Abstract
The endophytic fungus Phomopsis liquidambaris is characterized as a plant growth-promoting agent under salt stress, but its mechanism is unknown. Herein, 1-aminocyclopropane-1-carboxylate deaminase (ACCD) from the strain was confirmed that it had the ability of utilizing 1-aminocyclopropane-1-carboxylate as the sole nitrogen source. The full-length ACCD gene was 1152 bp, which encodes a mature protein of 384 amino acids with a molecular mass of 41.53 kDa. The ACCD activity was 3.9-fold in 3 mmol L−1 ACC by qRT-PCR under salt stress comparing with no salt tress. Ethylene production was increased to 34.55–70.60% and reduced the growth of rice by 23–69.73% under salt stress. Inoculation of P. liquidambaris increased root-shoot length, fresh and dry weight, and overall growth of stressed rice seedlings. ACC accumulation, ACC synthase and ACC oxidase activities increased in salt-treated rice seedlings, while they were significantly reduced when P. liquidambaris was inoculated into rice by qRT-PCR. It therefore can be concluded that P. liquidambaris can be used as a plant growth promoting fungus against salt stress and other biotic or abiotic stresses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Salt stress is the most damaging environmental hazard and a major obstacle to crop productivity worldwide, resulting huge economic losses (Ashraf and Ali 2008). The levels of salt stress significantly affect the biosynthesis of ethylene in plants (Siddikee et al. 2011a, b). Ethylene is an important growth hormone that mediates a wide range of plant responses (Arshad and Frankenberger 2002) such as seed germination, fruit ripening, flowering, senescence, root initiation, shoot and root growth, epinastic curvature, and secondary metabolite biosynthesis (Yuan et al. 2016). Low levels of ethylene (as low as 10 µl L−1) appear to enhance root initiation and growth, but plants need relatively higher amounts (approximately 100 µl L−1) of ethylene at the senescence or ripening (Abeles 1992). Under stress conditions, low levels of ethylene appear to act as signaling molecules and quickly transfer signals for defense. Because ethylene is gaseous in nature and rapidly spreads throughout the plant system, it activates the expression of genes encoding defensive enzymes in each cell to protect itself. However, stress causes over-production of ethylene (as high as 25 µl L−1), which is also responsible for the inhibition of root elongation, immature senescence, and death irrespective of developmental stages (Siddikee et al. 2011a, b; Zhang et al. 2018).
Numerous studies reported that plant growth promoting microorganisms (PGPM) facilitate plant growth by various direct and indirect mechanisms. Among these mechanisms, lowering of plant ethylene levels by enzymatic method has been well documented (Kohler et al. 2006). 1-aminocyclopropane-1-carboxylic acid deaminase (ACCD)-producing PGPM is an environmentally safe and cost-effective strategy to overcome salinity stress. PGPM colonize the roots and lower the stress effect by reducing ethylene through mechanisms that include utilization of 1-aminocyclopropane-1-carboxylic acid (ACC), the immediate precursor of ethylene, as a nitrogen and carbon source for its growth (Todorovic and Glick 2008) or by triggering systemic defense responses in plants (Indiragandhi et al. 2008). Numerous reports indicated that plants were inoculated with ACCD-producing PGPR were dramatically more resistant to the deleterious effects of stress ethylene; synthesized as a consequence of stressful conditions such as heavy metals (Burd et al. 2000), flooding (Grichko and Glick 2001), drought (Zahir et al. 2007), and high salt contents (Siddikee et al. 2010, 2011a, b) resulted in longer roots and decreased the inhibition of plant growth (Glick et al. 2007). However, little information is available regarding fungal ACCD and its role in plant growth promotion under stress such as Penicillium citrinum (Jia et al. 2000), Trichoderma asperellum and T. atroviride (Viterbo et al. 2010). However, there is no report on endophytic fungi with ACCD activity and their inoculation effect on plant growth under stress conditions. As endophytic fungi live within the inner tissues of plants without causing visible disease symptoms and help plants to grow and survive (Yuan et al. 2007, 2010) by various direct or indirect mechanisms (Wang et al. 2015); the use of endophytic fungi with ACCD activity may prove useful to facilitate plant growth and increase plant tolerance to salt stress. P. liquidambaris is an endophytic fungus isolated from the inner bark of Bischofia polycarpa (Chen et al. 2013a, 2013b). When applied to the rice, P. liquidambaris can establish mutualistic symbiosis, enhance nitrogen uptake and metabolism in low-N conditions, and thus promote growth and yield. Colonization of P. liquidambaris in peanut showed that it could induce nodulation and increase nitrogen fixation through activating auxin signaling (Zhou et al. 2014; Zhang et al. 2018). The application of P. liquidambaris could have enormous potential in agriculture and industry. ACCD-producing PGPM mainly colonize the root of the plant and can directly utilize ACC as a nitrogen and carbon source.
Based on this information above, we first investigated whether the endophytic fungus P. liquidambaris had ACCD activity by evaluating its ability to utilize ACC as the sole N in vitro. Then, the salt-stress effects on the growth of rice seedlings and the rice seedlings inoculated with P. liquidambaris were evaluated by comparative analysis of ethylene production, relative enzyme activities and reduced root-shoot length and dry mass. Our study provided a potential plant growth-promoting agent for P. liquidambaris under salt stress, which may greatly increase its use in agriculture.
Materials and methods
Fungal strain and rice seeds
Phomopsis liquidambaris was isolated from the inner bark of Bischofia polycarpa (Shi et al. 2004). The fungus was stored at 4 °C on potato dextrose agar (PDA, containing 200 g L−1 potato extract, 20 g L−1 glucose, and 20 g L−1 agar, pH 7.0). The rice cultivar was a japonica subspecies of Oryza sativa L. “Wuyunjing 7,” which was a common cultivar grown in the Jiangsu Province of south-eastern China.
The cloning and sequencing of ACCD gene from P. liquidambaris.
ACC was used as a nitrogen source for P. liquidambaris. The activity was measured according to the reported protocol (Siddikee et al. 2010). P. liquidambaris was inoculated into Jensen’s Nitrogen Free medium (JNF) supplemented with ACC (Sigma-Aldrich Co., St Louis, MO, USA).
Amplification, cloning and sequencing of ACCD gene from P. liquidambaris
Protein sequences of ACCD of Aspergillus flavus (XM_002378519.1), Neosartorya fischeri (XP_001265664.1), Penicillium citrinum (AB038511.1), Trichoderma asperellum (FJ751936), Fusarium oxysporum, and Gibberella zeae PH-1 (XP_385209.1) were obtained from the National Center for Biotechnology Information’s (NCBI) database and aligned using the ClustalW program (Thompson et al. 1994). Primer sets generated by iCODEHOP were chosen based on the conserved protein sequences AYGGNK and AFITDPVYEGKS, (Fig. 1 and ST. 1) and obtained from Invitrogen (Springen, Nanjing, China).
Phylogenetic tree analyses of nucleotide sequence of the ACCD gene (A) and deduced amino acid sequence of ACC deaminase protein (B) from P. liquidambari. Unweighted pair group method with arithmetic mean (UPGMA) Tree constructed using MEGA 7.0 software for the nucleotide and amino acid sequences of the ACCD gene of P. liquidambaris with nucleotide and amino acid sequences of ACCD genes retrieved from NCBI to show the phylogenetic relationship
Fungus genomic DNA was prepared using AxyPrep Multisource Genomic DNA Miniprep Kit (Axygen Biosciences, Union City, CA, USA) following the manufacturer's instruction. The PCR reaction (50 µL) contained 5 µL 10 × PCR buffer, 3 µL 25 mM MgCl2, 1 µL 10 mM dNTP, 1 µL of each primer (25 mM), 2 µL (5.0–10.0 ng) of total DNA, 0.5 U Taq polymerase, and 36.5 µL dd-H2O. DNA were amplified on 96-well thermal cycler (Applied Biosystems, Foster City, CA, USA) and program consisted of an initial denaturation at 95 °C for 5 min followed by 30 cycles of 94 °C for 60 s, 58 °C for 60 s, and 72 °C for 60 s with a final extension of 10 min. Samples (10 µL) of the PCR products were separated on a 1% agarose gel in TAE buffer (90 mM L−1 Tris-acetate, 2 mM l−1 EDTA, pH 8.3) containing 0.5 µg of ethidium bromide mL−1 at 120 V for 25 min., visualized under a UV transilluminator and photographed using the gel documentation system (Alpha Innotech Corp). The PCR products were purified using a DNA Gel Extraction Kit (Axygen Bioscience Inc.). Recovered fragments were cloned into the pMD 19-T vector and Escherichia coli DH5a (TaKaRa) competent cells and sent for sequencing to Invitrogen Corp. (Springen, Nanjing, China). Downstream flanking sequences of ACCD gene of P. liquidambaris were obtained by nested-PCR using different nested gene-specific primers (ST. 1 and SF. 2) and Universal GenomeWalker Kit (Clontech, Mountain View, CA, USA). The upstream flanking sequence was obtained using the same kit with another set of nested specific primers (ST. 1 and SF. 2). The GenBank accession number of the full sequence of P. liquidambaris ACCD gene is KX762324.
Quantitative assay of ACC deaminase activity
According to the previous reports, the activity of ACCD was analyzed (Siddikee et al 2010). Fungi were cultured in PDA for 72 h at 28 °C at 200 rpm, harvested by centrifugation at 6000 × g for 10 min at 4 °C and washed three times with sterilized ddH2O. Fungal pellet were transferred to 10 mL JNF medium supplemented with an aliquot of 60 µL of 0.5 M ACC as the sole nitrogen source to induce ACCD activity. Fungi were harvested again by centrifugation and washed by suspending in 5 mL of 0.1 mol L−1 Tris–HCl, pH 7.0. The pellet was re-suspended in 600 µL 0.1 mol L−1 Tris-HCl, pH 8.5 and 30 µL toluene and vortexed at the highest setting for 30 s. A 100 µL aliquot of the toluenized mycelia was set aside and stored at 4 °C for protein assay at a later time. Two hundred µL toluenized mycelia and 20 µL of 0.5 mol L−1 ACC were taken in a fresh 1.5 mL microcentrifuge tube and incubated at 30 °C for 15 min. Then, 1 mL of 0.56 mol L−1 HCl were added, vortexed for mixing and centrifuged at 12,000 × g for 5 min at room temperature. One ml of the supernatant was separated and vortexed together with 800 µL of 0.56 mol L−1 HCl. Again, 300 µL of the 2, 4-dinitrophenyl hydrazine reagent was added, vortexed and incubated for 30 min at 30 °C. Absorbance was measured at 540 nm immediate after adding 2 mL of 2 mol L−1 NaOH. All sample were carried out in triplicate. ACCD activity was evaluated quantitatively by measuring the amount of α-ketobutyrate produced by the deamination of ACC and expressed as μmol of α-ketobutyrate mg protein−1 h−1. Protein concentrations were use the bovine serum albumin (BSA) as standard (Lowry et al. 1951).
Ethylene production and growth assay of rice seedling under different levels of salt stress
The rice seeds were shocked in 70% ethanol for 15 min, rinsed twice with sterile water, sterilized for 25 min in 0.1% HgCl2, and rinsed six times in sterile distilled water (Yang et al. 2015). The sterilized seeds and the last washing water were placed on PDA/LB plates and incubated for 5 days at 28 °C as sterility checks (Feng et al. 2006). The sterilized seeds were transferred to petri dishes (20 cm diameter, 100 grains per dish), treated with 80 mL of sterilized deionized water and incubated in a growth cabinet {29 °C during the day and 25 °C during the night, 16/8 h photoperiod at 250 µmol m−2 s−1, relative humidity [RH] = 70%} for 3–4 days for germination. Germinated rice seeds were transplanted into pots (25 cm diameter, 35 cm high) containing sterilized 1.2 kg vermiculite and incubated in a growth chamber in the above-mentioned conditions. 14 day-old seedlings were exposed to different levels of salt stress (100, 150, and 200 mmol L−1 NaCl) for 7 days with three split applications to avoid osmotic shock. (I) Seeds and seedlings not treated with salt were used as negative controls-1 (NC1), (II) seeds and seedlings treated with AVG (an inhibitor of ACS activity) and salt were used as negative controls-2 (NC2), (III) seeds and seedlings treated with salt were used as positive controls (PC). The experiment was set with three replications.
Based on the previous reports and slight modification, the ethylene emission of rice seedlings was measured (Siddikee et al. 2011a, b). 14 day-old rice seedlings were irrigated using sterilized deionized water containing 50, 125, 150, or 200 mmol L−1 NaCl to create different levels of salt stress and incubated in a growth chamber at 28 °C, RH 70%, and light intensity 250 µmol m−2 s−1 for 6 h. Seedlings (30 for each treatment) were uprooted and washed using the respective concentrations of saline water to remove soil from the roots and placed inside a 40 × 7 cm glass tube. The amount of ethylene emission was expressed as pmol L−1 of ethylene g−1 dry weight h−1 by comparing the standard curve generated with pure ethylene. 21 day-old seedlings (n = 10) from plastic pots for each treatment were carefully removed for morphological and physiological assay. The length and fresh weight of root-shoot were immediately recorded. The root and shoot of ten seedlings were separated and dried in an oven at 70 °C for approximately 3–4 days until a constant weight was obtained and recorded.
The effect of P. liquidambaris inoculation on the ethylene production and growth of rice seedlings under in-vitro salt stress conditions
P. liquidambaris was cultured in 1 L Erlenmeyer flasks containing 500 mL of sterilized PDB for 3 days at 160 rpm in an orbital shaker at 28 °C. 10 mL of culture broth was separated to collect mycelia. Approximately 3.0 g (equivalent to ± 0.30 g dry weight) fungal mycelia were washed twice with sterile distilled water, re-suspended in JNF medium supplemented with 0.5 mmol L−1 ACC as sole nitrogen source and incubated for 24 h at 30 °C at 200 rpm to induce ACCD activity. Mycelia were harvested, washed, and re-suspended in sterile distilled water and diluted to a final volume of 200 mL. This suspension was used as the fungal agent for the germinating seeds to facilitate colonization. The thoroughly sterilized (see previous section) seeds were randomly divided into two groups and transferred to petri dishes. For the inoculated group (E+), 80 mL of the above-described fungal agent and for the non-inoculated group (E−) 80 mL of sterilized deionized water was added to each dish (Tian et al. 2007). Seeds were germinated and grown for 3–4 days in growth cabinet, transplanted into pots and further grown in growth cabinet in the conditions already mentioned above. Rice seedlings were subjected to the following six treatments; (1) negative control (NC1): no-inoculation and no salt; (2) NC2: no inoculation, 0.03 mol L−1 MgSO4 salt and AVG; (3) positive control (PC): 0.03 mol L−1 MgSO4; (4) PC + P. liquidambaris inoculation (PS + E+): 0.03 M MgSO4 + inoculation; (5) 150 m mol L−1 NaCl stress (SS): seedlings exposed to 150 m mol L−1 NaCl stress; and (6) SS + P. liquidambaris inoculation (SS + E+): seedlings inoculated with fungi and exposed to 150 m mol L−1 NaCl stress. Salt stress was applied to 14 day-old seedlings in three split application to avoid osmotic shock. Fertilization was performed in the form of irrigation of Hoagland nutrient solution as necessary. Three biological replicates per treatment were established. Ethylene production from rice seedling was assayed as described above. Data related to growth of rice seedlings were recorded from 21 day-old seedlings as described above. Based on the data obtained, the salt tolerance index (STI) of rice seedlings was determined (Siddikee et al. 2011a) as: STI = DWS or DWH/DWC (where, DWS = dry weight of plant grown under salt stress, DWH = dry weight of plant grown under salt stress with inoculation of P. liquidambaris and DWC = dry weight of plant grown in control condition (without salt stress and inoculation of P. liquidambaris). Meanwhile, 18–20 seedlings from each treatment were stored at − 80 °C and subsequently used for physiological assay such as ACC content, ACS and ACO activity, chlorophyll content of the leaves and ACCD activity.
Assessment of the effect of rice seedlings inoculated with P. liquidambaris on ACC content and ethylene biosynthesis-related enzyme activities
Based on the existing methods, some modifications were made to measure the ACC concentration in the stems of rice seedlings (Siddikee et al. 2011b). One gram of shoot sample was immediately frozen in liquid nitrogen and ground using a mortar and pestle. ACC from frozen ground tissue was extracted in 5 mL 80% methanol containing butylated hydroxytoluene (BHT, 2 mg L−1) as an antioxidant and incubated at room temperature for 45 min. Samples were centrifuged at 2000 × g at 20 °C for 15 min and were re-suspended in 4 mL methanol and again centrifuged. The combined supernatants were evaporated to dryness under a vacuum in a rotatory evaporator. ACC was determined according to the existing detection scheme (Wachter et al. 1999). Residues were re-suspended in 2 mL distilled water and the upper aqueous phase (0.5 mL) obtained by extraction with dichloromethane was mixed with 0.1 mL HgCl2 (80 mM) in test tubes and sealed with rubber septa. Then, 0.2 mL NaCl solution (40 ml NaOH, 80 mL 12.5% NaCl solution, and 30 mL distilled water) was injected into the tubes, shaken, and incubated for 8 min. One milliliter of the gaseous portion was removed and assayed for ethylene by gas chromatography (GC).
Determination of ACC synthase (ACS) and ACC oxidase (ACO) activities for rice seedlings inoculated with P. liquidambaris
ACS and ACO was the key enzyme for ethylene biosynthesis. The enzyme extracts used to measure the activity of ACS and ACO in vitro were modified according to the existing reports (Siddikee et al. 2011b). ACS activity was assayed by incubating a 100 µL enzyme solution with 100 m mol L−1 2-[4-(2-hydroxyethyl)-1-piperazinyl] ethanesulfonic acid (HEPES)–KOH (pH 8.5), 5 µM PLP, 100 µmol L−1 S-adenosyl-l-methionine (SAM), and test chemicals at given concentrations in a total volume of 400 µL in test tubes and sealed with rubber septa. The reaction mixture was incubated for 15 min at 30 °C and the amount of ACC was determined as described above. For assaying the ACO activity, frozen tissues were pulverized in liquid nitrogen and homogenized in 2 mL g−1 of extraction buffer consisting of 100 m mol L−1 Tris-HCl (pH 7.2), 10% (w/v) glycerol, and 30 m mol L−1 sodium ascorbate. The homogenate was centrifuged at 25,000 × g for 15 min at 4 °C. The supernatant obtained was used for the in vitro ACO assay (Siddikee et al. 2011b). Enzyme activity was assayed at 30 °C for 15 min in 10 mL screw-cap tubes fitted with a Teflon-coated septum containing 1.5 mL of supernatant, 50 µmol L−1 FeSO4, 1 m mol L−1 ACC, and 5% (v/v) CO2. After the incubation period, the quantity of ethylene released into the headspace was determined by GC.
Analysis of ACCD gene from P. liquidambaris by qRT-PCR
Approximately 0.1 g of mycelia (wet weight) was harvested and washed using ddH2O. Mycelia were frozen in liquid nitrogen, ground to powder using mortar and pestle and total RNA was extracted using an E.Z.N.A fungal RNA Kit (Omega, Bio-Tek, Winooski, VT, USA). RNA concentration was measured at 260 and 280 nm using NanoDrop™ 8000 Spectrophotometer and adjusted. Total RNA was used as the template to synthesize the first strand complementary DNA (cDNA) using a PrimeScript RT Reagent Kit with gDNA Eraser (TAKARA, Tokyo, Japan). The product was characterized by electrophoresis on 1.5% agarose gels and 2 µL product used as a template for qRT-PCR. qRT-PCR was performed on the StepOne Real-time PCR system (Applied Biosystems), using SYBR Premix Ex Taq Kit (Tli RNaseH Plus) (TAKARA). The specific primer pairs used for quantitative measurement of the transcription of ACCD gene were designed according to their partial mRNA sequence (ST. 1 and SF. 2). Each 20 µL qRT-PCR reactions mixture contained 2 × SYBR Premix Ex Taq (TAKARA), 10.0 µM each gene-specific primer, 0.4 µL of 50 × ROX Reference Dye, and 2.0 µL of cDNA template. The qRT-PCR cycling conditions were 95 °C for 1 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 45 s, and 72 °C for 30 s. The specificity of amplification was verified by a standard melting curve. The primers used for amplification of the ITS gene of P. liquidambaris used as an internal control are presented in ST. 1. The relative abundance of mRNA was normalized against the levels of ITS Amplification data were analyzed using the delta-delta-Ct (ΔΔCT) method (Livak and Schmittgen 2001). Each sample was amplified in triplicate in each experiment.
The transcriptional level of ACO, ACS, and ACCD gene expression from plant tissues by qRT-PCR
Total RNA was extracted from the shoots of 15 day-old rice seedling using a Plant RNase Mini Kit (Qiagen, Hilden, Germany) and subjected to RT-PCR. RNA concentration assay, cDNA synthesisand qRT-PCR were performed as described above. The specific primer pairs used for quantitative measurement of the transcription of ACO and ACS genes were designed according to their conserved mRNA sequence in rice as follows: ACO (accession: LOC_Os04g10350), ACS (accession: M96672.1) and ACCD gene (accession: LOC_Os02g53330 and LOC_Os01g50060); and actin was used as an internal control (ST. 1). The relative abundance of mRNA was normalized against the levels of actin.
Bioinformatics and statistical analysis
The nucleotide sequences of the ACCD gene were translated into amino acid sequences using http://www.fr33.net/teranslator.php and http://www.expasy.org. Molecular weight and isoelectric point of the proteins were predicted using http://www.expasy.org. The conserved domains were predicted and analyzed using the conserved domain database (CDD) (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Phylogenetic trees of nucleotide sequences and amino acid sequences of the ACCD gene were constructed using the MEGA7.0. Statistical analysis was performed using SPSS software (version 18.0, Chicago, IL, USA). All graphs were generated using Origin software version 8.0. All data are reported as the average of three biological replicates ± standard deviation (SD). One-way ANOVA and LSD test were used to determine significant differences in means among treatments.
Results
P. liquidambaris utilizing ACC as the sole nitrogen on plate
P. liquidambaris grew well in the ACC-supplemented JNF medium whereas no growth was detected in control JNF (SF. 1A). Moreover, nifH gene which fix nitrogen did not amplified from genomic DNA of P. liquidambaris. The average ACCD activity of P. liquidambaris in liquid cultures with ACC as the sole nitrogen source was 7.43 µM α-ketobutyrate mg protein−1 h−1. The results suggested that the strain produced ACCD activity and had an ability of utilizing ACC.
The sequencing of the full-length ACCD gene from P. liquidambaris and phylogenetic classification
A fragment of 756 bp of ACCD gene was successfully amplified using degenerated primers (SF. 1B). Three PCR reactions were carried out sequentially to amplify flanking sequences of the ACCD gene from the genomic DNA of P. liquidambaris using nested gene-specific PCR primers (PL1, PL2, and PL3: ST. 1; SF. 2) and walker-adapter primers (AP1, AP2, and AP3). Although the primers PL1 and PL2 were designed to overlap but PL2 and PL3 were positioned 103 bases apart to facilitate the confirmation of product specificity by size comparison (SF. 1C; SF. 2). It was clear from the size relationships between the product bands of the secondary and tertiary reactions that the targeted sequences were amplified and showed the expected differential shift of 103 bp (SF. 1C). Similarly, the upstream flanking sequence was amplified. Cloning and sequencing of the secondary PCR product uncovered 135-bp and 261-bp long segments located upstream and downstream of the flanking regions, respectively. A 1152-bp long nucleotide sequence of the ACCD gene was sequenced and translated into a protein containing 384 amino acids (SF. 3). The open reading frame (ORF) of the P. liquidambaris ACCD gene encodes a 384 amino acid-containing protein, which contains 5 PLP binding sites with an expected theoretical isoelectric point (pI) is 6.80 and a molecular weight (Mw) is 41.53 kDa. A BLASTn search of the ACCD gene nucleotide sequence of P. liquidambaris showed 77, 74, and 73% homology with the fungi Neurospora tetrasperma FGSC2508, Pantoea sp. EcB2 or Polaromonas sp. EcB7, and Ralstonia sp. EcB11, respectively, and 73% with the bacterium Pseudomonas sp. EcB10. Whereas, the amino acid sequence of ACCD of P. liquidambaris showed 94, 92, and 81% homology with the fungi Diaporthe ampelina, Valsa mali, and Fusarium fujikuroi IMI58289, respectively, and 73% with the bacterium Pseudomonas sp. EcB10 (SF. 4). The conserved glutamate (Glu) and leucine (Leu) amino acid residues that distinguish ACCDs are marked at 302 and 327 with box (SF. 4). Amino acid sequences were aligned by fitting at the PLP binding site with the sequences showed highest identity retrieved from NCBI where 12 regions (underlined) presenting 100% homology with at least 4–10 bases (SF. 4). Phylogram (Fig. 1A) constructed based on nucleotide sequences of ACC gene, P. liquidambaris instead of clustering together with other fungi, clustered separately, but closer to bacteria. On the other hand, phylogeny based on amino acid sequence of ACCD indicated a very close relationship with fungi (Fig. 1B).
Ethylene production and growth assay of rice seedling under different levels of salt stress
Ethylene synthesis increased in rice seedlings with increasing levels of salt stress as compared to the negative control (Fig. 2). Salt stress also caused inhibition of root elongation, overall growth, immature senescence, and death irrespective of developmental stages (Fig. 2). Under 150 mmol L−1 NaCl stress, approximately 50.0% ethylene production increased simultaneously growth reduced almost same extent compared to the control. Thus, this level of salt stress was used in subsequent experiments (Fig. 2). These results indicated that ethylene emission increased greatly, which affected the growth of plant.
Effect of salt stress on ethylene biosynthesis and growth of rice seedlings. Salt stress was applied on 14 day-old seedlings for 6 h to measure ethylene emission after transferring the seedlings to vials and incubating for 4 h. Rice seedlings were grown another 7 days and growth-related data were recorded from 21 day-old seedlings with the following treatments: 0 (NC), 50, 125, 150, and 200 mM NaCl. The values are the means ± SD of three replicates. Bar graphs of each treatment followed by the same letter indicate no significant difference (p ≤ 0.05) according to LSD test
Effect of salt stress and ACCD-producing P. liquidambaris inoculation on ethylene production, ACC content, and ACS and ACO activities in rice seedlings
Ethylene biosynthesis increased in rice seedlings by 47.0% in PC and 54.59% in SS treatment compared to NC-1, whereas ACCD-producing P. liquidambaris inoculation reduced the ethylene emission by 52.5 and 49.68%, respectively (Fig. 3). Compared to NC-1, ACC content in rice seedlings increased by 52.40% in PC and 64.14% in SS treatment. AVG application reduced ACC content in rice seedlings by 45.88% in PC, while ACCD-producing P. liquidambaris inoculation reduced 47.35% in PC and 50.10% in SS compared to NC-1. The in vitro ACS activity was increased by 52.12 and 56.82% in PC and SS, respectively, compared to NC1 (Fig. 3) whereas, P. liquidambaris inoculation reduced by 36.42 and 40.87% compared to PC and SS. However, increased ACS activity was recorded in PC + E+ and SS + E+ treatment compared to NC1. On the other hand, AVG application remarkably reduced ACS activity by 91.96% in PC, but did not result in a complete inhibition. The in vitro ACO activity increased by 66.71% in PC and 85.09% in SS compared to NC1 (Fig. 3). ACCD-producing P. liquidambaris inoculation reduced ACO activity in PC and SS by 56.46 and 56.96%, respectively, compared to non-inoculated PC and SS whereas AVG application reduced by 50.76% in NC-2 compared to PC (Fig. 3). Therefore, ACCD-producing P. liquidambaris was more efficient in reducing ACC content, ACO activity and ethylene production in stressed rice seedlings compared to AVG application.
Effect of salt stress with or without inoculation of P. liquidambaris on ethylene biosynthesis, ACC content, and ACC synthase and ACC oxidase activities in rice seedlings. Salt stresses were applied on 14 day-old seedling for 6 h and ethylene emission was measured after transferring the seedlings to vials and incubating for 4 h. The remaining seedlings were used to measure ACC content and ACC oxidase and ACC synthase activities. ACC synthase activity was determined from the amount of ACC formed with 100 µmol S-adenosyl-l-methionine as the substrate. ACC oxidase was determined by supplying 1 mmol of ACC as the substrate. Rice seedlings were grown in hydroponic conditions with the following treatments: NC-1 0 m mol L−1 NaCl, NC-2 AVG + 0.03 m mol L−1 MgSO4, PC 0.03 m mol L−1 MgSO4, PC + E+ 0.03 m mol L−1 MgSO4 + P. liquidambaris, SS 150 m mol L−1 NaCl, and SS + E+ 150 m mol L−1 NaCl + P. liquidambaris. Each value represents the mean of three replicates; bars indicate standard deviation. Bar graphs of each treatment followed by the same letter indicate no significant difference (p ≤ 0.05) according to LSD test
Analysis of P. liquidambaris ACCD gene and plant ACS, ACO and ACCD gene expression by qRT-PCR under salt stress
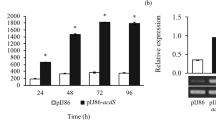
Like ACS and ACO activity, ACS and ACO gene expression also increased in salt-stressed rice seedlings when compared to other treatments (Fig. 4). As expected, ACCD-producing P. liquidambaris inoculation in rice seedlings challenged with salt stress reduced relative ACS, ACO gene expression compared to that in un-inoculated corresponding seedlings. Similarly, seedling treated with AVG, which inhibited ACS activity, showed lower ACS and ACO gene expression comparing with control. However, P. liquidambaris inoculated treatments showed higher relative ACCD gene-related gene expression compared to un-inoculated treatments. Relative fungal ACCD gene expression was increased 3.9-fold in ACC-supplemented JNF medium, while no growth was detected without nitrogen sources (Fig. 4).
Quantitative reverse-transcriptase-PCR analysis of ACC oxidase, ACC synthase, and ACC deaminase-related gene expression in rice seedlings subjected to NC-1 0 m mol L−1 NaCl, NC-2 AVG + 0.03 m mol L−1 MgSO4, PC 0.03 m mol L−1 MgSO4, PC + E+ 0.03 m mol L−1 MgSO4 + P. liquidambaris, SS 150 m mol L−1 NaCl, and SS + E+ 150 m mol L−1 NaCl + P. liquidambaris. Each value represents the mean of three replicates; bars indicate standard deviation
Effect of rice seedlings inoculated with P. liquidambaris on growth and modulation of biomass allocation of rice seedlings under salt stress
ACCD-producing P. liquidambaris inoculated seedlings remained green and continued to grow normally, similar to control seedlings whereas, salt stress seedlings showed bleaching and yellowish leaves as a result growth was significantly better (1.80-fold) (Table 1). Interestingly, growth of rice seedlings was less hampered in PC than in SS treatment. Similarly, the length and dry weight of roots and shoots were higher in PC + E+ than in SS + E+ treatment. Root growth was relatively more affected compared to that of shoots in the presence of a growth inhibitory level of salt (Tables 1 and 2). In relation with leaf color, the content of chl a, chl b, total chl and the ratios of a and b were estimated from the leaf of rice seedlings as a function of six treatments (Table 2). The concentration of chl a exhibited significantly greater variation in relation to different treatments. As predicted, the lowest chlorophyll content and a/b ratio were recorded in salt-stressed rice seedlings and the highest was in seedlings inoculated with P. liquidambaris (Table 2).
In NC1, dry matter in shoots and roots was approximately 60.31% and 39.69%, respectively whereas it was 62.61% and 37.39%, respectively in PC and 62.86 and 37.14%, respectively in SS treatment (Table 2). However, AVG application in PC treatment balanced dry matter allocation to roots and shoots to 38.02 and 61.98%, respectively, whereas P. liquidambaris inoculation was more efficient in balancing the dry matter allocation in roots and shoots of PC (39.80 and 60.20%, respectively) and SS (39.58 and 60.42%, respectively) treatments (Table 2). The tolerance index of ACCD-producing P. liquidambaris inoculated plants (Average STI = 1.45) was significantly higher than un-inoculated SS (STI = 0.58) and PC (STI = 0.71). The data presented in Tables 1 and 2 provide further evidence that ACCD-producing P. liquidambaris inoculation increased dry matter allocation toward roots by reducing the negative effect of stress ethylene on root cells of salt-stressed plants, consequently increasing salt adaptation.
Discussion
Identification, cloning, and characterization of the ACCD gene from P. liquidambaris
It is now established that microorganisms containing ACCD can break cyclic amino acid ACC into α-ketobutyrate and NH3 and utilize it as carbon and nitrogen sources for their growth (Glick et al. 1999). P. liquidambaris grew well in ACC-supplemented JNF medium indicating its ability to cleave the cyclopropane ring of ACC and utilize it as a nitrogen and carbon source whereas no growth was observed in control JNF (SF. 1A). Similar results of fungi and PGPR have been reported (Viterbo et al. 2010). Degenerated primers successfully amplified a 756-bp of ACCD gene (SF. 1B) and further confirmed its identification by cloning, sequencing, and BLASTn search. In some studies, degenerate primers were used to amplify a 531 bp fragment, and almost the same conserved region was considered in the design. The 1152 bp nucleotide sequence of ACCD gene was amplified by nested PCR (SF. 3) (Viterbo et al. 2010). However, some studies have sequenced the 1118 bp fragment in t. asperellum and the 1233 bp fragment in P. citrinum. The ORF of ACCD gene of Liquidambar formosana encodes a protein of 383 amino acids, which contains five PLP binding sites. The expected molecular weight is 41.53 kDa. The ORFs of T. asperellum and P. citrinum encode proteins of 348 and 360 amino acids with expected molecular weight of 37 kDa (Viterbo et al. 2010) and 39.2 kDa (Jia et al. 2000), respectively. An alignment of amino acid sequences of ACCD of P. liquidambaris showed highest 94% similarity with fungi D. ampelina and 73% with bacteria Pseudomonas sp. (SF. 4). It has been reported that 52% similarity of amino acid sequences between P. citrinum and Pseudomonas sp. and 54% homology between T. asperellum and P. putida. Glu and Leuresidues are conserved in true ACCD and essential for ACCD activity (Jia et al. 2000; Viterbo et al. 2010). The conserved Glu and Leu amino acid residues were located at the position of 302 and 327, respectively in P. liquidambaris. (SF. 4), but at Glu297 and Leu322 in Pseudomonas sp. and at Glu309 and leu334 in T. asperellum (Viterbo et al. 2010). Amino acid sequence alignment further showed that 4–10 amino acids of 12 regions matched 100% (SF. 4) and the six regions of 9–11 residues showed more than 80% homology (Jia et al. 2000). Phylogenetic tree analyses of the nucleotide sequence of the ACCD gene and deduced amino acid sequence of ACCD of P. liquidambaris were compared with those of the ACCD gene and ACCD retrieved from NCBI to ascertain their phylogenetic relationship (Fig. 1). The phylogram (Fig. 1A) of the ACCD gene gene showed that P. liquidambaris clustered separately, but closer to bacteria instead of clustering together with the other fungi. On the other hand, the phylogram of amino acid sequence of ACCD of P. liquidambaris showed a close relation with other fungi (Fig. 1B). This result was consistent with the phylogeny of fungal ACCD protein clustered together and commented that closely related strains possess similar ACCD (Nasimento et al. 2014). Therefore, it could be assumed that the ACCD gene was horizontally transferred in P. liquidambaris. ACCD activity of P. liquidambaris was comparable to those in T. asperellum T203 and T. atroviride (Viterbo et al. 2010). Another research reported that ACC synthase activity accumulated in the cells of P. citrinum and then induced ACCD activity (Jia et al. 2000). While this was not verified in our study, we assumed that ACCD activity of P. liquidambaris could be induced by ACC. Maybe, there are some differencesin the level of ACCD activity in different organisms (Glick 2005).
Effect of salt stress and P. liquidambaris inoculation on ethylene production, root elongation, ACC content, ACS and ACO activity and related gene expression in rice seedlings
Ethylene synthesis increased with levels of salt stress (Fig. 2) like other different biotic and abiotic stresses. Similar to reports were stressed seedlings showed bleaching and yellowish leaves, probably because of ethylene overproduction, while ACCD-producing P. liquidambaris inoculated plants remained green and continued normal growth toward completing their life cycle, similar to control seedlings (Siddikee et al. 2010, 2011a, b). However, ethylene sensitivity differed from species to species (Zapata et al. 2007) and even depends on environmental conditions such as light (Mortensen 1989), salinity O3, and CO2 (Morgan and Drew 1997). AVG, which was known to inhibit ACS activity, should reduce ACC concentration, ACO activity, and stress level ethylene production (Madhaiyan et al. 2007). Accumulating evidence also indicates that PGPM producing-ACCD colonize roots and reduce stress effects by reducing ethylene by utilizing ACC, the immediate precursor of ethylene, as the nitrogen and carbon source (Siddikee et al. 2010, 2011a, b).
Various biotic and abiotic stresses, including salinity, increase the concentrations of ACC and enzyme activities involved in ethylene metabolism such as ACS and ACO (Abeles et al. 1992; Siddikee et al. 2011b). As a rate-limiting enzyme, ACS is the major target for the regulation of ethylene production under stresses (Wang et al. 2002). AVG application should have completely inhibited ACS activity but this was not the case. However, the efficiency of P. Liquidambaris was lower than previously reported (AVG. Yi et al. 1999), IAA stimulated de novo synthesis of ACS. Liquidambar formosana parasitized in the root and leaf tissues of rice (Yang et al. 2015) produced IAA (Chen et al. 2011), which increased the total amount of IAA (Glick et al. 1999) and induced the synthesis of ACS. Similar to our observations, it has been reported that AVG reduces ACC content and ACO activity more efficiently than PGPR, which produces ACCD (Fig. 3) (Madhaiyan et al. 2007). This might be due to differences in mechanism and efficiency between AVG and P. liquidambari. AVG mainly inhibits ACS activities, which converts SAM (S-adenosyl methionine) to ACC, reducing ACC concentration, followed by ACO activity, which converts ACC to ethylene. Therefore, AVG needs to spread throughout plant system or needs to be systematic to completely inhibit ACS activity. On the other hand, ACCD-producing P. liquidambaris breaks ACC into α-ketobutyrate and NH3 and uses the products as carbon and nitrogen sources for its growth. Therefore, P. liquidambaris might get more benefit from plant association as it colonized inside plant and directly use ACC, thereby offering greater advantages in reducing ACC content, ACO activity. On the other hand, ACCD-producing PGPM mainly colonize the root surface of the plant and break ACC when released from the roots as exudates (Duan et al. 2009) and ACC exudations are proportional to ACC production (Bayliss et al. 1997). ACCD-producing PGPM reduced ACO activity was reported in canola, pea, red pepper and tomato (Madhaiyan et al. 2007; Barnawal et al. 2014; Siddikee et al. 2011a, b).
ACS and ACO gene expression increased in salt-stressed rice seedlings when compared to other treatments (Fig. 4). AVG treatment in rice seedlings reduced the relative ACS gene expression probably through a feedback mechanism and ACO by reducing the amounts of ACC, the substrate for ACO. Relative ACS gene expression was lower in P. liquidambaris inoculated rice seedlings compared to that in the corresponding control (Fig. 4), probably because of different functional ACS genes in plants (Tao et al. 2015). P. liquidambaris inoculation reduced the relative ACO gene expression in stressed rice seedlings probably by reducing the amounts of ACC indicating reduced ethylene synthesis at the molecular level (Fig. 4). Stress caused increased ethylene production, which is correlated with increased expression of both ACS (Park et al. 1992) and ACO genes (Woodson et al. 1992) and ACCD-producing Methylobacterium inoculation reduced ACO-related gene expression in diseased tomato (Yim et al. 2014). Some research demonstrated that ACCD-producing P. polymyxa inoculation suppressed the genes responsible for ethylene production and induced the genes involved in growth in Arabidopsis thaliana (Timmusk and Wagner 1999).
Sequence homology is not sufficient to define a gene as ACCD encoding sequences (Glick 2005) which might be the reason of absence of ACCD activity in the tissues of rice seedlings (data not shown). Similarly, a putative ACCD from tomato does not cleave the cyclopropane ring of ACC rather utilizes d-cysteine as a substrate (Todorovic and Glick 2008). However, the relative ACCD gene gene expression was higher in salt-stressed rice seedlings inoculated with P. liquidambaris compared to that in the corresponding un-inoculated control (Fig. 4), which might be due to the addition of the ACCD gene from P. liquidambaris in the tissues extract. However, relative fungal ACCD gene expression was 3.9-fold in ACC-supplemented JNF medium (Fig. 4). Real-time PCR was used to detect 3.5-fold ACC induced gene by 3 mM ACC and Northern blot was used to induce ACD gene expression (Viterbo et al. 2010; Jia et al. 2000).
Effect of salt stress and P. liquidambaris inoculation on growth and development, chl content and biomass allocation in rice seedlings
P. liquidambaris inoculated salt-stressed rice seedlings grew significantly better than AVG treated seedlings, indicating that P. liquidambaris was more efficient than AVG (Table 1; Fig. 3), especially in reducing ethylene production through ACCD activity. Similar results of PGPR production by ACCD have been reported. Exogenous application of ethylene or ACC or increased production of ethylene decrease root growth, while treatment with ethylene inhibitors such as AVGand ACCD-producing microbes promotes root growth (Madhaiyan et al. 2007; Siddikee et al. 2010, 2011a, b). Based on our results and previous reports, it is evident that ACCD-producing P. liquidambaris can promote root elongation and increase dry matter accumulation under salt stress (Sergeeva et al. 2006).
The weight ratio of Chla and Chlb (Chl a/b ratio) is an indicator of the functional pigment equipment and light adaptation of the photo-synthetic apparatus (Lichtenthaler et al. 1981). Chlorophyll content and ratio of a/b was low in salt-stressed seedlings compared to those in control (Table 2). Therefore, a/b ratio could be used as an indicator of stress effect. However, ACCD-producing P. liquidambaris reversed the pattern of chl content and ratio of a/b (Table 2) which suggesting the modulation of salt stress effect. Reduced effect might be the steady state level of signaling through the reduced level of stress-induced ethylene production by ACCD activity, resulting undisturbed photosynthesis, improved root growth, mineral nutrition and, in general, health of the plants.
It was assumed that higher dry weight would mean longer and stronger roots and shoots as well as plants that would be able to better withstand salt stress. Therefore, to clearly understand the effect of salt stress and ACCD-producing P. liquidambaris inoculation on rice seedlings, comparisons were made between salt-stressed plants (with/without inoculation) with control for dry biomass allocation to roots and shoots. Results showed that salinization decreased the dry matter root/shoot ratio compared to that in non-stressed plants (Table 2) which might be due to the ACC and ethylene concentrations are typically higher in the roots than in the shoots, thereby reducing physiological and metabolic activity and photosynthate allocation toward the roots. The photoassimilates produced under salt stress are used to support growth, maintenance and osmotic adjustment, resulting competition for the limited photoassimilate supplies among different physiological processes and sink organs (Munns and Termaat 1986). However, P. liquidambaris and AVG application balanced dry matter allocation and P. liquidambaris inoculation was more efficient in balancing the dry matter allocation in roots and shoots. These results are consistent with earlier studies and the tolerance index of ACCD-producing P. liquidambaris-inoculated plants (Average STI = 1.45) was significantly higher when compared to that of non-inoculated stressed plants (STI = 0.58), providing additional evidence that the increased STI might be due to the beneficial effect of P. liquidambaris (Table 2) (Siddikee et al. 2011a, b). The data presented in Table 2 provide further evidence that ACCD-producing P. liquidambaris inoculation increases dry matter allocation toward roots by reducing the negative effect of stress ethylene on root cells of salt-stressed plants, consequently increasing salt adaptation. ACCD-producing microbes enhanced the salt tolerance of canola, tomato, wheat and red pepper (Cheng et al. 2007; Zahir et al. 2009; Siddikee et al. 2011a, b).
In conclusion, salt stress increased ACS and ACO activities as well as ACC content and ethylene production in rice seedlings, but inoculation of ACCD-producing P. liquidambaris significantly reduced ACC content, ACO activity and ethylene production resulting enhanced salt tolerance and growth compared to AVG treatment which clearly indicating that ACCD playing essential role in salt stress condition. In addition, P. liquidambaris inoculation in stressed rice seedlings increased chl a content, accumulation of biomass and balanced biomass allocation toward root-shoot which indicates that it re-stabilize metabolic processes toward normal growth and development, which might be the result of the balance of ethylene level by ACCD activity and steady state level of signaling. The promotion of plant growth even in the presence of growth inhibitory level of salt by P. liquidambaris is a consequence of the reduced stress level of ethylene synthesis. Therefore, the result of this study proved the effectiveness of ACCD-producing P. liquidambaris which break ACC into NH3 and α-ketobutyrate reduced ACC and ethylene production over AVG treatment, the inhibitor of ACS which convert S-adenosyl-methionine (SAM) to ACC, the immediate precursor of ethylene. However, further studies are needed to uncover the effectiveness of ACCD-producing P. liquidambaris in promoting growth under salt stress in field conditions.
Change history
14 March 2022
A Correction to this paper has been published: https://doi.org/10.1007/s00203-022-02821-0
Abbreviations
- ACC:
-
1-Aminocyclopropane-1-carboxylic acid
- ACCD:
-
ACC deaminase
- ACS:
-
ACC synthase
- ACO:
-
ACC oxidase
- SAM:
-
S-adenosyl methionine
- AVG:
-
Aminoethoxyvinylglycine
- PGPM:
-
Plant growth promoting microorganisms
References
Abeles FB, Morgan PW, Saltveit ME (1992) Ethylene in plant biology. Academic Press, San Diego, pp 113–114
Arshad M, Frankenberger WT (2002) Ethylene: agricultural sources and application. Kluwer Academic/Plenum Publisher, NY, USA
Ashraf M, Ali Q (2008) Relative membrane permeability and activities of some antioxidant enzymes as the key determinants of salt tolerance in canola (Brassica napus L.). Environ Exp Bot 63:266–273
Barnawal D, Bharti N, Maji D, Chanotiya CS, Kalra A (2014) ACC deaminase-containing Arthrobacter protophormiae induces NaCl stress tolerance through reduced ACC oxidase activity and ethylene production resulting in improved nodulation and mycorrhization in Pisum sativum. J Plant Physiol 171:884–894
Bayliss C, Bent E, Culham DE, MacLellan S, Clarke AJ, Brown GL, Wood JM (1997) Bacterial genetic loci implicated in the Pseudomonas putida GR12-2R3-canola mutualism: identification of an exudate-inducible sugar transporter. Can J Microbiol 43:809–818
Burd GI, Dixon DG, Glick BR (2000) Plant growth-promoting bacteria that decrease heavy metal toxicity in plants. Can J Microbiol 46:237–245
Chen Y, Peng Y, Dai CC, Ju Q (2011) Biodegradation of 4-hydroxybenzoic acid by Phomopsis liquidambari. Appl Soil Ecol 51:102–110
Chen Y, Wang HW, Li L, Dai CC (2013a) The potential application of the endophyte Phomopsis liquidambaris to the ecological remediation of long-term cropping soil. Appl Soil Ecol 67:20–26
Chen Y, Xie XG, Ren CG, Dai CC (2013b) Degradation of N-heterocyclic indole by a novel endophytic fungus Phomopsis liquidambari. Bioresour Technol 129:568–574
Cheng Z, Park E, Glick BR (2007) 1-Aminocyclopropane-1-carboxylate deaminase from Pseudomonas putida UW4 facilitates the growth of canola in the presence of salt. Can J Microbiol 53:912–918
Duan J, Muller KM, Charles TC, Vesely S, Glick BR (2009) 1-Aminocyclopropane-1-carboxylate (ACC) deaminase genes in rhizobia from southern Saskatchewan. Microb Ecol 57:423–436
Feng Y, Shen D, Song W (2006) Rice endophyte Pantoea agglomerans YS19 promotes host plant growth and affects allocations of host photosynthates. J Appl Microbiol 100:938–945
Glick BR (2005) Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol Lett 251:1–7
Glick BR, Patten CL, Holguin G, Penrose DM (1999) Biochemical and genetic mechanisms used by plant growth promoting bacteria. Imperial College Press, London, p 270
Glick BR, Cheng Z, Czarny J, Duan J (2007) Promotion of plant growth by ACC deaminase-producing soil bacteria. Eur J Plant Pathol 119:329–339
Grichko VP, Glick BR (2001) Amelioration of flooding stress by ACC deaminase-containing plant growth-promoting bacteria. Plant Physiol Biochem 39:11–17
Indiragandhi P, Anandham R, Kim KA, Yim WJ, Madhaiyan M, Sa TM (2008) Induction of defense responses in tomato against Pseudomonas syringae pv. tomato by regulating the stress ethylene level with Methylobacterium oryzae CBMB20 containing 1-aminocyclopropane-1-carboxylate deaminase. World J Microbiol Biotech 24:1037–1045
Jia YJ, Ito H, Matsui H, Honma M (2000) 1-Aminocyclopropane-1-carboxylate (ACC) deaminase induced by ACC synthesized and accumulated in Penicillium citrinum intracellular spaces. Biosci Biotech Bioch 64:299–305
Kohler J, Caravaca F, Carrasco L, Roldan A (2006) Contribution of Pseudomonas mendocina and Glomus intraradices to aggregates stabilization and promotion of biological properties in rhizosphere soil of lettuce plants under field conditions. Soil Use Manage 22:298–304
Lichtenthaler HK et al (1981) Photosynthetic activity, chloroplast ultrastructure, and leaf characteristics of high-light and low-light plants and of sun and shade leaves. Photosynthesis Res 2:115–141
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with folin-phenol reagent. J Biol Chem 193:265–275
Madhaiyan M, Poonguzhali S, Sa TM (2007) Characterization of 1-aminocyclopropane-1-carboxylate (ACC) deaminase containing Methylobacterium oryzae and interactions with auxins and ACC regulation of ethylene in canola (Brassica campestris). Planta 226:867–876
Morgan PW, Drew MC (1997) Ethylene and plant responses to stress. Physiol Plant 100:620–630
Mortensen LM (1989) Effect of ethylene on growth of greenhouse lettuce at different light and temperature levels. Sci Hortic 39:97–103
Munns R, Termaat A (1986) Whole-plant responses to salinity. Aust J Plant Physiol 13:143–160
Nascimento FX, Rossi MJ, Soares CRFS, McConkey BJ, Glick BR (2014) New insights into 1-aminocyclopropane-1-carboxylate (ACC) deaminase phylogeny, evolution and ecological significance. Plos One 9(6):e99168. https://doi.org/10.1371/journal.pone.0099168
Park KY, Drory A, Woodson WR (1992) Molecular cloning of an 1-minocyclopropane-1-carboxylate synthase from senescing carnation flower petals. Plant Mol Biol 18:377–386
Sergeeva E, Shah S, Glick BR (2006) Growth of transgenic canola (Brassica napus) expressing a bacterial 1-aminocyclopropane-1-carboxylate (ACC) deaminase gene on high concentrations of salt. World J Microbiol Biotechnol 22:277–282
Shi Y, Dai CC, Wu YC, Yuan ZL (2004) Study on the degradation of wheat straw by endophytic fungi. Acta Sci Circum 24:144–149
Siddikee MA, Chauhan PS, Anandham R, Han GH, Sa TM (2010) Isolation, characterization, and use for plant growth promotion under salt stress, of ACC deaminase-producing halotolerant bacteria derived from coastal soil. J Microbiol Biotechnol 20(11):1577–1584
Siddikee MA, Glick BR, Chauhan PS, Yim WJ, Sa TM (2011a) Enhancement of growth and salt tolerance of red pepper seedlings (Capsicum annuum L.) by regulating stress ethylene synthesis with halotolerant bacteria containing 1-aminocyclopropane-1-carboxylic acid deaminase activity. Plant Physiol Biochem 49:427–434
Siddikee MA, Chauhan PS, Sa TM (2011) Regulation of ethylene biosynthesis under salt stress in red pepper (Capsicum annuum L.) by 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase-producing halotolerant bacteria. J Plant Growth Regul. https://doi.org/10.1007/s00344-011-9236-6
Tao JJ, Chen HW, Ma B, Zhang WK, Chen SY, Zhang JS (2015) The role of ethylene in plants under salinity stress. Front Plant Sci 6:1059
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tian LS, Dai CC, Zhao YT, Zhao M, Yong YH, Wang XX (2007) The degradation of phenanthrene by endophytic fungi Phomopsis sp. single and co-cultured with rice. China Environ Sci 27:757–762
Timmusk S, Gerhart E, Wagner H (1999) The plant-growth-promoting rhizobacterium Paenibacillus polymyxa induces changes in Arabidopsis thaliana gene expression: a possible connection between biotic and abiotic stress responses. MPMI 12(11):951–959
Todorovic B, Glick BR (2008) The interconversion of ACC deaminase and D-cysteine desulfhydrase by directed mutagenesis. Planta 229(1):193–205
Viterbo A, Landau U, Kim S, Chernin L, Chet I (2010) Characterization of ACC deaminase from the biocontrol and plant growth-promoting agent Trichoderma asperellum T203. FEMS Microbiol Lett 305(1):42–48
Wachter R, Fischer K, Gabler R, Kuhnemann F, Urban W, Bogemann GM, Voesenek LACJ, Blom CWPM, Ullrich CI (1999) Ethylene production and ACC accumulation Agrobacterium tumefaciens induced plant tumours and their impact on tumour and host stem structure and function. Plant Cell Environ 22:1263–1273
Wang KLC, Li H, Ecker JR (2002) Ethylene biosynthesis and signaling networks. Plant Cell 14:131–151
Wang HW, Zhang W, Su CL, Zhu H, Dai CC (2015) Biodegradation of the phytoestrogen luteolin by the endophytic fungus Phomopsis liquidambari. Biodegradation. https://doi.org/10.1007/s10532-015-9727-4
Yang B, Wang XM, Ma HY, Yang T, Jia Y, Zhou J, Dai CC (2015) Fungal endophyte Phomopsis liquidambaris affects nitrogen transformation processes and related microorganisms in the rice rhizosphere. Front Microbiol 6:982
Yi HC, Joo S, Nam KH, Lee JS, Kang BG, Kim WT (1999) Auxin and brassinosteroid differentially regulate the expression of three members of the 1-aminocyclopropane-1-carboxylate synthase gene family in mung bean (Vigna radiata L.). Plant Mol Biol 41:443–454
Yim WJ, Kim KY, Lee YW, Sundaram SP, Lee Y, Sa TM (2014) Real time expression of ACC oxidase and PR-protein genes mediated by Methylobacterium spp. in tomato plants challenged with Xanthomonas campestris pv. Vesicatoria. J Plant Physiol 171:1064–1075
Yuan ZL, Dai CC, Li X, Tian LS, Wang XX (2007) Extensive host range of an endophytic fungus affects the growth and physiological functions in rice (Oryza sativa L.). Symbiosis 43:21–28
Yuan ZL, Zhang CL, Lin FC (2010) Role of diverse non-systemic fungal endophytes in plant performance and response to stress: progress and approaches. J Plant Growth Regul 29:116–126
Yuan J, Sun K, Deng-Wang M-Y, Dai C-C (2016) The mechanism of ethylene signaling induced by endophytic fungus Gilmaniella sp. AL12 mediating sesquiterpenoids biosynthesis in Atractylodes lancea. Front Plant Sci 7:361
Zahir ZA, Munir A, Asghar HN, Shaharoona B, Arshad M (2007) Effectiveness of rhizobacteria containing ACC-deaminase for growth promotion of pea (Pisum sativum) under drought conditions. J Microbiol Biotechnol 18:958–963
Zahir AZ, Ghani U, Naveed M, Nadeem SM, Asghar HN (2009) Comparative effectiveness of Pseudomonas and Serratia sp. containing ACC-deaminase for improving growth and yield of wheat (Triticum aestivum L.) under salt-stressed conditions. Arch Microbiol 191:415-424
Zapata PJ, Botella MA, Pretel MT, Serrano M (2007) Responses of ethylene biosynthesis to saline stress in seedlings of eight plant species. Plant Growth Regul 53:97–106
Zhang W, Lu LY, Hu LY, Cao W, Sun K, Sun QB, Siddikee A, Shi RH, Dai CC (2018) Evidence for the involvement of auxin, ethylene and ROS signaling during primary root inhibition of Arabidopsis by the allelochemical benzoic acid. Plant Cell Physiol 59:1889–1904
Zhou J, Kang L, Wang HW, Yang T, Dai CC (2014) Liquid laccase production by Phomopsis liquidambaris accelerated phenolic acids degradation in long-term cropping soil of peanut. Acta Agricult Scand Sect-B Soil Plant Sci 64:683–693
Acknowledgements
We thank Miss Xiao-Mi Wang from Nanjing Institute of Soil Science, Academy of Science (CAS) for her technical assistance and her help in measuring ethylene produced by salt-stressed rice seedlings using GC. We are grateful to the National Natural Science Foundation of China (31570491), a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, Research Fund of State Key Laboratory of Soil and Sustainable Agriculture, Nanjing Institute of Soil Science, Chinese Academy of Science (Y412201435) for their financial support.
Author information
Authors and Affiliations
Contributions
CCD and MAS designed the experiment. MAS, MIZ, MW and WZ conducted the study, processed the data, performed data analysis, and wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical statement
Our work complies with the ethical rules applicable for this journal.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Siddikee, M.A., Zereen, M.I., Wu, M. et al. Phomopsis liquidambaris reduces ethylene biosynthesis in rice under salt stress via inhibiting the activity of 1-aminocyclopropane-1-carboxylate deaminase. Arch Microbiol 203, 6215–6229 (2021). https://doi.org/10.1007/s00203-021-02588-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-021-02588-w