accession number NAIMCC-B-03560.
ACCD enzyme activity under salinity stress and confirmation through Fourier transform infrared (FTIR)
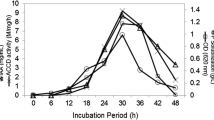
Isolate NKA32 showed variation in ACCD activity in the range of 50.25–167.36 μmol α-ketobutyrate per mg of cellular protein per hour under 0–1200 mM salt stress (Fig. 1). The highest ACCD activity exhibited by B. altitudinis NKA32 was 167.36 μmol α-ketobutyrate mg protein−1 h−1 at 600 mM NaCl, followed by 144.69 μmol α-ketobutyrate mg protein−1 h−1 under 300 mM NaCl stress, 132.41 μnmol α-ketobutyrate mg protein−1 h−1 under non-saline condition, 85.96 μmol α-ketobutyrate mg protein−1 h−1 at 900 mM NaCl and 50.25 μmol α-ketobutyrate mg protein−1 h−1 at 1200 mM salt stress (Fig. 1A). The ACCD activity by the isolate at different salt concentrations was further confirmed by FTIR spectrum analysis (Fig. 1B). A clear peak of 3305.3 and 1656.9 cm−1 at 0 mM NaCl concentration, 3297 and 1658.7 at 300 mM, 3381.7 and 1658.5 cm−1 at 600 mM, 3298.6 and 1658.3 cm−1 at 900 mM NaCl was obtained in the spectrum, confirming the existence of a ketonic group and an amino functional group, both identified as α-ketobutyrate (Singh and Jha 2022). The absorbance from 3273 to 3422 cm−1 indicates the N–H/O–H/C–H stretching of amines, hydroxyls and alkyl groups (Ghosh et al. 2022). The bands from 1640 to 1800 cm−1 indicate the stretching vibration of C = O, N–H and C–N groups (Haque et al. 2023). The molecular structure of alpha-ketobutyrate was indicated by this spectrum. Figure 1B depicts the FTIR spectrum of the reaction mixture of ACCD and ACC, which also produced peaks at the same frequencies. The presence of amines (N–H) and ketone (C = O) groups under different salinity stress showed the ability of the isolate to utilize ACC and cleave it into amines and ketones.
A Quantitative estimation of ACC deaminase in the bacterial cell under 0–1200 mM NaCl stress. When applying Duncan’s multiple range test (DMRT), various letters in rows indicate the values are significantly (P ≤ 0.05) different from one another. Values are presented as means ± standard deviations (S.D.; n = 5). B FTIR spectra analysis of alpha-ketobutyrate in NKA32 under different salt stress (0, 300, 600, and 900 mM NaCl)
PGP traits of the identified endophytic bacteria under salinity stress
Isolate NKA32 was screened for plant growth-promoting attributes under salinity stress. Quantitative analysis of PGP activities was performed in cell-free extract, the data are shown in Table 1. The ability of NKA32 to dissolve mineral phosphates was demonstrated by the formation of a clear zone around the bacterial colony on tricalcium phosphate-containing NBRIP medium. The phosphate solubilizing potential of the isolate increased significantly by 24.26% from non-saline to 300 mM salt stress. At 1200 mM NaCl, the solubilizing potential of NKA32 decreased by 97.75% (P ≤ 0.05) as compared to the control. The presence of an orange halo around the NKA32 colonies on CAS agar medium indicated the production of siderophore. Furthermore, the highest production was obtained at 300 mM NaCl, which was significantly (P ≤ 0.05) increased by 18.84% in comparison to control. The quantitative estimation of IAA production by NKA32 showed that phytohormone production increased up to a certain salt level (upto 300 mM NaCl) while gradually decreasing thereafter. The production increased by 16.17% (P ≤ 0.05) at 300 mM NaCl, while decreasing by 99.45% at 1200 mM NaCl as compared to unstressed conditions. Similarly, Zn solubilizing potential was also maintained up to 300 mM NaCl and declined above this, with no activity at 1200 mM salt stress.
Bacterial physiology under salinity stress
Endogenous accumulation of sodium ions and proline content was higher under salt stress compared to non-saline conditions. Salt-tolerant endophyte NKA32 demonstrated 227% and 564% (P ≤ 0.05) enhancement in proline and sodium ion accumulation at 900 mM as compared to no salt stress. The reduction in proline synthesis was around 47.5% and Na+ ion accumulation reduced by 27% at 1200 mM salt stress as compared to 900 mM NaCl. Further, the halotolerant endophyte NKA32 produced a significant amount of trehalose and exhibited DPPH scavenging activity in salt-amended media. Results showed that trehalose production and DPPH scavenging activity were highest at 900 mM salt stress. The isolate also produced trehalose and showed DPPH free radical scavenging activity at 300 and 600 mM NaCl but was insignificant (P ≤ 0.05) in non-saline condition. The trehalose production increased by 35% and DPPH scavenging activity by 24% at 900 mM salt stress as compared to unstressed conditions. Moreover, the production of trehalose and DPPH scavenging activity decreased above this stress and the lowest production was at 1200 mM salt stress (Table 1).
The ability of selected endophytic bacteria to colonize roots
The colonization ability of selected bacterium within root tissues was examined by quantifying number of colonies re-isolated from tissue homogenates of treated and untreated plants grown under 10 and 90 mM NaCl (Table S2). Ten days following inoculation, the selected endophytic bacteria effectively colonized its host plant tissues, with bacterial population of 1.31 × 104 and 3.49 × 105 CFU per gram of fresh plant roots under 90 mM and 10 mM NaCl, respectively. On the other hand, under given salt stress (90 mM and 10 mM), no bacterial cells were found in untreated plant roots (Table S2). Using SEM, images of the roots of rice were examined at 10th day after inoculation to confirm the location of the selected bacterium (Fig. 2) under different treatments. No bacterial cells were seen on the root surfaces (Fig. 2A, C) or in the tissues in un-inoculated seedlings at varying salt concentrations (10 and 90 mM NaCl), whereas inoculated plants under 10 mM NaCl stress showed large number of bacterial cells localized in root cortex of rice seedlings (Fig. 2B). Conversely, under 90 mM salt stress, comparatively lower number of bacterial cells were detected in root cortex of inoculated seedlings (Fig. 2D).
SEM imaging of the endophytic bacterial locations in the inoculated rice root tissues. A Untreated plants under 10 mM NaCl, the tissues of non-inoculated host plants did not contain any endophytic bacteria; B Treated plants under 10 mM NaCl, a large number of bacterial cells were detected in the intracellular spaces of root tissues; C Untreated plants under 90 mM NaCl, no bacterial cells were found in root tissues of rice; D Inoculated plants under 90 mM salt stress, few bacterial cells observed in root cortex and a few in the xylem vessel. Yellow arrows represent bacterial cells in plants roots, scale bar, 2.00 µm at 5,5000 magnifications
Growth parameters and yield
In pot experiments, inoculated plants showed significant differences in the growth parameters of rice plants under stress and non-stress conditions (Fig. S3). Plants grown in saline soil showed lesser growth as compared to ST-PGPE inoulated plants in non-saline and saline soil. For example, shoot length, root length, fresh weight, dry weight and effective tiller number decreased by 31%, 51%, 38% and 72% respectively, in saline soil (without NKA32 treatment) as compared to non-saline control. However, plants treated with NKA32 under non-saline conditions (T2), showed increased shoot length, root length, fresh weight, dry weight and effective tiller number by 22%, 23%, 11%, 50% and 22% respectively, as compared to non-saline control plants (T1). Similarly, under saline soil conditions, the shoot length, root length, fresh weight, dry weight and effective tiller number of rice plants increased significantly (P ≤ 0.05) by 37%, 86%, 94%, 56% and 104% respectively, when treated with NKA32 (T4) over stressed control plants (T3).
In terms of productivity, grain yield is highly dependent on number of panicles per plant. Salinity had a significant impact on number of panicles, number of seeds and total seed weight with a reduction of 57%, 56%, and 42% respectively, in plants growing in saline soil over the non-saline control. Bacterial treated plants in non-saline soil exhibited an increase by 29%, 27%, 22% for panicles, total seeds and seed weight respectively, in comparision to non-saline control. Similarly, in saline soil, rice plants treated with NKA32 showed higher grain yield in comparision to the uninoculated salt stressed plants (Table 2). However, no significant (P ≤ 0.05) changes in shoot length, root length, dry weight, and seed weight were observed in treatmemt T4 (plants treated with NKA32 grown in saline soil) as compared to treatment T1 (non-saline control).
Determination of biochemical and salt tolerance properties of plants
The changes in photosynthetic pigments of rice with and without bacterial inoculation under non-saline and saline soil conditions are illustrated in Table 2. Rice plants grown in saline soil without bacterial treatment showed reduced photosynthetic pigments: chl a (78%), chl b (49%), and carotenoid (60%) in comparison to uninoculated plants that were grown in non-saline soil. However, rice plants inoculated with B. altitudinis NKA32 considerably increased the levels of photosynthetic pigments. In non-saline soil, higher amount of chl a, chl b and carotenoids (3.82, 1.95 and 1.27 mg/g FW) were recorded in NKA32 treated plants, in comparision to untreated plants (2.52, 1.12 and 0.86 mg/g FW). Similarly, under saline conditions, bacterial treated (T4) plants showed significant enhancement in chl a (169%), chl b (76%) and carotenoid (100%) as compared to uninoculated plants (T3) (P < 0.05; Table 2).
The biochemical parameters such as protein, carbohydrates, phenol, flavonoids and RWC were measured to examine the effect of ST-PGPE bacteria on rice growth (Fig. 3). In contrast to control conditions, plants treated with NKA32 did not suffer salinity-induced stress, resulting in improved growth. In saline soil, the concentration of plants’ protein and carbohydrate significantly (P ≤ 0.05) decreased as compared to non-saline control. Inoculation with NKA32 demonstrated maximum ability to enhance protein (28%) and carbohydrate (12%) under non-saline conditions. Similarly, in the saline soil, the isolate significantly (P ≤ 0.05) enhanced protein (23%) and carbohydrates (26%) as compared to saline control (Fig. 3A). RWC, total flavonoid, and total phenolic content of leaves of salt-stressed rice plants were determined, and increased in the treated plants (Fig. 3B, C). Rice plants with bacterial inoculation showed maximum value for leaf’s RWC (238%), TFC (4.25 mg/g FW) and TPC (2.23 mg/g FW) in non-saline soil as compared to non-saline control. In addition, B. altitudinis NKA32 inoculation under salinity stress also increased the leaves’ RWC, TPC, and TFC by 34%, 156% and 182% when compared to uninoculated rice plants.
Effects of B. altitudinis strain NKA32 on protein and carbohydrate (A), total phenolic and total flavonoid content (B), relative water content (C), superoxide dismutase and ascorbate peroxidase (D), DPPH scavenging and H2O2 scavenging (E), proline and MDA content (F) on rice under normal and saline soil. Treatments: T1 = seedlings grown in non-saline soil, T2 = seedlings treated with NKA32 under non-saline soil, T3 = seedlings grown in saline soil, T4 = seedlings treated with NKA32 under saline soil. Data are presented as means ± standard deviations (S.D.; n = 3). Different letters after values indicate that there is a significant difference at P ≤ 0.05, as determined by Duncans multiple range tests (DMRT)
To evaluate the potential of NKA32 in lowering salinity stress, various salt-tolerance properties were tested. It was found that bacterial-treated plants under salinity stress showed higher SOD and APX activity when compared to non-saline control. Compared to saline control, the endophyte increased SOD activity (29%) and APX activity (66%) of rice plants grown in saline soil (Fig. 3D). Although antioxidant potential was found to be lower in treated and untreated non-saline soil as compared to T3 and T4 treatement. Further, hydroxyl scavenging and free radical scavenging activity of plants were also enhanced more under saline soil. In treatment T4 (bacterial inoculated seedlings grown in saline soil) the influence of bacterial treatment on H2O2 scavenging activity and DPPH free radical scavenging activity was found to be 63% and 25% higher respectively, as compared with uninoculated plants in saline soil (T3) (Fig. 3E). Similarly, in the presence of stress the proline content of leaves increased. Although NKA32 inoculated plants further increased the level of proline in the presence of stress. Bacterial treated plants in saline soil (T4) caused the highest accumulation of proline (47.54 μmol/g FW) (Fig. 3F). However, the lowest amount of proline (11.50 and 15.12 μmol/g FW) was found in non-saline treated and untreated plants. The MDA content of leaves of salt-stressed rice plants was determined and it was found that the treated plants showed less oxidative damage. The results showed that MDA content was 147% higher under saline soil (T3) as compared to non-saline control (T1). B. altitudinis NKA32 inoculation reduced the production of MDA and it was more efficient in decreasing MDA levels (54%) under T4 treatment in comparison to control in saline soil (Fig. 3F).
Ethylene evolution analysis
In comparison to the plants grown in non-saline soil, the rice plants exposed to saline soil produced a much higher amount of ethylene (Fig. 4). The generation of ethylene was considerably lower in NKA32 inoculated plants under saline conditions as compared to untreated plants. The results show maximum to minimum ethylene production of 872.87, 248.51, 207.29 and 180.23 pmol ethylene/g FW/h under untreated seedlings grown in saline soil (T3), seedlings treated with NKA32 grown in saline soil (T4), seedlings treated with NKA32 grown in non-saline soil (T2), and untreated seedlings grown in non-saline soil (T1) respectively. NKA32 treated seelings grown in saline soil significantly inhibited ethylene overproduction by about 72% as compared to untreated seedlings grown in saline soil. Ethylene production was insignificant between T1 and T2 treaments. The amount of ethylene production in treated and untreated plants under saline and non-saline conditions was confirmed by GC-FID (Fig. S4). Pure ethylene produced a peak at 1.914 min retention time (Fig. S4A). The crude extract from all treatments showed predominant peak at the same retention time with some changes as shown in Fig. S4 (B,C,D and E). In the GC-FID graph, Fig S4 BCDE shows ethylene peaks at 1.952, 1.954, 1.953 and 1.950 min retention time respectively under treatment T1, T2. T3 and T4. The changes in retention time from pure ethylene to treatments might be due to the handling error.
Effect of bacterial inoculation on ethylene concentration under saline and non-saline condition. Treatments: T1 = seedlings grown in non-saline soil, T2 = seedlings treated with NKA32 under non-saline soil, T3 = seedlings grown in saline soil, T4 = seedlings treated with NKA32 under saline soil. Each value represents a mean of five replicates. Error bar indicates the standard error of mean (± SE). Different letters after values indicate a significant difference at a P value of 0.05, as determined by Dun-cans multiple range tests (DMRT)
Correlation and cluster analysis of NKA32 plant growth-promoting traits and its effect on rice growth under salinity stress
The Pearson correlation coefficient results (P < 0.05) reveal the impact of the increasing concentration of NaCl (mM) on the potential of bacteriium NKA32 for ion accumulation, osmolyte production, and PGP traits. The findings demonstrate a positive correlation (correlation coefficient r ranging from 0.86 to 0.94) between salinity, ion accumulation, and proline production by NKA32. This underscores the significant role of the bacterium in enhancing stress tolerance. On the other hand, there was negative correlation between salinity and ACCD, DPPH, IAA, siderophore, trehalose and phosphate solubilisation (correlation coefficient r range -0.53 to -0.91). The dark red scale shows a strong negative correlation while dark blue represents a positive relationship with salinity (Fig. 5A).
A Heat map showing the relationship between quantitative statistical parameters and salinity. The values are means of three replicates. B Cluster heat map based on Pearson and Ward for determining distance and clustering to show the effect of NKA32 on growth parameters and salt tolerance properties of rice under saline and non-saline conditions (pot study). Treatments: T1 = seedlings grown in non-saline soil, T2 = seedlings treated with NKA32 under non-saline soil, T3 = seedlings grown in saline soil, T4 = seedlings treated with NKA32 under saline soil. The tested parameters were designated as: TPC- total phenolic content, TFC- total flavonoid content, FW- fresh weight, Carbo- carbohydrate, RWC- relative water content, CHL- chlorophyll, SL- shoot length, RL- root length, DW- dry weight, MDA- malondialdehyde, APX- ascorbate peroxidase, SOD- superoxide dismutase, DPPH- 1,1-diphenyl-2-picryl hydrazyl
The cluster analysis was used to determine the relationship between the ST-PGPE bacterial treatments used in the saline and and non-saline soil and the assessed growth parameters, biochemical traits and salt-tolerance properties of rice plants (Fig. 5B). The dendrograms showed two clusters with significant differences between treated and untreated plants under saline and non-saline conditions. The two clusters were treated and untreated plants under non-saline condition (T1 and T2) with higher growth parameters, photosynthetic pigments, grain yield, protein, carbohydrate and RWC and treated and untreated plants under saline condition (T3 and T4) with higher biochemical (TPC and TFC) and salt tolerance properties (SOD, APX, proline and DPPH). The clustering analysis further demonstrated that untreated plants grown in saline soil (T3) showed lesser growth parameters, yield, biochemical traits, salt tolerance parameters, while higher MDA and ethylene content compared to T1, T2 and T4 treatments. The heat map concludes that the growth parameters of all plants grown under saline conditions were suppressed in comparision to plants grown in non-saline soil. However, plants treated with NKA32 showed enhanced growth parameters under saline as well as non-saline conditions.
Discussion
It is well known that plant growth-promoting endophytic bacteria improve plant health and confer resistance against biotic and abiotic stresses (Khare et al. 2018). In this study, endophyte NKA32 identified as B. altitudinis was isolated from roots of rice growing in saline soil and selected based on salt-tolerance and PGP traits in lab conditions. The ability of NKA32 to produce ACCD, IAA, siderophore and solubilize P and Zn under both saline and non-saline conditions was confired by qualitative and quantitative analysis. The bacterium B. altitudinis NKA32 showed excellent PGP properties such as the production of IAA and siderophore, solubilization of phosphate and Zn at 300 mM NaCl, but further increase in the salt concentration resulted in decline of PGP activities. The bacterium NKA32 was isolated from saline habitat, which makes it a potent candidate for plant growth promotion under saline conditions. ACCD production by NKA32 up to 600 mM NaCl can also be an important stress alleviation strategy to improve plant growth in saline conditions. Although, B. altitudins has been reported as an ACCD producer (Yue et al. 2019, 2022; Oviya et al. 2023), but to the best of our knowledge there are no reports regarding this bacterium as rice root endophyte that too in saline soil conditions. Yue et al. (2019, 2022) reported that wheat endophytic PGPB strain B. altitudinis WR10 can solubilize minerals (P and Zn), produce siderophore, IAA and ACCD under different abiotic stresses and tolerate up to 12% NaCl. Recently, Oviya et al. (2023) found that nodule endophyte B. altitudinus TBB5A from groundnut produced ACCD and solubilized phosphate under 3% salt stress.
Production of ACCD by PGPB endophytes, especially under saline conditions plays an important role in microbe and plant survival as well as improving growth and productivity (Afridi et al. 2019). The FTIR results indicate the presence of functional groups belonging to ACCD under saline and non-saline conditions. The isolate’s ACCD activity appears to be related to its adjustment to salt-affected regions. Previously, researchers reported the same functional groups in ACCD-producing ST-PGPR strains of Enterobacter sp. Burkholderia sp Aneurinibacillus aneurinilyticus, Paenibacillus sp., Pseudomonas aeruginosa and Bacillus subtilis (Gupta and Panday 2019; Gupta et al. 2023). Further, it was reported that the selected isolate could colonise plant tissues, particularly primary roots and the zone of elongation under saline conditions, suggesting the ability to establish itself even under osmotic stress. The in vitro study showed that under 90 mM NaCl stress, the selected isolate successfully colonized inside the root region. This might be due to the production of IAA. Bacillus spp. may interact with plants through the production of IAA for the colonization mechanism (Sood 2019; Mahgoub et al. 2021). In addition, this occurs due to the the ability of endophytic PGPB to solubilize phosphate and Zn and transfer the nutrients to the host plant during their initial colonization of the root region (Rajkumar et al. 2009; Sahu et al. 2021). Herpell et al. (2023) reported that Paraburkholderia dioscoreae Msb3, a yam phyllosphere symbiont, colonized the tomato phyllosphere and stimulated plant growth through its ACCD activity. The present study indicated that the ACCD activity of NKA32 along with other PGP traits under salinity stress may be associated with endophytic colonization and plant–microbe interactions both under saline and non-saline conditions. However, the detailed mechanism and the complex signalling between plant and microbes need to be explored and for this the present study can be a hallmark.
ST-PGPE are also known to synthesize or accumulate compatible solutes, produce antioxidants and regulate ion homeostasis, which play an important role in protection against excessive osmolarity (Fan et al. 2020; Bhutani et al. 2022; Cruz Barrera et al. 2020). These metabolites have been strongly linked to the ability of some microorganisms to survive in saline environments (Choudhary et al. 2023). In this study, production of proline, trehalose, and DPPH scavenging activity by bacterial cells increased up to 900 mM salt stress, while above this concentration solutes level, antioxidant activity and ion accumulation got reduced. This suggests that these metabolites may help in osmoregulation (Fatima and Arora 2021). Mahmoud et al. (2020) found that B. pumilus S2, Bacillus mojavensis S1, and Pseudomonas fluorescens S3 cells accumulated more proline when they were exposed to salt stress. Orozco-Mosqueda et al. (2019) found the synergistic effect of ACCD and trehalose in the protection of tomato plants against salt stress and enhanced growth and productivity. The positive correlation between ACCD, proline, trehalose, DPPH scavenging activity, sodium ion uptake along with PGP traits of NKA32 at applied salt concentration, suggests that the isolate has the capability of plant growth promotion even under saline conditions.
The application of bacterium as bioinoculant to rice in pot trial revealed that NKA32 effectively improved plant growth and development in saline soil. Herein, saline soil had significantly higher Na+ content (74.34 mg/kg) in comparison to normal soil (8.4 mg/kg) that negatively impacted the growth and development of plants. Several studies have reported that high salt concentration creates ion toxicity of Na+ and Cl− affecting the uptake of nutrients like Ca+ and K+ that are important for maintaining ion balance in plants (Arora et al. 2020; Mishra et al. 2021). Ion toxicity due to high Na+ content also damages the photosynthetic apparatus and disturbs the photosynthesis mechanism by blocking the photosystem II reaction centers, electron transport chain and oxygen evolving complex in plants (Acosta-Motos et al. 2017). Bacterium NKA32 exhibiting ACCD activity, IAA production, and the ability to mineralize nutrients, as well as salt tolerance, induced plant growth promotion in saline conditions. IAA production by PGPB endophytes, specifically in saline soil, enhances germination, promotes root growth and elongation by limiting sodium ion uptake and increasing the solubilization of mineral nutrients (Rodríguez Coca et al. 2023). According to Gupta et al. (2023), apart from improving plant health, IAA increases antioxidant activity, photosynthetic pigments, and inhibits lipid peroxidation under salinity stress. Plants’ uptake of nutrients like P, Zn, K, and Fe has also been linked to salt-stress mitigation. These nutrients play an important role in stomatal action, osmoregulation, cell expansion, enzyme activity, membrane stability, and neutralization of non-diffusible negatively charged ions (Fatima and Arora 2021). Win et al. (2018) found that ACCD producing Bacillus sp. limited the uptake of sodium ions in tomato plants due to their nutrient chelation strategy. In this study, higher biomass accumulation was found in endophyte-inoculated plants, which might be due to reduced sodium uptake, and ion compartmentalization by maintaining the Na+/K+ balance in cells. Even though we did not measure sodium levels in plants, we directly established the ability of NKA32 to chelate sodium ions endogenously (tested in in vitro condition). This explains that how the bacterium protects the plant (rice) even under very high soil salinity level (~ 74 mg/kg). Furthur, osmotic stress could have been balanced through accumulation of osmoregulants such as proline, both in bacteria and stressed plants. As indicated by the study, NKA32 inoculation (in saline soil) showed significant impact on the protein, carbohydrate, and RWC. Soluble sugar and protein contents protect cells from the harmful effects of salt stress and help to maintain ionic balance, whereas RWC indicates plants’ adaptation to salinity (Liu et al. 2022). Tian et al. (2017) reported that photosynthetic system II and fresh weight of wheat decreased adversely due to reduction in RWC. Furthermore, in the present study, non-enzymatic antioxidants such as phenols, flavonoids and proline were produced maximum in NKA32-treated plants grown under saline soil. Ahmed et al. (2021) reported that inoculation with ST-PGPE results in accumulation of the proline in rice plants to lessen the impact of salinity stress. It is also reported that increased accumulation of flavonoids and phenolics in ST-PGPB treated plants exposed to salinity stress may aid in the suppression of ROS and decomposed H2O2 to avert oxidative stress (Asif et al. 2023). Yoolong et al. (2019) reported that Streptomyces inoculation to rice suppressed salt-induced damage by increasing proline levels, chlorophyll content, K+/Na+ ratio, RWC and ROS.
In the present study, DPPH scavenging, hydroxyl radical scavenging, SOD and APX activity were higher in treated and untreated plants under saline soil as compared to unstressed plants. Excessive ROS production causes the breakdown of nucleic acids, lipids, and proteins under salinity stress, resulting in loss of cellular strength, spillage, and dehydration (Abd_Allah et al. 2018). In response to salinity stress, mustard and maize plants showed increased SOD, APX, DPPH scavenging, and OH radical scavenging activity (Khan et al. 2023). Although salt stress enhanced antioxidant enzymes, inoculation with B. subtilis or P. fluorescens further increased antioxidant activity and energized the removal of toxic ROS (El-Esawi et al. 2019). SOD activity is seen to function as a catalytic agent for scavenging free radicals (Das and Roychoudhury 2014), while APX isoenzyme facilitates the reaction that converts H2O2 to H2O, with ascorbate acting as an electron donor (Sofo et al. 2015).
Ji et al. (2020) found that rice seedlings inoculated with ACCD-producing ST-PGPB mitigated the negative effects of salt stress by controlling phytohormone (ethylene) and ROS accumulation, promoting ion homeostasis, increasing the expression of stress-respone. Present study found that under saline soil conditions, NKA32-treated rice plants had a lower rate of MDA synthesis, whereas non-inoculated rice plants had a significantly (P < 0.05) higher rate of MDA synthesis. Elevated MDA levels might be due to enhanced Na+ concentration in saline soil, which harmed cell membrane. In plants under salt stress, maintaining an osmotic imbalance is thought to be a defensive strategy against oxidative stress. The inoculation of salt-tolerant bacterial strain reduced the negative effect of salinity due to reduced Na+ uptake by plants, resulting in lesser metabolic disruption in the plants (Shahid et al. 2022). Previously, it has been shown that the inoculation of Bacillus decreased the production of ROS and the amount of MDA under salt stress (Kazerooni et al. 2021). Shahid et al. (2022) reported the role of salt-tolerant PGPB endophytes in the maintenance of plant redox homeostasis.
Under salt stress, the shoot growth of rice plants was found to be slow in untreated plants. This was most likely due to the negative effect of ethylene stress, which initiates a series of processes that result in leaf yellowing, organ senescence, abscission of leaves, and premature death (Zahir et al. 2009; Orozco-Mosqueda et al. 2019). Generally, ethylene is found at very low concentrations (0.01 μl/l) within plant tissues and governs plants’ root and shoot development (Reid 1995), however under abiotic stresses, elevated level (20–25 g/l) of ethylene is produced from the surplus of ACC (Moon et al. 2022). When a plant is stressed by salinity, it synthesizes ACC (precursor of ethylene), increasing the endogenous concentration of ethylene. Increased ACC production, on the other hand, would benefit the bacterium because the ACC exuded from plants under salt stress is sequestered by the surrounding microbes (that express ACCD) and hydrolyzed to ammonia and α-ketobutyrate, which maintains the equilibrium between the internal and external ACC levels (Neshat et al. 2022; Haroon et al. 2023). Decreased ACC inhibits stress-induced ethylene formation in host plants and promotes plant growth. According to Penrose and Glick (2003), this could be due to the ACCD activity of B. altitudinis (> 20 nmol of α-ketobutyrate), which is optimum for stress-induced ethylene levels and effective plant growth promotion assay. Kumar et al. (2021) reported that ST-PGPE B. pumilus strain JPVS11 was found to reduce ethylene in rice by the activity of ACCD, and subsequently promote photosynthetic pigments, proline, and antioxidants. Recently, it was reported that bacterial strains with ACCD activity significantly reduced stress-induced ethylene levels (38% and 22%) in Phaseolus vulgaris and Capsicum annum (Gupta and Pandey 2020; Choudhury et al. 2021). In present study, ACCD–producing B. altitudinis NKA32 caused approximately 72% reduction in ethylene under salt stress, which is greater than the previous records available on ACCD producing endophytic bacteria. Higher germination rate, chlorophyll content, grain yield and biomass of rice plants were also observed when treated with ACCD-positive NKA32 under salinity stress. The reason could be the increased degradation of ACC, along with other PGP traits exhibited by bacteria.
The study highlights the exceptional potential of B. altitudinis NKA32 as an endophytic bacterium for promoting the growth and resilience of rice plants in saline conditions. The ACCD production by NKA32, even at high salinity levels, played an important role in alleviating plant stress by modulating ethylene levels. This capability not only enhances plant growth but also contributes significantly to stress tolerance. Additionally, the observed DPPH scavenging activity by B. altitudinis NKA32 showcases its antioxidant ability, further underlining its role in mitigating the harmful effects of ROS induced by salinity stress. The combination of ACCD and antioxidant activities as well as Na+ ion accumulation showcases B. altitudinis NKA32 as a promising candidate for sustainable agricultural practices, offering a holistic approach to enhance plant health and productivity in challenging environments. Exploration of root colonization potential of B. altitudinis NKA32 across varying salt concentrations provided insights into the adaptive strategies of the bacterium.
Conclusion
Endophytes have proved to be important microbes that provide numerous advantages to their host plants. The current study suggests that the endophytic bacterium had significant effect on stress tolerance traits and growth parameters of O. sativa L. The application of ST-PGPE to the plants resulted in enhanced biochemical and salt tolerance properties along with significant reduction in ethylene stress and MDA content, which are growth inhibitors. The study unfolds mechanism of action of a ST-PGPE strain under salt stress, however, modern omic-based techniques can provide better insights into their signaling pathways and related responses. Futher, field trials are required to be done before exploring the potential of B. altitudinis NKA32 as an effective bioinoculant for enhancing productivity of saline agro-ecosystems in a sustainable manner.
Data availability
The article contains all of the publication-related data.
References
Abd Allah EF, Alqarawi AA, Hashem A, Radhakrishnan R, Al-Huqail AA, Al-Otibi FO, Malik JA, Alharbi RI, Egamberdieva D (2018) Endophytic bacterium Bacillus subtilis (BERA 71) improves salt tolerance in chickpea plants by regulating the plant defense mechanisms. J Plant Interact 13(1):37–44. https://doi.org/10.1080/17429145.2017.1414321
Acosta-Motos JR, Ortuño MF, Bernal-Vicente A, Diaz-Vivancos P, Sanchez-Blanco MJ, Hernandez JA (2017) Plant responses to salt stress: adaptive mechanisms. Agron 7(1):18. https://doi.org/10.3390/agronomy7010018
Afridi MS, Mahmood T, Salam A, Mukhtar T, Mehmood S, Ali J, Khatoon Z, Bibi M, Javed MT, Sultan T, Chaudhary HJ (2019) Induction of tolerance to salinity in wheat genotypes by plant growth promoting endophytes: involvement of ACC deaminase and antioxidant enzymes. Plant Physiol Biochem 139:569–577. https://doi.org/10.1016/j.plaphy.2019.03.041
Ahmed S, Heo TY, Roy Choudhury A, Walitang DI, Choi J, Sa T (2021) Accumulation of compatible solutes in rice (Oryza sativa L.) cultivars by inoculation of endophytic plant growth-promoting bacteria to alleviate salt stress. Appl Biol Chem 64:68. https://doi.org/10.1186/s13765-021-00638-x
Al Mahmud J, Bhuyan MB, Anee TI, Nahar K, Fujita M, Hasanuzzaman M (2019) Reactive oxygen species metabolism and antioxidant defense in plants under metal/metalloid stress. In: Hasanuzzaman M, Hakeem K, Nahar K, Alharby H (eds) Plant abiotic stress tolerance. Springer, Cham, pp 221–257
Arnon DI, McSwain BD, Tsujimoto HY, Wada K (1974) Photochemical activity and components of membrane preparations from blue–green algae. I. Coexistence of two photosystems in relation to chlorophyll a and removal of phycocyanin. Biochimica Et Biophysica Acta (BBA) Bioenergetics 357(2):231–245. https://doi.org/10.1016/0005-2728(74)90063-2
Arora NK, Mishra J, Singh P, Fatima T (2024) Salt‐tolerant plant growth-promoting Pseudomonas atacamensis KSS‐6 in combination with organic manure enhances rice yield improves nutrient content and soil properties under salinity stress. J Basic Microbiol. https://doi.org/10.1002/jobm.202300767
Arora NK, Fatima T, Mishra J, Mishra I, Verma S, Verma R, Verma M, Bhattacharya A, Verma P, Mishra P, Bharti C (2020) Halo-tolerant plant growth promoting rhizobacteria for improving productivity and remediation of saline soils. J Adv Res 26:69–82. https://doi.org/10.1016/j.jare.2020.07.003
Asif S, Jan R, Kim N, Asaf S, Lubna Khan MA, Kim EG, Jang YH, Bhatta D, Lee IJ, Kim KM (2023) Halotolerant endophytic bacteria alleviate salinity stress in rice (Oryza sativa L.) by modulating ion content, endogenous hormones, the antioxidant system and gene expression. BMC Plant Biol 23(1):494. https://doi.org/10.1186/s12870-023-04517-z
Bates LS, Waldren RA, Teare ID (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207. https://doi.org/10.1007/BF00018060
Bergey DH (1994) Bergey’s manual of determinative bacteriology. Lippincott Williams & Wilkins
Bhutani N, Maheshwari R, Sharma N, Kumar P, Dang AS, Suneja P (2022) Characterization of halo-tolerant plant growth promoting endophytic Bacillus licheniformis MHN 12. JGEB 20(1):113. https://doi.org/10.1186/s43141-022-00407-3
Chang W, Chen W, Hu Y, Wang Z (2024) Bacillus altitudinis LZP02 improves rice growth by reshaping the rhizosphere microbiome. Plant Soil 498(1):279–294. https://doi.org/10.1007/s11104-023-06435-3
Chompa SS, Zuan A, Amin AM, Hun TG, Hamzah A, Nabayi A (2024) Carrier based liquid bioformulation of salt-tolerant PGPR Bacillus species for prolonged survivability. Sains Malaysiana 53(5):1055–1065. https://doi.org/10.17576/jsm-2024-5305-07
Choudhury AR, Choi J, Walitang DI, Trivedi P, Lee Y, Sa T (2021) ACC deaminase and indole acetic acid producing endophytic bacterial co-inoculation improves physiological traits of red pepper (Capsicum annum L.) under salt stress. J Plant Physiol 267:153544. https://doi.org/10.1016/j.jplph.2021.153544
Cruz Barrera M, Jakobs-Schoenwandt D, Persicke M, Gómez MI, Ruppel S, Patel AV (2020) Anhydrobiotic engineering for the endophyte bacterium Kosakonia radicincitans by osmoadaptation and providing exogenously hydroxyectoine. World J Microbiol Biotechnol 36:1–16. https://doi.org/10.1007/s11274-019-2780-0
Das K, Roychoudhury A (2014) Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front Environ Sci 2:53. https://doi.org/10.3389/fenvs.2014.00053
Das A, Chaudhuri D, Ghate NB, Chatterjee A, Mandal N (2014) Phytochemical analysis, antioxidant and anticancer potential of leaf extracts from edible greater yam, Dioscorea alata L., from North-East India. Int J Phytopharm 5(2):109–119
Dif G, Belaouni HA, Yekkour A, Goudjal Y, Djemouai N, Peňázová E, Čechová J, Berraf-Tebbal A, Eichmeier A, Zitouni A (2022) Performance of halotolerant bacteria associated with Sahara-inhabiting halophytes Atriplex halimus L. and Lygeum spartum L. ameliorate tomato plant growth and tolerance to saline stress: from selective isolation to genomic analysis of potential determinants. World J Microbiol Biotechnol 38(1):16. https://doi.org/10.1007/s11274-021-03203-2
El-Esawi MA, Al-Ghamdi AA, Ali HM, Alayafi AA (2019) Azospirillum lipoferum FK1 confers improved salt tolerance in chickpea (Cicer arietinum L.) by modulating osmolytes, antioxidant machinery and stress-related genes expression. Environ Exp Bot 159:55–65. https://doi.org/10.1016/j.envexpbot.2018.12.001
El-Tarabily KA (2008) Promotion of tomato (Lycopersicon esculentum Mill.) plant growth by rhizosphere competent 1-aminocyclopropane-1-carboxylic acid deaminase-producing streptomycete actinomycetes. Plant Soil 308:161–174. https://doi.org/10.1007/s11104-008-9616-2
Etesami H, Alikhani HA, Hosseini HM (2015) Indole-3-acetic acid (IAA) production trait, a useful screening to select endophytic and rhizosphere competent bacteria for rice growth promoting agents. MethodsX 2:72–78. https://doi.org/10.1016/j.mex.2015.02.008
El-shakh ASA, Kakar KU, Wang X, et al. (2015) Controlling bacterial leaf blight of rice and enhancing the plant growth with endophytic and rhizobacterial Bacillus strains. Toxicol Environ Chem 97(6):766–785
Fan D, Subramanian S, Smith DL (2020) Plant endophytes promote growth and alleviate salt stress in Arabidopsis thaliana. Sci Rep 10:12740. https://doi.org/10.1038/s41598-020-69713-5
Fasim F, Ahmed N, Parsons R, Gadd GM (2002) Solubilization of zinc salts by a bacterium isolated from the air environment of a tannery. FEMS Microbiol Lett 213(1):1–6. https://doi.org/10.1111/j.1574-6968.2002.tb11277.x
Fatima T, Arora NK (2021) Pseudomonas entomophila PE3 and its exopolysaccharides as biostimulants for enhancing growth, yield and tolerance responses of sunflower under saline conditions. Microbiol Res 244:126671. https://doi.org/10.1016/j.micres.2020.126671
Fatima T, Mishra I, Verma R, Arora NK (2020) Mechanisms of halotolerant plant growth promoting Alcaligenes sp. involved in salt tolerance and enhancement of the growth of rice under salinity stress. 3Biotech 10:361. https://doi.org/10.1007/s13205-020-02348-5
Fatnassi IC, Chiboub M, Saadani O, Jebara M, Jebara SH (2015) Impact of dual inoculation with Rhizobium and PGPR on growth and antioxidant status of Vicia faba L. under copper stress. C R Biol 338(4):241–254. https://doi.org/10.1016/j.crvi.2015.02.001
Glick BR (2014) Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res 169(1):30–39. https://doi.org/10.1016/j.micres.2013.09.009
Glick BR, Karaturovíc DM, Newell PC (1995) A novel procedure for rapid isolation of plant growth promoting pseudomonads. Can J Microbiol 41(6):533–536. https://doi.org/10.1139/m95-070
Ghosh A, Pramanik K, Bhattacharya S, Mondal S, Ghosh SK, Maiti TK (2022) A potent cadmium bioaccumulating Enterobacter cloacae strain displays phytobeneficial property in Cd-exposed rice seedlings. Curr Res Micro Sci 3:100101. https://doi.org/10.1016/j.crmicr.2021.100101
Gowtham HG, Singh B, Murali M, Shilpa N, Prasad M, Aiyaz M, Amruthesh KN, Niranjana SR (2020) Induction of drought tolerance in tomato upon the application of ACC deaminase producing plant growth promoting rhizobacterium Bacillus subtilis Rhizo SF 48. Microbiol Res 234:126422. https://doi.org/10.1016/j.micres.2020.126422
Gupta S, Pandey S (2019) ACC deaminase producing bacteria with multifarious plant growth promoting traits alleviates salinity stress in French bean (Phaseolus vulgaris) plants. Front Microbiol 10:1506. https://doi.org/10.3389/fmicb.2019.01506
Gupta S, Pandey S (2020) Enhanced salinity tolerance in the common bean (Phaseolus vulgaris) plants using twin ACC deaminase producing rhizobacterial inoculation. Rhizosphere 16:100241. https://doi.org/10.1016/j.rhisph.2020.100241
Gupta S, Pandey S, Kotra V (2023) Assessing the role of ACC deaminase-producing bacteria in alleviating salinity stress and enhancing zinc uptake in plants by altering the root architecture ofrench bean (Phaseolus vulgaris) plants. Planta 258:3. https://doi.org/10.1007/s00425-023-04159-3
Haroon U, Munis MFH, Liaquat F (2023) Biofilm formation and flocculation potential analysis of halotolerant Bacillus tequilensis and its inoculation in soil to mitigate salinity stress of chickpea. Physiol Mol Biol Plants 29:277–288. https://doi.org/10.1007/s12298-023-01280-1
Haque MA, Simo, Prodhan MY, Ghosh S, Hossain MS, Rahman A, Sarker UK, Haque MA (2023) Enhanced rice plant (BRRI-28) growth at lower doses of urea caused by diazinon mineralizing endophytic bacterial consortia and explorations of relevant regulatory genes in a Klebsiella sp strain HSTU-F2D4R. Arc Microbiol 205(6):231
Herpell JB, Alickovic A, Diallo B, Schindler F, Weckwerth W (2023) Phyllosphere symbiont promotes plant growth through ACC deaminase production. ISME J 17(8):1267–1277. https://doi.org/10.1038/s41396-023-01428-7
Honma M, Shimomura T (1978) Metabolism of 1-aminocyclopropane-1-carboxylic acid. Agric Biol Chem 42(10):1825–1831. https://doi.org/10.1080/00021369.1978.10863261
Irigoyen JJ, Einerich DW, Sánchez-Díaz M (1992) Water stress induced changes in concentrations of proline and total soluble sugars in nodulated alfalfa (Medicago sativa) plants. Physiol Plant 84(1):55–60. https://doi.org/10.1111/j.1399-3054.1992.tb08764.x
Inostroza NG, Barra PJ, Wick LY, Mora ML, Jorquera MA (2017) Effect of rhizobacterial consortia from undisturbed arid‐and agro‐ecosystems on wheat growth under different conditions. Lett Appl Microbiol 64(2):158–163. https://doi.org/10.1111/lam.12697
Jhuma TA, Rafeya J, Sultana S, Rahman MT, Karim MM (2021) Isolation of endophytic salt-tolerant plant growth-promoting rhizobacteria from Oryza sativa and evaluation of their plant growth-promoting traits under salinity stress condition. Front Sustain Food Syst 5:687531. https://doi.org/10.3389/fsufs.2021.687531
Ji J, Yuan D, Jin C (2020) Enhancement of growth and salt tolerance of rice seedlings (Oryza sativa L.) by regulating ethylene production with a novel halotolerant PGPR strain Glutamicibacter sp. YD01 containing ACC deaminase activity. Acta Physiol Plant 42:42. https://doi.org/10.1007/s11738-020-3034-3
Kalita P, Tapan BK, Pal TK, Kalita R (2013) Estimation of total flavonoids content (TFC) and anti oxidant activities of methanolic whole plant extract of Biophytum sensitivum Linn. JDDT 3(4):33–37. https://doi.org/10.22270/jddt.v3i4.546
Kakar KU, Ren X-l, Nawaz Z, et al. (2016) A consortium of rhizobacterial strains and biochemical growth elicitors improve cold and drought stress tolerance in rice (Oryza sativa L.). Plant Biol 18(3):471–483. https://doi.org/10.1111/plb.12427
Kazerooni EA, Maharachchikumbura SS, Adhikari A, Al-Sadi AM, Kang SM, Lee IJ (2021) Rhizospheric Bacillus amyloliquefaciens protects Capsicum annuum cv. Geumsugangsan from multiple abiotic stresses via multifarious plant growth-promoting attributes. Front Plant Sci 12:669693
Khan I, Hussan S, Chattha MU (2023) Acetic acid mitigates salinity-induced toxic effects in wheat by maintaining photosynthetic efficiency, antioxidant activities, ionic homeostasis, and synthesis of stress-protection hormones and osmolytes. Gesunde Pflanzen 75:979–992. https://doi.org/10.1007/s10343-022-00759-3
Khare E, Mishra J, Arora NK (2018) Multifaceted interactions between endophytes and plant: developments and prospects. Front Microbiol 9:2732. https://doi.org/10.3389/fmicb.2018.02732
Kiani R, Arzani A, Mirmohammady Maibody SAM (2021) Polyphenols, flavonoids, and antioxidant activity involved in salt tolerance in wheat, Aegilops cylindrica and their amphidiploids. Front Plant Sci 12:64622. https://doi.org/10.3389/fpls.2021.646221
Kimura M (1980) Simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Kumar A, Singh S, Mukherjee A, Rastogi RP, Verma JP (2021) Salt-tolerant plant growth-promoting Bacillus pumilus strain JPVS11 to enhance plant growth attributes of rice and improve soil health under salinity stress. Microbiol Res 242:126616. https://doi.org/10.1016/j.micres.2020.126616
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 70 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Kushwaha P, Srivastava R, Pandiyan K, Singh A, Chakdar H, Kashyap PL, Saxena AK (2021) Enhancement in plant growth and zinc biofortification of chickpea (Cicer arietinum L.) by Bacillus altitudinis. J Soil Sci Plant Nutr 21:922–935. https://doi.org/10.1007/s42729-021-00411-5
Li X, Geng X, Xie R (2016) The endophytic bacteria isolated from elephant grass (Pennisetum purpureum Schumach) promote plant growth and enhance salt tolerance of Hybrid Pennisetum. Biotechnol Biofuels 9:190. https://doi.org/10.1186/s13068-016-0592-0
Litalien A, Zeeb B (2020) Curing the earth: a review of anthropogenic soil salinization and plant-based strategies for sustainable mitigation. Sci Total Environ 698:134235. https://doi.org/10.1016/j.scitotenv.2019.134235
Liu C, Mao B, Yuan D, Chu C, Duan M (2022) Salt tolerance in rice: Physiological responses and molecular mechanisms. Crop J 10(1):13–25. https://doi.org/10.1016/j.cj.2021.02.010
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275. https://doi.org/10.1016/S0021-9258(19)52451-6
Mahmoud OMB, Hidri R, Talbi-Zribi O, Taamalli W, Abdelly C, Djébali N (2020) Auxin and proline producing rhizobacteria mitigate salt-induced growth inhibition of barley plants by enhancing water and nutrient status. South Afr J Bot 128 209–217. https://doi.org/10.1016/j.sajb.2019.10.023
Mahgoub HAM, Fouda A, Eid AM (2021) Biotechnological application of plant growth-promoting endophytic bacteria isolated from halophytic plants to ameliorate salinity tolerance of Vicia faba L. Plant Biotechnol Rep 15:819–843. https://doi.org/10.1007/s11816-021-00716-y
Minh LT, Khang DT, Ha PT, Tuyen PT, Minh TN, Quan NV, Xuan TD (2016) Effects of salinity stress on growth and phenolics of rice (Oryza sativa L.). Int Lett Nat Sci 57:1–10. https://doi.org/10.56431/p-f1p658
Mishra P, Mishra J, Arora NK (2021) Plant growth promoting bacteria for combating salinity stress in plants–Recent developments and prospects: a review. Microbiol Res 252:126861. https://doi.org/10.1016/j.micres.2021.126861
Moon YS, Ali S (2022) Possible mechanisms for the equilibrium of ACC and role of ACC deaminase-producing bacteria. Appl Microbiol Biotechnol 106:877–887. https://doi.org/10.1007/s00253-022-11772-x
Mukhtar T, Rehman SU, Smith D, Sultan T, Seleima MF, Alsadon AA, Amna Ali S, Chaudhary HJ, Solieman TH, Ibrahim AA (2020) Mitigation of heat stress in Solanum lycopersicum L. by ACC-deaminase and exopolysaccharide producing Bacillus cereus: effects on biochemical profiling. Sustainability 12(6):2159. https://doi.org/10.3390/su12062159
Musson G, McInroy JA, Kloepper JW (1995) Development of delivery systems for introducing endophytic bacteria into cotton. Biocontrol Sci Technol 5(4):407–416. https://doi.org/10.1080/09583159550039602
Nawaz A, Shahbaz M, Asadullah Imran A, Marghoob MU, Imtiaz M, Mubeen F (2020) Potential of salt tolerant PGPR in growth and yield augmentation of wheat (Triticum aestivum L.) under saline conditions. Front Microbiol 11:2019. https://doi.org/10.3389/fmicb.2020.02019
Neshat M, Abbasi A, Hosseinzadeh A (2022) Plant growth promoting bacteria (PGPR) induce antioxidant tolerance against salinity stress through biochemical and physiological mechanisms. Physiol Mol Biol Plants 28:347–361. https://doi.org/10.1007/s12298-022-01128-0
Nowell J, Parules J (1980) Preparation of experimental tissue for scanning electron microscopy, Vol. 2. AMSO-HARE, Chicago, IL
Odlare M, Pell M, Svensson K (2008) Changes in soil chemical and microbiological properties during 4 years of application of various organic residues. Waste Manag 28(7):1246–1253
Orozco-Mosqueda MDC, Duan J, DiBernardo M, Zetter E, Campos-García J, Glick BR, Santoyo G (2019) The production of ACC deaminase and trehalose by the plant growth promoting bacterium Pseudomonas sp. UW4 synergistically protect tomato plants against salt stress. Front Microbiol 10:1392. https://doi.org/10.3389/fmicb.2019.01392
Oviya G, Rangasamy A, Ariyan M (2023) Halotolerant nodule rhizobial and passenger endophytes alleviates salinity stress in groundnut (Arachis hypogaea L.). J Plant Growth Regul 42:6620–6635. https://doi.org/10.1007/s00344-023-10919-y
Parejo I, Viladomat F, Bastida J, Rosas-Romero A, Saavedra G, Murcia MA, Jiménez AM, Codina C (2003) Investigation of Bolivian plant extracts for their radical scavenging activity and antioxidant activity. Life Sci 73(13):1667–1681. https://doi.org/10.1016/S0024-3205(03)00488-0
Patel M, Vurukonda SSKP, Patel A (2023) Multi-trait halotolerant plant growth-promoting bacteria mitigate induced salt stress and enhance growth of Amaranthus Viridis. J Soil Sci Plant Nutr 23:1860–1883. https://doi.org/10.1007/s42729-023-01143-4
Penrose DM, Glick BR (2003) Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol Plant 118(1):10–15. https://doi.org/10.1034/j.1399-3054.2003.00086.x
Rajkumar M, Ae N, Freitas H (2009) Endophytic bacteria and their potential to enhance heavy metal phytoextraction. Chemosphere 77(2):153–160. https://doi.org/10.1016/j.chemosphere.2009.06.047
Rasheed F, Sehar Z, Fatma M (2022) Involvement of ethylene in reversal of salt stress by salicylic acid in the presence of sulfur in mustard (Brassica juncea L.). J Plant Growth Regul 41:3449–3466. https://doi.org/10.1007/s00344-021-10526-9
Reid MS (1995) Ethylene in plant growth, development, and senescence. In: Davies PJ (ed) Plant hormones. Springer, Dordrecht, pp 486–508. https://doi.org/10.1007/978-94-011-0473-9_23
Rodríguez Coca LI, García González MT, Gil Unday Z, Jiménez Hernández J, Rodríguez Jáuregui MM, Fernández Cancio Y (2023) Effects of sodium salinity on rice (Oryza sativa L.) cultivation: a review. Sustainability 15(3):1804. https://doi.org/10.3390/su15031804
Romero-Munar A, Aroca R (2023) A non-K+-solubilizing PGPB (Bacillus megaterium) increased K+ deprivation tolerance in Oryza sativa seedlings by up-regulating root K+ transporters. Plant Physiol Biochem 196:774–782. https://doi.org/10.1016/j.plaphy.2023.02.027
Roy Choudhury A, Trivedi P, Choi J, Madhaiyan M, Park JH, Choi W, Walitang DI, Sa T (2023) Inoculation of ACC deaminase producing endophytic bacteria down-regulates ethylene induced PR-signaling in red pepper (Capsicum annum L.) under salt stress. Physiol Plant 175(2):13909. https://doi.org/10.1111/ppl.13909
Sagar A, Sayyed RZ, Ramteke PW, Sharma S, Marraiki N, Elgorban AM, Syed A (2020) ACC deaminase and antioxidant enzymes producing halophilic Enterobacter sp. PR14 promotes the growth of rice and millets under salinity stress. Physiol Mol Biol Plants 26:1847–1854. https://doi.org/10.1007/s12298-020-00852-9
Saghafi D, Ghorbanpour M, Shirafkan Ajirloo H (2019) Enhancement of growth and salt tolerance in Brassica napus L. seedlings by halotolerant Rhizobium strains containing ACC-deaminase activity. Plant Physiol Rep 24:225–235. https://doi.org/10.1007/s40502-019-00444-0
Sahu PK, Singh S, Singh UB, Chakdar H, Sharma PK, Sarma BK, Teli B, Bajpai R, Bhowmik A, Singh HV, Saxena AK (2021) Inter-genera colonization of Ocimum tenuiflorum endophytes in tomato and their complementary effects on na+/k+ balance, oxidative stress regulation, and root architecture under elevated soil salinity. Front Microbiol 12:744733. https://doi.org/10.3389/fmicb.2021.744733
Sarker PK, Karmoker D, Shohan MUS (2023) Effects of multiple halotolerant rhizobacteria on the tolerance, growth, and yield of rice plants under salt stress. Folia Microbiol 68:55–72. https://doi.org/10.1007/s12223-022-00997-y
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal biochem 160(1):47–56. https://doi.org/10.1016/0003-2697(87)90612-97
Shahid M, Shah AA, Basit F, Noman M, Zubair M, Ahmed T, Naqqash T, Manzoor I, Maqsood A (2020) Achromobacter sp. FB-14 harboring ACC deaminase activity augmented rice growth by upregulating the expression of stress-responsive CIPK genes under salinity stress. Braz J Microbiol 51:719–728. https://doi.org/10.1007/s42770-019-00199-8
Shahid M, Al-Khattaf FS, Danish M, Zeyad MT, Atef Hatamleh A, Mohamed A, Ali S (2022) PGPR Kosakonia Radicincitans KR-17 increases the salt tolerance of radish by regulating ion-homeostasis, photosynthetic molecules, redox potential, and stressor metabolites. Front Plant Sci 13:919696. https://doi.org/10.3389/fpls.2022.919696
Sheoran P, Basak N, Kumar A, Yadav RK, Singh R, Sharma R, Kumar S, Singh RK, Sharma PC (2021) Ameliorants and salt tolerant varieties improve rice-wheat production in soils undergoing sodification with alkali water irrigation in Indo-Gangetic Plains of India. Agric Water Manag 243:106492. https://doi.org/10.1016/j.agwat.2020.106492
Siddikee MA, Zereen MI, Wu M, Zhang W, Dai CC (2021) Phomopsis liquidambaris reduces ethylene biosynthesis in rice under salt stress via inhibiting the activity of 1-aminocyclopropane-1-carboxylate deaminase. Arch Microbiol 203:6215–6229. https://doi.org/10.1007/s00203-021-02588-w
Singh RP, Jha PN (2022) Transposon mutagenesis of ACC deamination gene alters the proteomic analysis of wheat plant under non-saline and saline stress. JPP 13(1):39–53. https://doi.org/10.1007/s42485-022-00083-4
Sofo A, Scopa A, Nuzzaci M, Vitti A (2015) Ascorbate peroxidase and catalase activities and their genetic regulation in plants subjected to drought and salinity stresses. Int J Mol Sci 16(6):13561–13578. https://doi.org/10.3390/ijms160613561
Sood G, Kaushal R, Panwar G, Dhiman M (2019) Effect of indigenous plant growth-promoting rhizobacteria on wheat (Triticum aestivum L.) productivity and soil nutrients. Commun Soil Sci Plant Anal 50(2):141–152. https://doi.org/10.1080/00103624.2018.1556282
Sunita K, Mishra I, Mishra J, Prakash J, Arora NK (2020) Secondary metabolites from halotolerant plant growth promoting rhizobacteria for ameliorating salinity stress in plants. Front Microbiol 11:567768. https://doi.org/10.3389/fmicb.2020.567768
Tian Y, Ungerer P, Zhang H, Ruban AV (2017) Direct impact of the sustained decline in the photosystem II efficiency upon plant productivity at different developmental stages. J Plant Physiol 212:45–53. https://doi.org/10.1016/j.jplph.2016.10.017
Weatherley P (1950) Studies in the water relations of the cotton plant. I. The field measurement of water deficits in leaves. New Phytol 49(1):81–97
Win KT, Tanaka F, Okazaki K, Ohwaki Y (2018) The ACC deaminase expressing endophyte Pseudomonas spp. enhances NaCl stress tolerance by reducing stress-related ethylene production, resulting in improved growth, photosynthetic performance, and ionic balance in tomato plants. Plant Physiol Biochem 27:599–607. https://doi.org/10.1016/j.plaphy.2018.04.038
Xing J, Wang G, Zhang Q, Liu X, Gu Z, Zhang H, Chen YQ, Chen W (2015) Determining antioxidant activities of lactobacilli cell-free supernatants by cellular antioxidant assay: a comparison with traditional methods. PLoS ONE 10(3):0119058. https://doi.org/10.1371/journal.pone.0119058
Yoolong S, Kruasuwan W, Thanh Phạm HT, Jaemsaeng R, Jantasuriyarat C, Thamchaipenet A (2019) Modulation of salt tolerance in Thai jasmine rice (Oryza sativa L. cv KDML105) by Streptomyces venezuelae ATCC 10712 expressing ACC deaminase. Sci Rep 9(1):1275. https://doi.org/10.1038/s41598-018-37987-5
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67(5):1613. https://doi.org/10.1099/ijsem.0.001755
Yue Z, Shen Y, Chen Y, Liang A, Chu C, Chen C, Sun Z (2019) Microbiological insights into the stress-alleviating property of an endophytic Bacillus altitudinis WR10 in wheat under low-phosphorus and high-salinity stresses. Microorganisms 7(11):508. https://doi.org/10.3390/microorganisms7110508
Yue Z, Chen Y, Wang Y, Zheng L, Zhang Q, Liu Y, Hu C, Chen C, Sun MK, Z, (2022) Halotolerant Bacillus altitudinis WR10 improves salt tolerance in wheat via a multi-level mechanism. Front Plant Sci 13:941388. https://doi.org/10.3389/fpls.2022.941388
Zahir ZA, Ghani U, Naveed M, Nadeem SM (2009) Asghar HN (2009) Comparative effectiveness of Pseudomonas and Serratia sp containing ACC-deaminase for improving growth and yield of wheat (Triticum aestivum L.) under salt-stressed conditions. Arch Microbiol 191:415–424. https://doi.org/10.1007/s00203-009-0466-y
Zhou X, Liu X, Zhao J, Guan F, Yao D, Wu N, Tian J (2021) The endophytic bacterium Bacillus koreensis 181–22 promotes rice growth and alleviates cadmium stress under cadmium exposure. Appl Microbiol Biotechnol 105:8517–8529. https://doi.org/10.1007/s00253-021-11613-3
Acknowledgements
Authors acknowledge National Centre for Microbial Resource (NCMR), Pune, India for conducting 16s rRNA sequencing of bacteria, University Sophisticated Instrumentation Centre (USIC), BBA University Lucknow, for SEM analysis and Advanced Instrumentation Research Facility (AIRF), JNU, India for GC-FID analysis. Chanda Bharti thanks UGC-JRF Grant (No.- F.14-34/2011 (CPP-II) for financial support. NKA is thankful to Biotechnology Industry Research Assistance Council (BIRAC), India for the Grant (No. BFD/RO/B.04/0346/2021-22).
Author information
Authors and Affiliations
Contributions
NKA conceptualized the idea, supervised, reviewed and edited the manuscript. CB performed the experiment, data analysis, figure preparation, and written original draft of manuscript. Writing, reviewing and editing were performed by TF, PM, PV, AB, and BA. The manuscript has been read thoroughly and approved by all authors.
Corresponding author
Ethics declarations
Conflict of interest
There are no significant financial or non-financial interests that the authors need to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bharti, C., Fatima, T., Mishra, P. et al. Salt-tolerant endophytic Bacillus altitudinis NKA32 with ACC deaminase activity modulates physiochemical mechanisms in rice for adaptation in saline ecosystem. Environmental Sustainability 7, 231–249 (2024). https://doi.org/10.1007/s42398-024-00316-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42398-024-00316-w