Abstract
Purpose
Accurate segmentation of articular cartilage from MR images is crucial for quantitative investigation of pathoanatomical conditions such as osteoarthritis (OA). Recently, deep learning-based methods have made significant progress in hard tissue segmentation. However, it remains a challenge to develop accurate methods for automatic segmentation of articular cartilage.
Methods
We propose a two-stage method for automatic segmentation of articular cartilage. At the first stage, nnU-Net is employed to get segmentation of both hard tissues and articular cartilage. Based on the initial segmentation, we compute distance maps as well as entropy maps, which encode the uncertainty information about the initial cartilage segmentation. At the second stage, both distance maps and entropy maps are concatenated to the original image. We then crop a sub-volume around the cartilage region based on the initial segmentation, which is used as the input to another nnU-Net for segmentation refinement.
Results
We designed and conducted comprehensive experiments on segmenting three different types of articular cartilage from two datasets, i.e., an in-house dataset consisting of 25 hip MR images and a publicly available dataset from Osteoarthritis Initiative (OAI). Our method achieved an average Dice similarity coefficient (DSC) of \(92.1\pm 0.99\%\) for the combined hip cartilage, \(89.8\pm 2.50\%\) for the femoral cartilage and \(86.4\pm 4.13\%\) for the tibial cartilage, respectively.
Conclusion
In summary, we developed a new approach for automatic segmentation of articular cartilage from MR images. Comprehensive experiments conducted on segmenting articular cartilage of the knee and hip joints demonstrated the efficacy of the present approach. Our method achieved equivalent or better results than the state-of-the-art methods.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Hip and knee osteoarthritis (OA) is one of the leading causes of musculo-skeletal diseases. Magnetic resonance imaging (MRI) is widely recognized as the imaging technique of choice for the assessment of OA due to its excellent soft tissue contrast and no ionizing radiation. To get objective, quantitative, reproducible analysis, an accurate and high-quality cartilage segmentation from MR images is crucial. Manual segmentation of articular cartilage is tedious, subjective, and labor-intensive. Hence, development of automatic segmentation methods has drawn more and more attentions.
Automatic methods have been proposed for articular cartilage segmentation. There exist methods using statistical shape models (SSM) [5] and graph optimization [16]. For example, Fripp et al. [5] introduced a three-stage SSM-based method for automated segmentation of knee cartilage. Their method starts with the segmentation of hard tissues, followed by extraction of bone–cartilage interfaces (BCI) and finally segmentation of articular cartilage based on an expectation-maximization Gaussian mixture model. Xia et al. [16] developed an arc-weighted graph optimization method for automatic hip cartilage segmentation.
With the introduction of publicly available MR image dataset for the knee such as the Osteoarthritis Initiative (OAI) data and the MICCAI grand challenge “Segmentation of Knee Images 2010” (SKI10) data [6], significant progresses have been achieved in segmentation of knee cartilage. For example, Shan et al. [14] proposed a multi-atlas segmentation strategy with non-local patch-based label fusion. Lee et al. [8] introduced a three-stage segmentation scheme consisting of multiple-atlas building, local weighted vote and graph-cut-based region adjustment.
Recently, deep learning-based methods, especially methods based on convolutional neural networks (CNNs), have made tremendous progress in medical image segmentation tasks. There exist CNNs-based methods developed for automatic segmentation of knee cartilage. Prasoon et al. [12] were the first to use triplanar-CNNs for articular cartilage segmentation. Liu et al. [9] introduced a fully automatic segmentation pipeline combining a semantic segmentation CNN with 3D simplex deformable models (DefModel). Ambellan et al. [1] presented a method for automated segmentation of knee bones and cartilage from MR images, combining a priori knowledge of anatomical shape with CNNs.
Compared with cartilage of the knee, hip cartilage is much thinner [11]. Moreover, caused by high curvature of the joint, voxels composing the hip cartilage are severely affected by partial volume effect. It is particularly challenging to distinct the individual cartilage plates (femoral and acetabular) in weight-bearing areas without the use of leg traction devices or contrast agents [2]. Thus, the acetabular cartilage and the femoral cartilage of the hip are usually treated as one combined bulk cartilage. For example, Siversson et al. [15] developed a multi-template-based label fusion technique for automatic segmentation of the combined acetabular and femoral cartilage. Schmaranzer et al. [13] presented a study to compare CNNs-based segmentation of the combined femoral and acetabular cartilage with manual segmentation.
In this paper, based on the recently introduced nnU-Net [7] model, we propose a two-stage method for automatic segmentation of articular cartilage. Our contributions can be summarized as follows:
-
1.
We develop and validate a two-stage method for accurate segmentation of articular cartilage, where results obtained from the first stage are used to compute intermediate features for cartilage segmentation refinement at the second stage;
-
2.
We propose to use entropy maps and distance maps to guide the cartilage segmentation refinement;
-
3.
We design and conduct comprehensive experiments on datasets of both hip and knee cartilage to evaluate the efficacy of the present method.
Methods
Overview of the present method
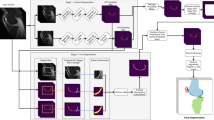
Figure 1 presents a schematic illustration of the two-stage method for automatic cartilage segmentation. At both stages, nnU-Net [7] is used. Specifically, at the first stage, nnU-Net is used to get segmentation of both hard tissues and articular cartilage. Based on the initial segmentation, we generate distance maps, which are computed from the hard tissue segmentation, as well as entropy maps, which encode the uncertainty information about the initial cartilage segmentation. At the second stage, both distance maps and entropy maps are concatenated to the original image. We then crop a sub-volume around the cartilage region determined by the initial cartilage segmentation, which is used as the input to another nnU-Net for segmentation refinement.
Below we present the details for segmentation of the knee cartilage. Similar pipeline has also been developed for the hip cartilage. For the description as presented below, we use X and G to represent the original MR image and the corresponding ground-truth label, respectively. P and L are used to represent a probability map and a predicted label, respectively. E and D are used to represent the entropy maps and the distance maps, respectively.
Stage one
At stage one, we train a 3D nnU-Net model \({F_1}\) to get a coarse segmentation of both hard tissues and articular cartilage. After training, the model takes an input 3D volume \({X_1} \in {R^{H \times W \times D}}\) and outputs a probability map after the last softmax layer: \({P_1} \in {R^{H \times W \times D \times C}}\), where H, W, D, C represent the height, the width, the depth and the number of classes, respectively. The predicted coarse labels \({L_1}\) are then obtained as follows.
where c represents class index; \(p_1^{(h,w,d,c)}\) represents each voxel in the probability map \({P_1}\); and \(l_1^{(h,w,d)}\) represents the voxel in the label map \({L_1}\).
Intermediate data computing
After the initial segmentation at stage one, we observe that segmentation of the hard tissues is good enough but not for the articular cartilage, especially around the boundary regions. In order to further refine the cartilage segmentation, we propose to use entropy maps, which are computed from the initial segmentation of the articular cartilage, and distance maps, which are computed from the initial segmentation of the hard tissues, to guide the refinement. The rationale behind such a strategy is that (a) high entropy values usually indicate high segmentation uncertainty; and (b) previous work [5] demonstrated that bone–cartilage interfaces (BCI) encoded a priori knowledge about the cartilage distribution. The entropy map \({E^c}\) is defined as follows:
where \({e^{(h,w,d,c)}}\) represents a voxel in the entropy map \({E^c}\).
The distance map \({D^c}\) is calculated as follows:
where \(d^{\left( h,w,d,c \right) }\) represents a voxel in the distance map \({D^c}\); \(\partial L_{1}^{c}\) represents the boundary voxels of \(L_{1}^{c}\). The boundary is defined as voxels with at least one neighbor that is not part of the respective segmentation label. In this paper, we only compute the bone distance maps and combine them into one volume to obtain a combined distance map D.
Stage two
At stage two, we first concatenate both the combined distance map and the entropy maps to the original image. We then crop the concatenated data to a sub-volume around the cartilage region by adding a randomly selected margin of 10–15 voxels to the boundary box determined by the initial cartilage segmentation. The cropped data are then used as the input to another nnU-Net model \({F_2}\) to get refined segmentation map \(L_{2}\) of the articular cartilage.
Loss function
Training a deep neural network is challenging. As the matter of gradient vanishing, final loss cannot be efficiently back propagated to shallow layers, which is more difficult for 3D cases when limited number of annotated data is available. To address this issue, nnU-Net [7] incorporates deep supervision mechanism. Specifically, three branch classifiers at three different resolutions are injected into the network in addition to the classifier of the main network. Thus, the total loss is a sum of losses computed at each resolution.
Let \(L_D\), \(L_{CE}\) be the Dice loss [4] and the cross-entropy loss, respectively. The loss function used for training nnU-Net is defined as:
where \(G_i\) represents the ground truth labels at the ith resolution. Accordingly, \(P_i\) represents the probability map of the classifier at the ith resolution.
In Eq. 4, the ground truth labels at a lower resolution are obtained by downsampling from the labels at the higher resolution.
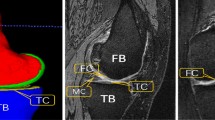
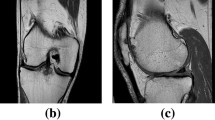
Qualitative comparison of the results obtained from different methods on segmenting femoral cartilage (top two rows) and hip cartilage (bottom two rows). a and g: ground truth label; b and h: results from 3D U-Net (DSC of femoral cartilage: 90.4%; DSC of hip cartilage: 85.8%); c and i: results from nnU-Net (DSC of femoral cartilage: 92.5%; DSC of hip cartilage: 92.1%); d and j results from our method (DSC of femoral cartilage: 93.8%; DSC of hip cartilage: 92.9%); e and k entropy map; f and l: color-coded surface distance between the prediction of our method and ground truth segmentation (unit: mm).
Implementation details
All methods reported in this study were implemented in Python using PyTorch [10] framework and were trained and tested on a desktop with a 2.1GHz Intel Xeon(R) Silver 4110 and a NVIDIA GeForce GTX 2080Ti graphics card with 11GB GPU memory. We trained our network from scratch, and the parameters were updated by the stochastic gradient descent (SGD) algorithm (nesterov momentum = 0.99, weight decay=0.00003). The batch size was 2 and the initial learning rate was 0.01 and decayed with poly learning rate policy. We trained the network for a total of 150,000 iterations. Data augmentations as implemented in the nnU-net framework [7] were used to enlarge the training samples.
Experiments and results
Datasets description and evaluation metrics
We designed and conducted comprehensive experiments on two MR image datasets to evaluate the efficacy of the present method. Specifically, the first dataset consists of 507 3D MR volumes of the knee from the OAI databaseFootnote 1. Each volume was acquired with Double-Echo Steady-Stage (DESS) sequence. Manual segmentation of both hard tissues and knee cartilage was supplied by experienced users at Zuse Institute Berlin (ZIB)Footnote 2. We refer this dataset as OAI-ZIB dataset, which is publicly available [1]. The second dataset consists of 25 delayed gadolinium-enhanced MR images of cartilage (dGEMRIC) of the hip, which has been previously used in [13]. Slice-by-slice manual segmentation of both hard tissues and hip cartilage was done by experienced clinicians [13]. For details about these two datasets, we refer readers to [1] and [13].
Volume-based metrics such as Dice similarity coefficient (DSC) and Jaccard coefficient (JC) as well as distance-based metrics like average symmetry surface distance (ASSD) and Hausdorff distance (HD) are used to evaluate the performance of different methods.
Ablation study
We first investigate the influence of different components of the present method on the segmentation performance. We conduct the ablation study on the OAI-ZIB dataset. In total, 254 volumes out of 507 volumes are used for training and the remaining data are used for validation. The results are present in Table 1. From this table, one can see that (1) either distance maps or entropy maps help to boost the performance of the nnU-Net in segmenting both femoral and tibial cartilage; and (2) the best results are obtained when both maps are used.
Validation study
We designed and conducted two studies to compare the present method with the state-of-the-art methods. First, we conducted a twofold cross-validation study using the same protocol as introduced in [1], which then allowed us to compare our method with the method introduced in [1] on segmenting both hard tissues and cartilage of the knee. The results are present in Table 2. From this table, one can observe that both methods achieved equivalent performance. Overall, an average DSC of \(93.15\%\) was achieved by their method, while our method achieved an average DSC of \(93.35\%\).
We additionally conducted a second study to compare the present method with the state-of-the-art methods such as 3D U-Net [3] and nnU-Net [10]. We conducted this study on both the OAI-ZIB dataset and the hip dataset. For the OAI-ZIB dataset, we took 254 volumes out of 507 volumes for training and the remaining data for validation. For the hip dataset, we used 20 cases for training and the remaining 5 cases for validation. The results are presented in Table 3. From this table, one can see that (1) nnU-Net achieved consistently better results than 3D U-Net, demonstrating the efficacy of nnU-Net; and (2) our method achieved the best results, with an average DSC of \(89.8\pm 2.50\%\), \(86.4\pm 4.13\%\) and \(92.1\pm 0.99\%\), when segmenting femoral cartilage, tibial cartilage, and hip cartilage, respectively. Using Wilcoxon signed-ranks test (SPSSAU, Version 21.0, www.spssau.com) and taking 0.05 as the significance level, it was found that the differences between the proposed method and the other two methods on segmentation of both femoral and tibial cartilage were statistically significant (p-value < 0.001). Figure 2 shows qualitative comparison of the present method with 3D U-Net and nnU-Net.
Discussion
In this paper, we proposed a two-stage method for automatic segmentation of articular cartilage. Starting from the initial segmentation obtained at stage one, we computed both entropy maps and distance maps, which were then used to refine the cartilage segmentation at the second stage. We designed and conducted comprehensive experiments on two datasets to validate the efficacy of the present method on segmenting articular cartilage of both the hip and the knee. Our method achieved an average DSC of \(92.1\pm 0.99\%\) for the combined hip cartilage, \(89.8\pm 2.50\%\) for the femoral cartilage and \(86.4\pm 4.13\%\) for the tibial cartilage, respectively. We further investigated the effectiveness of different components of the present method. Our experimental results confirmed that both distance maps and entropy maps helped to improve the segmentation performance.
The performance of the present approach is compared with the state-of-the-art articular cartilage segmentation methods [1, 8, 9, 13,14,15,16]. The comparison results are summarized in Tables 4 and 5. Due to the fact that different datasets are used in evaluation of different methods, direct comparison of different methods is difficult. Thus, the comparison results in Tables 4 and 5 should be interpreted cautiously. Nevertheless, as shown in Tables 4 and 5, our approach is better than most of the existing work including atlas-based methods [8, 14, 15], graph optimization method [16], and CNN-based methods [1, 9, 13]. One possible explanation why the present approach achieves better results than the existing atlas-based or graph optimization methods is that these methods are difficult to handle large variations of shape and appearance, leading to limited performance. By contrast, by learning hierarchies of relevant features directly from training data, our method can handle large variations of shape and appearance. Our method also achieves better results than other existing CNN-based methods such as those introduced in [9, 13]. Possible explanations include (a) our method is based on nnU-Net which is a state-of-the-art CNN architecture, and (b) we additionally use distance maps and entropy maps to guide the cartilage segmentation refinement. This has been demonstrated by the results shown in Tables 1 and 3.
It is worth to compare the present method with the method reported in [1] as both methods have been evaluated on the publicly available OAI-ZIB dataset. As shown in Table 2, although these two methods achieved equivalent accuracy when evaluated on the same dataset, there are significant differences between our approach and the method reported in [1]. Specifically, their method involves a complicated pipeline consisting of following steps: (a) at the first step, 2D CNN is used to get the masks of both femoral bone and tibial bone; (b) at the second step, SSM adjustment is used to regularize the results of the first step by fitting SSMs to the bone masks; (c) at the third step, 3D CNN is used to segment small MRI subvolume at the bone surfaces as given by the second step; and (d) at the last step, SSM post-processing uses regions pre-defined on SSMs to enhance the results of 3D CNN. By contrast, our method is based on a two-stage pipeline. At both stages, we use the same nnU-Net architecture. We compute the distance maps and the entropy maps from the initial segmentation of the first stage and then use both maps to guide the segmentation refinement at the second stage. This is also the reason why at testing, our method is significantly faster than theirs. As shown in Table 4, it takes only 0.28min for our method to finish one case, while their method needs 9.37min to finish one case.
In summary, we presented a fully automatic method for segmenting articular cartilage and validated our method on two datasets of both the hip and the knee. The strength of the present approach lies in the combination of the state-of-the-art deep learning architecture model with self-context including distance maps and entropy maps.
References
Ambellan F, Tack A, Ehlke M, Zachow S (2019) Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: Data from the osteoarthritis initiative. Med Image Anal 52:109–118
Cheng Y, Guo C, Wang Y, Bai J, Tamura S (2012) Accuracy limits for the thickness measurement of the hip joint cartilage in 3-d MR images: simulation and validation. IEEE Trans Biomed Eng 60(2):517–533
Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O (2016) 3d u-net: learning dense volumetric segmentation from sparse annotation. In: International conference on medical image computing and computer-assisted intervention. pp. 424–432. Springer
Drozdzal M, Vorontsov E, Chartrand G, Kadoury S, Pal C (2016) The importance of skip connections in biomedical image segmentation. In Deep learning and data labeling for medical applications, pp 179–187. Springer
Fripp J, Crozier S, Warfield SK, Ourselin S (2007) Automatic segmentation of the bone and extraction of the bone-cartilage interface from magnetic resonance images of the knee. Phys Med Biol 52(6):1617
Heimann T, Morrison BJ, Styner MA, Niethammer M, Warfield S (2010) Segmentation of knee images: a grand challenge. In Proceedings of MICCAI Workshop on Medical Image Analysis for the Clinic. pp 207–214. Beijing, China
Isensee F, Jaeger PF, Kohl SA, Petersen J, Maier-Hein KH (2021) nnu-net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 18(2):203–211
Lee JG, Gumus S, Moon CH, Kwoh CK, Bae KT (2014) Fully automated segmentation of cartilage from the MR images of knee using a multi-atlas and local structural analysis method. Med Phys 41(9):092303
Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R (2018) Deep convolutional neural network and 3d deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn Reson Med 79(4):2379–2391
Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L, Desmaison A, Köpf A, Yang E, DeVito Z, Raison M, Tejani A, Chilamkurthy S, Steiner B, Fang L, Bai J, Chintala S (2019) Pytorch: An imperative style, high-performance deep learning library. arXiv preprint arXiv:1912.01703
Pedoia V, Majumdar S, Link TM (2016) Segmentation of joint and musculoskeletal tissue in the study of arthritis. Magn Reson Mater Phys Biol Med 29(2):207–221
Prasoon A, Petersen K, Igel C, Lauze F, Dam E, Nielsen M (2013) Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. In International conference on medical image computing and computer-assisted intervention. pp 246–253. Springer
Schmaranzer F, Helfenstein R, Zeng G, Lerch TD, Novais EN, Wylie JD, Kim YJ, Siebenrock KA, Tannast M, Zheng G (2019) Automatic MRI-based three-dimensional models of hip cartilage provide improved morphologic and biochemical analysis. Clinical Orthop Relat Res 477(5):1036
Shan L, Zach C, Charles C, Niethammer M (2014) Automatic atlas-based three-label cartilage segmentation from MR knee images. Med Image Anal 18(7):1233–1246
Siversson C, Akhondi-Asl A, Bixby S, Kim YJ, Warfield SK (2014) Three-dimensional hip cartilage quality assessment of morphology and Dgemric by planar maps and automated segmentation. Osteoarth Cartil 22(10):1511–1515
Xia Y, Chandra SS, Engstrom C, Strudwick MW, Crozier S, Fripp J (2014) Automatic hip cartilage segmentation from 3d MR images using arc-weighted graph searching. Phys Med Biol 59(23):7245
Acknowledgements
The work was partially supported by the Key Program of the Medical Engineering Interdisciplinary Research Fund of Shanghai Jiaotong University via project YG2019ZDA22 and YG2019ZDB09, and by the Natural Science Foundation of China via project U20A20199. Osteoarthritis Initiative is a public-private partnership comprised of five contracts (N01-AR-2-2258; N01-AR-2-2259; N01-AR-2-2260; N01-AR-2-2261; N01-AR-2-2262) funded by the National Institutes of Health, a branch of the Department of Health and Human Services, and conducted by the OAI Study Investigators. Private funding partners include Merck Research Laboratories; Novartis Pharmaceuticals Corporation, GlaxoSmithKline; and Pfizer, Inc. Private sector funding for the OAI is managed by the Foundation for the National Institutes of Health. This manuscript was prepared using an OAI public use dataset and does not necessarily reflect the opinions or views of the OAI investigators, the NIH, or the private funding partners.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
Informed consent was obtained from all individuals included in the study.
Ethics approval
Institutional ethics approval was obtained for the use of clinical data in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Z., Chen, K., Liu, P. et al. Entropy and distance maps-guided segmentation of articular cartilage: data from the Osteoarthritis Initiative. Int J CARS 17, 553–560 (2022). https://doi.org/10.1007/s11548-021-02555-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-021-02555-2