Abstract
Knee osteoarthritis is a major diarthrodial joint disorder with profound global socioeconomic impact. Diagnostic imaging using magnetic resonance image can produce morphometric biomarkers to investigate the epidemiology of knee osteoarthritis in clinical trials, which is critical to attain early detection and develop effective regenerative treatment/therapy. With tremendous increase in image data size, manual segmentation as the standard practice becomes largely unsuitable. This review aims to provide an in-depth insight about a broad collection of classical and deep learning segmentation techniques used in knee osteoarthritis research. Specifically, this is the first review that covers both bone and cartilage segmentation models in recognition that knee osteoarthritis is a “whole joint” disease, as well as highlights on diagnostic values of deep learning in emerging knee osteoarthritis research. Besides, we have collected useful deep learning reviews to serve as source of reference to ease future development of deep learning models in this field. Lastly, we highlight on the diagnostic value of deep learning as key future computer-aided diagnosis applications to conclude this review.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Knee Osteoarthritis (OA) is a whole joint disease (Loeser et al. 2012) caused by a multifactorial combination of biomechanical (Englund 2010), biochemical (Sokolove and Lepus 2013), systemic and intrinsic (Warner and Valdes 2016) risk factors. Often, the disease is associated with joint pain and progressive structural destruction of articular cartilage; and causes permanent physical impairment to the patients. In a recent literature update on OA epidemiology, knee OA has shown high prevalence rate across the globe (Vina and Kwoh 2018). Besides, various studies have highlighted the harmful effect of knee OA on our economies in terms of countries’ GDP losses (Hiligsmann et al. 2013), direct healthcare cost burden (Palazzo et al. 2016) and annual productivity cost of work loss (Gan et al. 2016; Sharif et al. 2017).
To date, the pathophysiology of knee OA is still not fully comprehended and there is a lack of effective cure to treat or halt the progression of knee OA (Favero et al. 2015). Given that cartilage degradation is reversible at early stage, morphological alterations in articular cartilage and subchondral bone are identified as two cardinal characteristics at the onset of knee OA development cycle. Studies have focused on biomarkers such as changes of cartilage volume, thickness and surface curvature (Collins et al. 2016) to quantify underlying morphological alternations. Ultimately, the goals are to capture the knee OA progression pattern, to develop effective disease modifying OA drug (DMOAD) and to design effective regenerative-based therapy/treatment (Zhang et al. 2016).
Literally, MR imaging technology is the central modality to analyze the progression and incidence of knee OA due to its’ ability to protrude soft tissue property of knee joint (Eckstein and Peterfy 2016). Based on the literature, MR imaging sequences such as dual energy steady state (DESS), fast low angle shot (FLASH), spoiled-gradient echo (SPGR), gradient recalled echo (GRE), turbo spin-echo (TSE), fast spin-echo (FSE), spin-echo spectral attenuated inversion recovery (SPAIR) and T1-weighted imaging sequence with fat suppression (FS) or water excitation (WE) are commonly used in cartilage imaging to produce high resolution knee images. Figure 1 shows several MR imaging sequences used in knee OA research. Selection of MR imaging sequence is vital in consideration of a few factors such as signal-to-noise ratio, contrast-to-noise ratio and scanning time.
Knee segmentation (see Fig. 2) plays paramount role to extract biomarkers from MR image (Eckstein and Wirth 2011). The biomarkers contain valuable information to characterize, stimuli and predict the incidence and progression of knee OA. Two longitudinal multicenter knee image datasets i.e. the Osteoarthritis Initiative (OAI) (Peterfy et al. 2008) and Multicenter Osteoarthritis Study (MOST) (Roemer et al. 2010) distribute free image data to researchers upon request. A smaller knee image dataset known as Pfizer Longitudinal Study (PLS) also offers up to 706 MR images from 155 subjects to support knee OA research. In addition, there are two reputable open competitions to promote joint evaluation on knee segmentation models: Segmentation of Knee Image 2010 (SKI10) by MICCAI (Heimann et al. 2010) and MRNet by Stanford University (Bien et al. 2018).
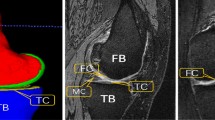
Development of 3D models of knee is prerequisite to knee OA quantification. a Knee bone model consists of patella (PB), femur (FB) and tibia (TB), b posterior view of bone model. c Knee cartilage model consists of patellae cartilage (PC), femoral cartilage (FC) and tibial cartilage (TC). d A left knee cartilage model with subregion labels i.e. medial and lateral femoral cartilage, MFC and LFC; and medial and lateral tibial cartilage, MTC and LTC
Below, we have compiled a list of existing reviews on segmentation techniques of musculoskeletal tissues (see Table 1). In summary, there are five highlights about these reviews:
-
1.
Majority of the reviews concentrated on cartilage segmentation, and covered a diverse semiautomatic and fully automatic segmentation models, properties of MR imaging sequence, as well as advantages and drawbacks of these imaging sequences;
-
2.
Deep learning on cartilage segmentation was first described by Ebrahimkhani et al. (2020), but was limited to five publications only;
-
3.
Comparison of performance among deep learning, semiautomatic and fully automatic segmentation models was not available;
-
4.
The last review on bone segmentation was performed by Aprovitola et al. (2016), which did not cover deep learning;
-
5.
None of current reviews provided any insight about the diagnostic value of deep learning in knee OA studies.
In recent years, deep learning (LeCun et al. 2015) becomes very popular in academia. Many reviews on deep learning has been published; covering various technical aspects such as architectures of deep learning variants (Dargan et al. 2019; Khan et al. 2020; Shrestha and Mahmood 2019), useful data repositories for deep learning practitioners (Sengupta et al. 2020), deep learning libraries and resources (Raghu and Schmidt 2020), as well as advantages, disadvantages and limitations of deep neural network models (Serre 2019). We also compiled existing reviews on deep learning in medical image analysis: (Greenspan et al. 2016; Hesamian et al. 2019; Litjens et al. 2017; Lundervold and Lundervold 2019; Maier et al. 2019; Shen et al. 2017; Singh et al. 2020; Zhou et al. 2019).
This review covers original research articles, book chapters, conference proceedings and symposium in knee cartilage and bone segmentation methods published from January 1st 1990 until now. The search was conducted via PubMed, IEEE Xplore, Science Direct, Google Scholar and arXiv. Keywords “Osteoarthritis”, “Image”, “Segmentation”, “Deep Learning”, “MRI”, “Cartilage”, and “Bone” were used during the review process. Figure 3 illustrates the taxonomy of knee cartilage and bone segmentation method in this review.
The review aims to contribute in the following aspects:
-
1.
This survey gathers existing reviews on musculoskeletal segmentation in Table 1 to provide an overview about the recent development trend of knee segmentation
-
2.
This survey collects existing reviews on technical aspects of deep learning and deep learning for medical image analysis. The objective is to promote bilateral knowledge transfer between deep learning and medical image analysis field by providing suitable source of reference at the ease of readers;
-
3.
Since knee OA was regarded as a “whole joint” disease, bone model plays an important role in successive cartilage segmentation. Thus, we performs an extensive survey on both knee cartilage and bone segmentation models;
-
4.
This is the first survey on deep learning-based knee bone segmentation used in knee OA research, and we put all related models under one special sub-section;
-
5.
The survey provides an updated list of deep learning-based knee cartilage segmentation models under one special sub-section;
-
6.
The survey compares the performance of classical and deep learning segmentation models in the perspective of statistical evaluation and quantification value of biomarkers;
-
7.
The survey highlights on diagnostic values of deep learning in knee OA research by including related studies in prediction, detection and simulation.
The rest of this review is organized as follows: Sect. 2 presents the pathogenesis of knee OA. Sections 3 and 4 reviews existing bone and cartilage segmentation models, respectively. Section 5 discusses performance of imaging biomarkers in quantitative morphometric analysis. Section 6 discusses the diagnostic value of deep learning in knee OA research.
2 Pathogenesis of knee osteoarthritis
Destruction of cartilage is mainly due to the loss of chondrocytes (Charlier et al. 2016) and alteration in extracellular matrix (Maldonado and Nam 2013). When the damage worsens, a broad secondary changes such as development of osteophytes, remodeling of subchondral bone, meniscal degeneration and formation of bone marrow lesions (BMLs) are triggered. In the following sub-sections, we elaborate the involvement of articular cartilage and subchondral bone during the pathogenesis of knee OA.
2.1 Articular cartilage
Human articular cartilage comprises of dense layer of highly specialized chondrocyte, matrix macromolecules such as collagen and proteoglycans, and water. Since cartilage has low metabolic activities, it is extremely vulnerable to shear stress on its surface. Under normal circumstance, chondrocytes plays imperative role to synthesize the turnover and proliferation of these macromolecules whenever tissue damage is detected. The symbiosis relationship helps to maintain a healthy cartilage condition. In case of knee OA, the homeostasis between synthesis and breakdown of degraded extracellular matrix components is disrupted, which adversely affects the cell ability to maintain and restore cartilaginous tissues (Man and Mologhianu 2014).
Aggrecanases and collagenases are components of matrix metalloproteinase (MMP) which are responsible to degrade the aggrecan, a causal proteoglycan in cartilage repair process. Under normal circumstance, tissue inhibitor of metalloproteinases (TIMPs) are activated to inhibit MMP activity (Kapoor et al. 2011). When the symbiosis is disrupted, proteoglycan aggregation and aggrecan concentration will decrease as a result of overwhelming MMP. At the same time, structural changes in collagenous frameworks cause swelling in aggrecan molecules and increase in water content (Roughley and Mort 2014). These deteriorations reduce the stiffness of matrix and ability of self-repair. Subsequently, death of chondrocytes would occur and contributes to significant cartilage damage. The destruction of matrix structure also leads to subchondral bone changes in terms of density, sclerosis, formation of cysts and osteophytes (Goldring and Goldring 2010).
2.2 Subchondral bone
Subchondral bone is a thin cortical lamellae located beneath the calcified cartilage layer where its role is to facilitate the force distribution and reduce the shear stress on articular cartilage. For instance, adaptive process of bone modelling and remodeling play vital role in maintaining the good responsiveness of joint. Bone remodeling comprises of bone resorption by osteoclasts at damaged bone site and generation of new bone by osteoblastic precursors on the resorbed surface. Likewise, bone modeling continues to drive change in bone architecture and volume via direct apposition to existing bone surface due to skeletal adaption to the stressor. Together with articular cartilage, both components are actively preserving the homeostasis of healthy joint environment (Li et al. 2013).
When the strain threshold is beyond the normal adaptive process, disruption to normal bone modelling and remodeling cause delay in new bone formation which weaken the bone structure (Stewart and Kawcak 2018). Subchondral bone volume and density changes are indicative biomarkers to the modification of bone architecture. As the pathological events progress, the subchondral bone plate becomes thicker, which affects the ability of articular cartilage to withstand mechanical loading. The deformation causes horizontal clefts within the deep zone of cartilage as well as other pathological features such as sclerosis, osteophytes, bone shape alternation and bone cysts (Barr et al. 2015). Even though our knowledge about the pathogenesis of knee OA is advancing, there are still much questions about the underlying mechanisms and their relationships which requires further clinical investigations (Sharma et al. 2013).
3 Knee bone segmentation
Knee OA-affected bone endures consistent loss of mineralization, causing it sensitives to structural deformation (Neogi 2012). Some radiologically visible structural changes such as osteophytes, bone marrow lesions (BMLs) and subchondral bone attrition (SBA) are good biomarkers for OA-related clinical trials. A study has reported that subchondral BMLs were apparent across knee regions with increased biomechanical loading (Hunter et al. 2006) whereas other studies showed that development of BMLs was related to cartilage loss (Davies-Tuck et al. 2010; Neogi et al. 2009; Wluka et al. 2009). Bone segmentation is needed to support the discovery and characterization of these biomarkers.
Specifically, purposes of knee bone segmentation are reflected in the following applications: to produce bone-cartilage interface (BCI) in order to extract cartilage tissue from bone surface (Fripp et al. 2007; Kashyap et al. 2016; Yin et al. 2010), to quantify and monitor the changes of bone shape and surface associated with structural deformations (Neogi et al. 2013), and to compute bone model to investigate the effect of biomechanical stress at different localized knee sites (Paranjape et al. 2019). An example of 2D bone structure and segmentation result is shown in Fig. 4. Because knee bone has regular shape and big anatomical size, bone segmentation is easier than cartilage segmentation. At the end of this section, complete lists of classical and deep learning-based knee bone segmentation models were provided in Tables 2 and 3, respectively.
The position and anatomical size of knee bones compared to other tissues give advantages to knee bone segmentation. a Sagittal view of knee joint with femur (FB), tibia (TB) and patella (PB) at the center of image, b knee bone is the biggest structure of knee joint with consistent shape (white arrows: FB; red arrows: TB), c segmented femur (FB) and tibia (TB) usually have better boundary delineation
3.1 Deformable model-based methods
An active contour is defined by a collection of points along the curve, \(X\left(s\right)=\left(X\left(s\right),Y\left(s\right)\right),s\in [\mathrm{0,1}]\), which is governed by either parametric or geometric information available in the image. Parametric (Cohen 1991; Kass et al. 1988) and geometric (Caselles et al. 1993, 1997; Malladi et al. 1995) deformable models differ in terms of the evolving curves and surface representations. For instance, parametric deformable models represent curves and surfaces explicitly in their parametric form as an energy minimizing and dynamic force formulation, whereas geometric deformable models represent the evolving curves and surfaces implicitly as a function of level set.
Active contours model (ACM) (Kass et al. 1988) is the benchmark deformable model in image segmentation. Deformation of active contour is equivalent to minimizing an energy function, \(\varepsilon \left(X\right)\), which comprises of internal and external spline force as shown below:
The formulation aims to identify a parameterized curve that minimizes the weighted sum of both spline force. The internal spline force, \({\varepsilon }_{int}\left(X\right)\), controls the elasticity of contour deformation based on contour tension and rigidity. The external spline force, \({\varepsilon }_{ext}\left(X\right)\), matches the boundary of deformable model toward the targeted object. On the other hand, evolution of geometric curves is independent of parameterization. The model relies on geometric measures such as the unit normal and curvature along the normal direction to form the representation function. Given a moving curve \(\gamma \left(p,t\right)=\left[X\left(p,t\right),Y(p,t)\right]\), where \(p\) is any parameterization, \(t\) is time, \(\mathrm{\rm N}\) is its inward unit normal and \(\upkappa\) is its curvature, evolution of curve can be described as:
where \(V\left(\upkappa \right)\) is known as a speed function that determines the speed of curve evolution.
In addition, primary deformable model has been extended by incorporating prior shape information. Some prominent extensions include statistical shape model (SSM) (Heimann and Meinzer 2009), active shape models (ASM) (Cootes and Taylor 1992) and active appearance models (AAM) (Cootes et al. 2001). Intuitively, these deformable models usually involve training to acquire details of shape variability or appearance feature about the targeted object. The infusion of priori knowledge can be performed via manual interaction such as placing a set of landmark points to form a point distribution model (PDM).
Due to its shape consistency and size advantages, deformable model was extensively used in knee bone segmentation. These deformable models include active contour model (ACM) (Guo et al. 2011; Lorigo et al. 1998; Schmid and Magnenat-Thalmann 2008), statistical shape model (SSM) (Fripp et al. 2007; Seim et al. 2010; Wang et al. 2014) and active appearance model (AAM) (Neogi et al. 2013; Williams et al. 2010b). Semiautomatic hybrid geodesic active contour was implemented in both Lorigo et al. (1998) and Guo et al. (2011). Classical active contours was sensitive to the location of contour placement (it would fail when the contour was placed too far from target object) and lacked the convergence to boundary concavities (Abdelsamea et al. 2015). Besides, it often leaked in the event of varied intensity across the trabecular bone areas and noisy boundary. Initially, texture information was added into the model to compensate the weakness of intensity gradient-based energy function (Lorigo et al. 1998). Then, a statistical overlap constrain was introduced to the active contour stopping function to overcome boundary leaking (Guo et al. 2011). A performance comparison between both models showed that the later (Dice Similarity Coefficient (DSC): 0.94) outperformed the former (DSC: 0.89).
Fripp et al. (2007) built three separate 3D SSM models for femur, tibia and patella with adapted initialization from existing atlas. The surface related to the atlas was propagated to the knee image through affine transform obtained from aligning the atlas to the knee image. In total, there were 2563, 10,242 and 10,242 correspondence points on the surface of femur, tibia and patella, respectively. Then, pose and shape parameters of the propagated surface were trained to estimate the pose and shape variation inside the SSM (Fripp et al. 2007). Similar automatic bone segmentation model was found in Seim et al. (2010), where SSMs of tibia and femur were generated. Since SSM could generate robust bone model, it was frequently used to extract BCI from surface of bone model. Another application of shape model was to predict the onset of radiographic knee OA. Neogi et al. (2013) trained the AAM by using 96 knees to learn the shape variation and graylevel texture of femur, tibia and patella. Then, these information were encoded as principal components. A total of 69, 66 and 59 principal components for femur, tibia and patella bone were created to generate the AAM models (Neogi et al. 2013).
3.2 Graph-based methods
Graph-based method treats an image as a graph, \(G=\left(V,E\right)\) where the pixel is denoted as node, \(v\in V\) and the relationship between two neighboring nodes is denoted by edge, \(e \in E\subsetneq V \times V\). Every edge is assigned by a weight, \(w\), which is extracted from value difference between two nodes \({v}_{i}\) and \({v}_{j}\). Retrospectively, graph segmentation started to gain attention after the normalized cut (Jianbo and Malik 2000) and \(s/t\) graph cuts (Boykov and Jolly 2001) were published. In this context, the partition of graph is known as a “cut”. A general graph-based binary segmentation will partition the graph into two subgraphs i.e. \({g}_{m}\) and \({g}_{n}\), where \({g}_{m}\cup {g}_{n}=V\) and \({g}_{m}\cap {g}_{n}=\varnothing\), by minimizing the degree of dissimilarity across \({g}_{m}\) and \({g}_{n}\). We calculate the dissimilarity as the total weight of the edges that have been removed:
where \(v_{{g_{m} }}\) and \(v_{{g_{n} }}\) are nodes in two disjoint subgraphs.
In practice, achieving an optimal “cut” is a non-trivial task. The solution requires minimizing an energy function, which is known to be NP-hard. Wu and Leahy (1993) proposed to divide the graph into \(K\) sub-graphs based on Max-flow min-cut theorem. The theorem defines a maximum flow from node \(s\) to node \(t\) is equivalent to the minimum cut value (minimal cost) that separates \(s\) and \(t\). In order to obtain the optimal solution, the algorithm would recursively search for every potential cut that divide the two nodes with minimal cost (Wu and Leahy 1993). Nonetheless, the algorithm is biased to segmenting small fraction of nodes. Normalized cut has been proposed by Jianbo and Malik (2000) to address this problem.
The \(s/t\) graph cuts for binary image segmentation was formulated based on maximum flow algorithm with addition seeds as hard constraint. Users need to mark some pixels as “foreground” or “background” to represent the designated terminal nodes \(s\) (source) and \(t\) (sink), respectively. The aim is to attain an optimal cut that severs the edges between these two types of terminal nodes. Hence, a minimal cost may correspond to a segmentation with a desirable balance of boundary and regional properties (Boykov and Funka-Lea 2006). The segmentation energy of graph cuts, \(E(A)\), is defined as:
where \(A = \left( {A_{1} , \ldots ,A_{p} , \ldots ,A_{{\left| {\mathcal{P}} \right|}} } \right)\) is label vector assigned to pixel \(p\) in \({\mathcal{P}}\), \(R_{p} ( \cdot )\) is the regional term of \(A\) and \(B_{{\left\{ {p,q} \right\}}}\) is the boundary term of \(A\). Coefficient \(\lambda \ge 0\) controls a relative importance of the regional term, \(R_{p} ( \cdot )\), versus boundary term, \(B_{{\left\{ {p,q} \right\}}}\). The regional term, \(R_{p} ( \cdot )\), assumes that the individual penalties for assigning pixel \(p\) to “foreground” and “background”, correspondingly. For example, \(R_{p} ( \cdot )\) may reflect on how the intensity of pixel \(p\) fits into a known intensity mode (e.g. histogram) of the “foreground” and “background”. Meanwhile, the boundary term measures the penalty of discontinuity between pixel \(p\) and \(q\).
Other prominent graph-based segmentation methods include random walks (Grady 2006), intelligent scissors (Mortensen and Barrett 1998) and Live Wire (Falcao et al. 2000). The segmentation model can be automatic or interactive. Several studies (Ababneh et al. 2011; Kashyap et al. 2018; Park et al. 2009; Shim et al. 2009b; Yin et al. 2010) have implemented graph cuts to extract knee bone from MR image. Shim et al. (2009b) has implemented semiautomatic graph cuts to segment femur, tibia and patella. To localize the search space during the segmentation, user needed to place scribbles on region of interest as hard constraint. The authors only compared the efficiency of graph cuts with manual segmentation and the model’s accuracy performance was not available (Shim et al. 2009b). Besides, classical semiautomatic graph cuts model depend heavily on seeds to initialize and refine the segmentation, which led to substantial amount of manual intervention.
In Park et al. (2009) and Ababneh et al. (2011), automatic graph cuts segmentation models were proposed. To replace manual seed deployment, extra priori information was required to complement the lack of discriminative power faced by classical graph cuts energy function. Park et al. (2009) proposed to incorporate shape obtained from shape template into their model, and decomposed translation, rotation and scale parameters for shape prior configuration. Then, branch-and-mincut algorithm was repetitively computed to optimize the decomposition. In addition, low intensities of bone tissues was taken as intensity prior to further improve the segmentation accuracy (Park et al. 2009). Ababneh et al. (2011) proposed a multi-stage bone segmentation framework. At initial stage, the image was divided into n × n square blocks and classified into background and non-background blocks based on a set of features extracted from training data. Then, these image blocks were treated as seeds to initiate the graph cuts algorithm while several GLCM-derived features were exported into the construction of energy function (Ababneh et al. 2011). Both improved graph cuts models have registered DSCs of 0.958 (Park et al. 2009) and 0.941 (Ababneh et al. 2011).
3.3 Atlas-based methods
An atlas is defined as a reference model with labels related to the anatomical structures. These labels contain useful priori information to describe certain anatomical structure. Example of priori information includes topological, shape and positional details of the structure, as well as spatial relationship between them. Atlas registration, selection and propagation are three fundamental steps in atlas-based segmentation. Given precise point-to-point correspondence from an image to pre-constructed atlas, the methods are capable of segmenting image with poor relation between regions and pixel’s intensities due to diffuse boundary or image noise. The process of coordinate mapping is known as registration. For an image, \(I\), and an atlas, \(A\), the correspondence is defined as a coordinate transformation \(T\) that maps any specific image coordinates, \(\mathfrak{x}\) in the domain of \(I\) onto the atlas, \(A\). The mapping is given below:
Overall, there are four atlas selection methods: single atlas, the best atlas, average-shape atlas and multiple atlas (Rohlfing et al. 2005). Single atlas method uses an individual segmented image. The selection can be random or based on certain criterion such as quality of image. The best atlas chooses the most desirable atlas from a set of atlases. In order to identify an optimal segmentation from the results of different atlases, one could check the image similarity by using normalized mutual information (NMI) and magnitude of deformation after registration. Compared to previous methods, the averaged shape atlas maps all original individual images onto a common reference to produce an average image. Then, the original images are mapped onto the first average to produce a new average. The mapping process occurs iteratively until convergence. Multiple atlases approach applies different atlases onto a raw image. Then, the segmentations are combined into a final segmentation based on “Vote Rule” decision fusion.
Several research groups have utilized multiple templates to perform knee bone segmentation (Dam et al. 2015; Lee et al. 2014; Shan et al. 2014). Due to the overlapping tibiofemoral boundary after segmentation, direct cartilage segmentation was challenging. Therefore, an initial bone segmentation would serve as shape prior to guide successive cartilage segmentation. Lee et al. (2014) applied non rigid registration to align all templates to target image and selected the best matched template. Then, a locally weighted vote approach with local structure analysis was deployed to generate label fusion. Instead of intensity similarity metrics, the new voting scheme used cartilage model as local reference to generate probability of correspondence to compute the target label. As a result, it was able to avoid poor accuracy attributed to magnetic field inhomogeneity (Lee et al. 2014). The bone segmentation model has reported an average surface distance error of 0.63 mm for femur and 0.53 mm for tibia, which were lower than other bone-cum-cartilage segmentation models.
Dam et al. (2015) introduced a multiatlas pre-registration as a source of training before kNN-based classification of cartilage voxels. During the training process, a rigid registration would transform a given atlas to a common training space to enable the determination of region of interest (ROI) for each anatomical structure and feature extraction (Dam et al. 2015). Based on a set a features from Folkesson et al. (2007), kNN classifier would classify the voxels within the ROI instead of whole image. The structure-wise ROI identification reduced the computation cost of classifiers but construction of atlas was a daunting task. Plus, segmentation accuracy would depend on several other factors, such as high quality registration to create precise structure-wise ROI and reliability of features used to perform classification. In this model, only tibia was segmented from knee images, which gave DSC of 0.975 on 30 training data.
3.4 Miscelleneous segmentation methods
Other knee bone segmentation models were found to be using edge and thresholding (Lee and Chung 2005), ray casting (Dodin et al. 2011), and level set (Dalvi et al. 2007; Gandhamal et al. 2017). Lee and Chung (2005) proposed a multi-stage knee bone segmentation model with a series of edge detection, thresholding and contrast enhancement to enhance the contrast of bone edges and extract bone boundary information. The information was incorporated into region grow algorithm to perform a final segmentation. Then, the model was evaluated by using 40 knees but the presentation of result was not clear (Lee and Chung 2005). Meanwhile, Dodin et al. (2011) has imported the ray casting algorithm, which was typically used to create solid geometry model in computer graphic, into knee bone segmentation. The algorithm decomposed knee image into multiple surface layers via Laplacian operators. At each surface layer, the algorithm relied on a set of localization points known as “observers” as input to project the ray pattern at different angles and derived the cylindrical pattern of bone. The model was able to capture local bone irregularities such as osteophytes, which might be taken as error by shape-based methods. Eventually, the model was validated on a larger data size of 161 images and has reported DSC of 0.94 for femur and 0.92 for tibia (Dodin et al. 2011).
In a similar fashion, Dalvi et al. (2007) and Gandhamal et al. (2017) have implemented level set in their knee bone segmentation models. Specifically, Dalvi et al. (2007) used a region growing algorithm to undersegment the knee bone and followed by segmentation refinement via Laplacian level set algorithm. It was validated by using sensitivity (Sens) and specificity (Spec) evaluation metrics on two healthy subjects. Gandhamal et al. (2017) proposed a hierarchical knee bone segmentation model. At preprocessing stage, the image intensity was transformed by a sigmoid-alike function to improve the contrast between soft and hard tissues. Then, two automatic seeds was identified to represent the femur and tibia, and used to initiate a novel distance-regularized level-set evolution (DRLSE) algorithm to extract the bone regions. Once the evolution completed in one slice, geometric centroids of the level set function would be updated and used in successive slices. The limitation of this level-set function, for instance, was that it would stop if the area of the earlier segmented bone region was less than 100 pixels. Therefore, the algorithm would perform badly in very small bone region as well as when the bone was separated into two regions.
In conclusion, there were two key highlights. First, segmentation models in this category were independent on any training dataset or user interaction, which were different from shape-, atlas-, graph-, and machine learning-based methods. To ensure the model remained automatic, the learning gap was filled by a variety of preprocessing and image property learning procedures instead. Second, different strategies have been adopted to attain the final bone segmentation based on modified image properties. While these models were able to overcome common anatomical features of bone, their suitability would depend heavily on tissue and image property of the image. Besides, some models required predefined threshold values. Consequently, it could be hard to generalize these models to dataset of larger size in comparison to modern machine learning techniques, especially deep learning.
3.5 Classical machine learning-based methods
General machine learning framework comprises of data and prediction algorithm. The data is a set of observations used during the training and testing while prediction algorithm learns descriptive data pattern to perform certain classification task. Classical machine learning uses a set of discriminative handcrafted features to describe the object of interest and fed to a classifier to assign image pixel to the most likely label. The family of machine learning is wide; comprising of supervised learning, unsupervised learning, semi-supervised learning and reinforcement learning. Noteworthy, supervised and unsupervised learning represent two dominant clusters of learning algorithms which can be applied to any machine learning.
Supervised learning studies the relationship between an input space \(x\) and a label (output) space \(y\). Given a set of labels \(\left\{\mathrm{0,1},..,L\right\}\), the model acquires the functional relationship between the input and label \(f:x\to y\), where the mapping \(f\) is a classifier by taking training data, \(\left({X}_{1},{Y}_{1}\right),\dots ,\left({X}_{n},{Y}_{n}\right)\in x\times y\), as source of learning. Supervised learning is commonly applied for regression and classification problems. Common supervised learning algorithms include:
-
1.
Decision Tree (Quinlan 1986): The algorithm is in a form of tree structure with branches and nodes. Each leaf node represents a class label and each branch represents the outcome. The algorithm will hierarchically sort attributes from the root of the tree until it reaches a leaf node.
-
2.
Naïve Bayes (Rish 2001): The algorithm applies Bayes’ Theorem, which assumes that features are statistical independent. The classification is performed based on conditional probability of an occurrence of an outcome derived from the probabilities imposed on it by the input variables.
-
3.
Support Vector Machine (Cortes and Vapnik 1995): A margin is defined as the distance between two supporting vectors which are separated by a hyperplane. Larger margin implies smaller classification errors. Thus, the algorithm aims to draw the most suitable margins in which the distance between each class and the nearest margin is maximized.
-
4.
Ensemble Learning (Rokach 2010): A method to aggregate multiple weak classifiers to construct a strong classifier. Important ensemble learning algorithms include boosting and bagging.
On the other hand, there is no labeled data in unsupervised learning. So unsupervised model draws inferences from input data based on similarities and redundancy reduction during the training. Clustering and association rule are two well-known types of unsupervised learning. Popular unsupervised learning algorithms include:
-
1.
k-Means (MacQueen 1967): This clustering algorithm groups data into k clusters based on their homogeneity. An individual mean value represents the center of each cluster. During the implementation, data values will be assigned to the most likely class label based on their proximity to the nearest mean with the least error function.
-
2.
Principal Component Analysis (Jolliffe 2002): This method aims to reduce of dimensionality of data by finding a set of mutually un-correlated linear low dimensional data representations which have largest variance. This linear dimensionality technique is useful in exploring the latent interaction between the variable in an unsupervised setting.
Bourgeat et al. (2007) extracted additional texture features from phase information of MR image to consider magnetic property of tissue. The features were fed into a multiscale support vector machine (SVM) framework. An image subsampling process would classify the voxels from coarse-to-fine representation. In total, 40,000 voxels were extracted from 4 training images. In order to maintain the segmentation quality, contour refinement was performed to eliminate jaggy boundary from the segmented bones (Bourgeat et al. 2007). Fabian et al. (2015) applied random forest (RF) classifier with bagged decision trees of 20 trees to segment the femur. Only 5% of the femur voxels and 5% of the non-femur voxels from each data were selected to train the classifier by using a set of features that included spatial location, volumetric mean, volumetric variance, volumetric entropy, skewness, kurtosis, edge and Hessian (Fabian et al. 2015). Nonetheless, the classification accuracy relied heavily on the quality of labeled data, which indicated the major limitation faced by classical machine learning.
3.6 Deep learning-based methods
Deep learning is a powerful machine learning model equipped with automatic hierarchical feature representation learning ability. General architecture comprises of input layer, hidden (feature extraction) layers and output (classification) layer (Goceri 2018). A comparison between classical machine learning and deep learning is shown in Fig. 5. Major architectures of deep learning (LeCun et al. 2015) are convolutional neural network (CNN), recurrent neural network (RNN), recursive neural network and unsupervised pretrained networks (UPN). Among these networks, CNN is well suited to image processing applications such as object detection, image classification and segmentation. During model training, the value of each node is estimated by parameterizing weights through convolutional filters and the objective function is then optimized via backpropagation.
Segmentation of knee bone by using a classical machine learning and b deep learning. Feature engineering of classical machine learning involves handpicked feature representations and mapping. On the other hand, deep learning uses multiple hidden layers to extract hierarchical feature representations
A list of deep learning-based knee bone segmentation is indicated here: (Almajalid et al. 2019; Ambellan et al. 2019; Cheng et al. 2020; Lee et al. 2018; Liu et al. 2018a; Zhou et al. 2018). In general, knee bone segmentation model adopts CNN architecture with some modifications. Liu et al. (2018a) built a 10-layers SegNet framework with discarded fully connected layer after the decoder network, to perform pixelwise semantic labelling on 2D knee image. The processed labels were fed to the marching cube algorithm to generate 3D simplex mesh. Then, the simplex mesh was sent to 3D simplex deformable process with each individual segmentation objects being separately refined based on the source image (Liu et al. 2018a). Lastly, performance of the model was compared to U-net. Because SegNet removed fully connected layer, the model has lower number of parameters. Besides, SegNet performed a nonlinear upsampling; hence, credential features and boundary delineation could be better reconstructed. Later, Zhou et al. (2018) extended the model into multiple tissue segmentation by using conditional RF to perform multiclass classification. The model has reported DSC accuracy of 0.97 for femur, 0.962 for tibia and 0.898 for patella (Zhou et al. 2018).
Ambellan et al. (2019) adopted the concept of slice-wise segmentation from Liu et al. (2018a) and added SSM as extra feature into their 2D/3D U-net based bone segmentation model. The purpose of SSM was to overcome the holes in segmentation masks due to poor intensity contrast or image artifacts as well as to remove false positive voxels from femur and tibia that were detected at the outside of typical range of osteophytic growth. By utilizing 60 training shapes, the model has attained excellent DSC of 0.986 for femur and 0.985 for tibia (Ambellan et al. 2019). Notwithstanding, the good performance was achieved at the expense of huge computational resources and localized training. For instance, general-purpose graphic cards with smaller memory were not capable to support the 3D convolution, so it wouldn’t be easy to extend the model to process larger dataset without suitable graphic card. Besides, the 3D model was trained on small subvolumes of 64 × 64 × 16 voxels along the bone contours to reduce computational burden and compensated the inability of SSM to provide osteophytic details. The training option, however, compromised the surrounding voxel intensity and texture feature.
In recognition of abovementioned limitations, Cheng et al. (2020) proposed a simplified CNN model, known as holistically nested network (HNN), to segment femur and patella bones. HNN eliminated the decoding path to form a forward-feeding network; and reduced the graphic card computational size. Plus, the network was trained on whole knee image by using a 1 × 1 convolution at first layer (to produce fine details such as edge) until a 32 × 32 convolution at fifth layer (to produce coarse details such as shape of bone; thereby, acquiring both local and global contextual information. At the end, a weighted fusion layer was developed to average the probability map at each layer and successively computed the final prediction (Cheng et al. 2020). Although the authors tried to perform a comprehensive validation against current state-of-art, it was hindered by the type of bone selection (immature bone vs mature bone; and different bone compartment) and the absence of public groundtruth. Moreover, it is noteworthy that training of deep learning model is computationally heavy in spite of its better robustness. According to Ambellan et al. (2019), implementation of deep learning model on large scale data image of 50,000 would consume 43 weeks on a single computational node, which explicitly highlighted the expansive cost of computation. Some researchers have simplified CNN architecture to reduce complexity, but the issue still required further analysis.
4 Knee cartilage segmentation
Knee cartilage segmentation from MR image produces cartilage model, which is used in a broad range of OA-related studies: imaging biomarkers analysis (Hafezi-Nejad et al. 2017; Schaefer et al. 2017; Shah et al. 2019; Williams et al. 2010a), classification and detection of knee OA progression (Ashinsky et al. 2017; Ashinsky et al. 2015; Chang et al. 2018; Almajalid et al. 2019a; Tiulpin et al. 2019), biomechanical modeling (Liukkonen et al. 2017b) and stimulation of cartilage degeneration (Liukkonen et al. 2017a; Mononen et al. 2019, 2016; Peuna et al. 2018). To date, cartilage segmentation remains an active research problem because knee cartilage has very thin structure at a few millimeters wherein some extremely thin areas are measured in submillimeter. Examples of knee cartilage geometry complexity are shown in Fig. 6. Moreover, femoral, tibial and patellae cartilage have distinctively different and changing shapes across the slides.
Illustrations of anatomical complexity demonstrated by knee cartilage. a Irregular cartilage structure with diffuse boundary (white arrowheads). b Thin cartilage structure (white arrowheads) adds more challenges to segmentation process. c Effusion between at patellofemoral cartilage (white arrows) adds ambiguity to segmentation accuracy. d Narrow tibiofemoral boundary (white arrows) hardens the segmentation of tibial and femoral cartilage
Some medical image analysis tools (Akhtar et al. 2007; Bonaretti et al. 2020; Duryea et al. 2016; Gan et al. 2014a; Iranpour-Boroujeni et al. 2011) were developed to preprocess knee image, segment the cartilage and quantify knee OA progression via imaging biomarkers. Established medical image analysis consultancy such as Imorphics (based in Manchester, UK), ArthroVision (based in Montreal, Canada) and Chondrometrics (based in Ainring, Germany) caters to the need of medical image analysis service. Complete lists of classical cartilage segmentation model were given in Table 4 (semiautomatic) and Table 5 (fully automatic). An updated list of deep learning-based knee cartilage segmentation models was given in Table 6.
4.1 Region-based methods
Region growing exploits the homogeneity property of neighboring pixels’ values. Often, users need to place an initial set of seed points \({S}_{i}=\left\{{S}_{1},{S}_{2},..,{S}_{n}\right\}\). Then, the algorithm will start to expand to neighboring pixels in search for homogenous pixels (Adams and Bischof 1994). Let \(T\) be the set of all pixels which are adjacent to at least one of the pixels in \({S}_{i}\)
where \(nb(x)\) is the set of immediate neighbors of the pixel \(x\). The search will continue updating the mean of corresponding region and expand until the similarity criterion is breached. Common similarity criterion includes intensity or image texture. Classical region growing is not good at coping with inhomogeneous image property landscape of knee image. As a result, researchers need to combine region growing with other image processing techniques in their segmentation models.
Pakin et al. (2002) applied region growing to obtain an initial segmentation of knee cartilage from image background. A subsequent two-class local clustering voting mechanism was introduced to determine the class of unlabeled regions based on their proximity to knee bone and contrast difference at the region boundaries. Lastly, surface mesh was generated to produce a 3D cartilage model (Pakin et al. 2002). While the model has reported an accuracy of 98.87%, it was validated on a MR image of knee only. Cashman et al. (2002) proposed a multistage region growing-based knee cartilage segmentation model. At preprocessing stage, image noise was filtered with median filter and background image was removed by using edge detection and thresholding. Then, a recursive region grow method would pre-segment the image to produce a bone-plus-cartilage region mask, where the bone region would be subtracted to leave-out cartilage (Cashman et al. 2002). Similar work was found at Riza et al. (2019).
Region growing was one of the earliest segmentation algorithm implemented in knee cartilage segmentation. Although it was simple to use, direct implementation was computational heavy. Further, knee images were infamous for weak tissue boundaries; causing potential under- or oversegmentation. Significant amount of manual intervention was required throughout the segmentation to fill in discontinued boundary during edge detection, to place seed points inside bone region during pre-segmentation and to eliminate any residual non-cartilage pixels adhering to the outside of cartilage. Furthermore, performance of region growing methods (Pakin et al. 2002; Riza et al. 2019) were not properly validated. These limitations question the applicability of region growing to be extended to large patient groups.
4.2 Deformable model-based methods
Application of traditional ACM in knee cartilage segmentation model could be found in Stammberger et al. (1999), Lynch et al. (2000), Duryea et al. (2007) and Brem et al. (2009). As highlighted in Sect. 3.1, traditional ACM was sensitive to initialization and suffered from poor convergence performance of the contour for concave boundaries. Some improvement works were proposed. Gradient Vector Flow (GVF) was integrated into ACM to resolve abovementioned problems but the model was still vulnerable to poor tissue contrast or blur cartilage boundary (Tang et al. 2006). Carballido-Gamio et al. (2005) used Bezier spline to control active contour formation. The Bezier spline was created by manually inserting control points inside the cartilage. Then, local information acquired from the initial control points would update the Bezier spline until it reached the articular surface. The model also took into consideration the disconnected cartilage boundary; hence, it was more robust to weak boundary problem (Carballido-Gamio et al. 2005). Meanwhile, Ahn et al. (2016) applied level set contour as an active contour. At initialization phase, 20 normal knee templates were used to estimate the initial contour in level set. The energy function was designed to consider local regions while spatial information from the templates was incorporated into the energy function to minimize the effect of noise (Ahn et al. 2016).
Apart from ACM, researchers also apply ASM for cartilage segmentation. The model requires user to place n landmark points, \(\left\{\left({x}_{1},{y}_{1}\right),\left({x}_{2},{y}_{2}\right),\dots ,\left({x}_{n},{y}_{n}\right)\right\}\) on cartilage boundary (see Fig. 7), which is arranged as a 2n element vector, \(X={({x}_{1},\dots ,{x}_{n},{y}_{1},\dots ,{y}_{n})}^{T}\). The landmarking process is repeated on a stack of \(N\) training images to produce a cloud of landmark points. These shapes are aligned in a common model coordinate frame by using the Procrustes algorithm. Then, principal component analysis (PCA) is implemented on the set of vectors \(\left\{{X}_{i}\right\}\). An affine transformation will be defining the position, \(\left({X}_{t},{Y}_{t}\right)\), orientation, \(\theta\), and scale, \(s\), of the model in knee image frame.
Active Shape Model Algorithm Summary: |
|---|
1. Calculate the mean of the data |
\(\stackrel{\sim }{X}= \frac{1}{N}\sum_{i=1}^{N}{X}_{i}\) |
2. Calculate the covariance of the data |
\(S= \frac{1}{N-1}\sum_{i=1}^{N}({X}_{i}-\stackrel{\sim }{X}){({X}_{i}-\stackrel{\sim }{X})}^{T}\) |
3. Compute the eigenvectors, \({\mathcal{P}}_{i}\) and corresponding eigenvalues, \({\lambda }_{i}\) of the covariance matrix. Each eigenvalue gives the variance of the data about the mean in the direction of the corresponding eigenvector |
4. Select the \(m\) largest eigenvalues where \(m\) is the number of mode of variation |
5. Approximate the linear shape model given the eigenvectors \(\left\{{\mathcal{P}}_{i}\right\}\) and shape parameters \(b\) |
\(X\approx \stackrel{\sim }{X}+\sum_{m}{\mathcal{P}}_{m}{b}_{m}\) |
6. Update the parameters pose parameters \(\left({X}_{t}, {Y}_{t},s,\theta \right)\) and shape parameters \(b\) |
7. Repeat until convergence |
Solloway et al. (1997) placed 42 around the cartilage boundary and 22 landmark points around the endosteal surface of femoral condyles to produce a 2D femoral cartilage ASM model. In total, 10 modes of variations were utilized to derive the mean shape approximation. The model has achieved CV of 2.8% for cartilage thickness measurement (Solloway et al. 1997). However, the shape variation flexibility was often constraint by the number of principal components extracted from the diagonal of the covariance matrix, which depend on the number of training shapes. As a result, traditional ASM suffered from over-restrictive shape variation and problematic re-initialization in knee cartilage segmentation.
To overcome these limitations, two studies have integrated additional feature information during the training of shape model. González and Escalante-Ramírez (2013) has tested Hermite transform, Haar- and Sym-5 wavelet transform on the x and y coordinate directions of the contour in an attempt to capture more information from the contours at different spatial resolutions. The Sym-5 wavelet transform based model has reported the best DSC score of 0.8329 with 16 training samples (González and Escalante-Ramírez 2013). In another study, González and Escalante-Ramírez (2014) has combined texture features from Local Binary Patterns (LBP) into ASM. The texture information from each landmark point was computed into LBP histogram. Unfortunately, the model only produced DSC of 0.8132. (González and Escalante-Ramírez 2014). Accordingly, different modified ASM models failed to demonstrate apparent accuracy attainment, instead these models continued to rely on handpicked feature and number of training samples. Although direct performance comparison between different deformable models is not available, neither a single deformable model nor modified deformable model seems to be able to cope with frequent changes in cartilage structure.
4.3 Graph-based methods
Graph cuts defines a segmentation as an optimization of energy cost function problem. Bae et al (2009) and Shim et al. (2009a) have applied classical graph cuts to segment knee cartilage of 20 and 10 subjects, respectively. User scribbles were utilized as hard constraint and the segmentation has reported good DSC accuracy of 94.3%. However, both works did not address the notorious smallcut problem and image noise problem suffered by the algorithm. Besides, classical graph cuts does not support multiclass segmentation.
To address abovementioned limitations, a hierarchical segmentation model known as Layered Optimal Graph Segmentation of Multiple Objects and Surfaces (LOGISMOS) was proposed by Yin et al. (2010). The model approximated the interaction between interacting surfaces of different objects by utilizing prior knowledge about knee structure, which was infused as multi-surface interaction constraint and multi-objective interactive constraint during graph construction. The former analyzed the relationship between bone and surrounding soft tissue inclusive of cartilage, while the latter analyzed the relationship between bone and cartilage. The design of cost function was essential to accommodate all pretrained information (Yin et al. 2010). Unfortunately, the cost function in original LOGISMOS failed to capture the regionally-specific appearance of the surrounding menisci, muscle bone and other anatomies; which caused certain intensity profile of normal cartilage areas to be mistaken as pathological case. An extension of LOGISMOS with Just Enough Interaction (JEI) was introduced in Kashyap et al. (2018) to rectify the cost function.
Another type of graph-based method, random walks models the segmentation problem as looking for solution to Dirichlet problem. In theory, a harmonic function that satisfies the boundary condition will minimize the Dirichlet integral. Thus, the probability of unlabeled pixel belongs to each label class could be computed by solving a system of linear equations.
Random Walks Algorithm Summary: |
|---|
1. Map the image intensity value, \(g\), to edge weights, \(w\), for two pixels \(i\) and \(j\) in lattice structure |
\({w}_{ij}= exp\left(-\beta {\left({g}_{i}-{g}_{j}\right)}^{2}\right)\) where \(\beta\) is a free parameter |
2. Organize the nodes into two sets, \({V}_{M}\) (labeled nodes) and \({V}_{U}\) (unlabeled nodes) such that \({{V}_{M}\cup V}_{U}=V\) and \({{V}_{M}\cap V}_{U}=\varnothing\) |
3. Obtain a set of \({V}_{M}\) labeled pixels with \(K\) labels via interactive or automatic approach |
4. Define the set of labels for the labeled pixels as a function |
\(Q\left({v}_{j}\right)= s,\forall {v}_{j}\in {V}_{M}\) where \(s\in {\mathbb{Z}},0<s<K\) |
5. Define the \(\left|{V}_{M}\right|\times 1\) vector for each label, \(s\), at node \({v}_{j}\in {V}_{M}\) as |
\({m}_{j}^{s}=\left\{\begin{array}{cc}1& if Q\left({v}_{j}\right)= s \\ 0& if Q\left({v}_{j}\right)\ne s\end{array}\right.\) |
6. Resolve the combinatorial Dirichlet problem for each label |
\({L}_{U}{x}^{s}= -{B}^{T}{m}^{s}\) |
7. Compute a final segmentation by assigning to each node, \({v}_{i}\), the label corresponding to \({max}_{s}({x}_{i}^{s})\), where the probabilities at any node will sum to unity,\(\sum_{s}{x}_{i}^{s}=1,\forall {v}_{i}\in V\) |
Because random walks is robust to weak boundary problem, it can overcome the diffuse boundary observed in pathological cartilage. Thorough analyses were conducted to analyze the performance of random walks for knee cartilage segmentation (Gan and Sayuti 2016; Gan et al. 2017,2019,2018,2014b, c). The model was evaluated against manual segmentation. As shown in Fig. 8, classical random walks relied heavily on seed points’ locations to provide local information about knee structure (Gan et al. 2017). Subsequently, an improved model was developed, which demonstrated DSC accuracy of 0.94, 0.91 and 0.88 for normal femoral, tibial and patellae cartilage, as well as 0.93, 0.88 and 0.84 for pathological femoral, tibial and patellae cartilage (Gan et al. 2019).
In summary, design of energy function plays an influential role in developing graph-based methods. A majority of the graph-based methods’ energy function in knee cartilage segmentation aimed to partition the graph through min-cut concept. However, minimization of the energy function was always a daunting task due to various issues such as smallcut and binary segmentation problem. Besides, incorporation of user interaction to provide priori knowledge was ubiquitous in both advanced graph model such as LOGISMOS and other simpler graph-based models. User-specific markers was manually inserted through scribbles or boundary points. The priori knowledge was essential to initialize and modify the segmentation (Bowers et al. 2008; Gan et al. 2017, 2019; Gougoutas et al. 2004) as well as to compensate the lacking of cost function (Kashyap et al. 2018). Consequently, graph-based methods were often plagued with overdependence on user interaction in order to attain desirable segmentation results.
4.4 Atlas-based methods
Different from previously discussed segmentation methods, atlas-based segmentation makes use of priori knowledge from labeled training images to segment the target image. Because the atlas is directly created by expert, the priori information is rich of discriminative details about the location, shape, object class, priori probabilities and topological details of target object. Given that the knee cartilages are sharing similar texture and spatial features, as well as ill-defined boundary with surrounding soft tissues, atlas-based methods is expected to excel in knee cartilage segmentation. Still, atlas-based methods are not without any disadvantage. Creation of atlas can be time- and resources consuming, and small number of atlas image can potentially lead to overfitting problem.
There are common four atlas selection methods, namely single atlas (which selects a reference image from a set of labeled images), the best atlas (which identify the most suitable labeled image from the set), averaged-shape atlas (which constructs an averaged atlas from a set of labeled images), and multiple atlases (which registers every individual labeled image to the test image independently). The preceding three selection approaches were not robust enough though, so most studies have employed the multiple atlas method in knee cartilage segmentation model (Carballido-Gamio and Majumdar 2011; Dam et al. 2015; Lee et al. 2014; Liu et al. 2015; Shan et al. 2012a, 2012b, 2014; Tamez-Peña et al. 2012). The main role of atlas was to provide spatial prior to guide an automatic multilayer knee cartilage segmentation model at initialization stage.
Among these works, Shan’s research group has conducted a pipeline of studies to investigate the most suitable atlas-assisted probabilistic classification segmentation structure (Shan et al. 2012a, 2012b, 2014). Priori information was transferred from the atlas to test image via non-rigid image registration. Interestingly, the concept was somehow similar to the classical machine learning-based methods, which we would discuss later. Compared to other atlas-based models, their final model (Shan et al. 2014) have registered the best DSC score of 0.856 for femoral cartilage and 0.859 tibial cartilage on a group of 155 subjects. A major disadvantage of atlas-based method, however, was its dependence on registration method and anatomical similarity between the atlas and the subject to achieve good performance.
4.5 Classical machine learning-based methods
Given its anatomical complexity, classification of cartilage is a daunting task indeed. Folkesson et al. (2007) has published one of the earliest classification model by using two binary kNN classifiers to segment the femoral and tibial cartilage. The feature learning was a computational heavy process. Approximately 500,000 training voxels for background, 120,000 voxels for tibia cartilage and 300,000 voxels for femoral cartilage were involved. To alleviate this issue, human knowledge prior about the location of the cartilage was pre-defined at the initialization stage (Folkesson et al. 2007). Even though the model reported DSC of 0.80, it became the benchmark for future classification-based knee cartilage segmentation model.
Numerous multi-stage/multi-level (Dodin et al. 2010; Lee et al. 2011; Öztürk and Albayrak 2016; Pang et al. 2015; Wang et al. 2014; Zhang et al. 2013) classification models have employed different approaches such as bone pre-segmentation to derive BCI (Lee et al. 2011; Wang et al. 2014), SVM based edge classification (Pang et al. 2015), utilization of four types of image contrast to extract rich features (Zhang et al. 2013) and subsampling of background voxels to enable feasible kNN classification (Öztürk and Albayrak 2016). Because different researchers were restricted to their own classification strategies, their models’ architectures varied significantly according to the image feature, spatial priors, imaging sequence type and cartilage type. Consequently, these models lacked the generalizability to unseen pathological features in knee image. Further, handpicked features were subjective to the training data; these concerns severely undermined the robustness of classification models.
For example, Öztürk and Albayrak (2016) applied central coordinate computation and one-versus-all classification. During the training, subsampling processes were adopted to eliminate abundant background voxels step-by-step and helped to increase computational feasibility. A total of 150 features were extracted. At the testing, separate kNN classifiers were used to classify femoral, tibial and patellae cartilage. Despite its complexity, the model merely achieved DSC of 0.826 for femoral cartilage, 0.831 for tibial cartilage and 0.726 for patellae cartilage. In Zhang et al. (2013), T1-weighted FS SPGR, T2/T1-weighted FIESTA, T2/T1-weighted IDEAL GRE waster and fat imaging sequence were used to exploit spectral correlation among different imaging sequences. But the model has reported very big results variance; depending on the inclusion of number of features (DSC ranged from 0.019 to 0.880) and classification models (DSC ranged from 0.456 to 0.880).
4.6 Deep Learning-based methods
Recently, artificial intelligence (AI), especially deep learning, has emerged as a popular research topic (Goceri and Goceri 2017). Deep learning uses convolutional filters to extract deep features and fed the concatenated feature vector into dense layer (see Fig. 9). Large number of studies (Ambellan et al. 2019; Norman et al. 2018a; Panfilov et al. 2019; Prasoon et al. 2013; Raj et al. 2018; Tack and Zachow 2019; Tan et al. 2019; Xu and Niethammer 2019) which used deep learning in knee cartilage segmentation were published. In particular, CNN architecture has received the most research attention. For example, a U-net architecture of 4 convolutional layers and kernel filter of 5 × 5 for 2D CNN and 5 × 5 × 5 for 3D CNN was adopted in Ambellan et al. (2019). In Prasoon et al. (2013), three CNN models were computed from xy-, -yz, and –zx planes respectively with a kernel size of 5 × 5. Meanwhile, a 3D CNN with 5 convolutional layers and 3 × 3 × 3 kernel filter was adopted in Tack and Zachow (2019).
Among these works, some have added extra improvements to enhance the accuracy of existing models. Given that U-net failed to segment low contrast areas, Ambellan et al. (2019) imported shape information from SSM to fill in holes and sub-holes in segmentation masks. A better segmentation accuracy of 85.6–89.9% (DSC) was reported at the expense of laborious SSM construction. On the other hand, Panfilov et al. (2019) attempted two regularization techniques, namely mix-up and unsupervised domain adaptation (UDA) to improve the robustness of their U-net model. Unfortunately, their investigation showed mixed results, and even performance deterioration when both mix-up and UDA were combined together.
Tan et al. (2019) introduced a deep learning segmentation framework which integrated collaborative multi-agent learning mechanism to label cartilage and discriminator to determine output cartilage label. A V-net with 3 convolutional layers and 2 × 2 kernel filters served as the fundamental of segmentation model. Their work has reported high accuracy of 0.900 ± 0.037 for femoral cartilage, 0.889 ± 0.038 for tibial cartilage and 0.880 ± 0.043 for patellae cartilage. Another deep learning segmentation, DeepAtlas (Xu and Niethammer 2019) proposed a joint learning mechanism from weakly supervised image registration and semi-supervised segmentation learning. The authors have adopted a U-net structure. During the learning mechanism, an anatomy similarity loss would compute the segmentation dissimilarity through matching segmentations between the target image and the warped moving image in order to guide the model training. Despite the authors claimed the DeepAtlas would benefit from fewer manual segmentations during model training, it suffered from lower accuracy performance i.e. DSC of 81.19 ± 3.47% and complex learning objective function.
Many deep learning-based segmentation models were validated on more than one dataset. For example, DeepAtlas was validated on the OAI dataset and OASIS-TRT dataset (brain MR images), while Tack and Zachow(2019) and Ambellan et al. (2019) utilized SKI10, OAI Imorphics and OAI Zuse Institute Berlin (ZIB) dataset to evaluate their models’ performance. However, a real problem in medical image analysis was the lack of large-scale annotated image data with high quality (Goceri 2019). To segment knee cartilage, research groups have to either train their CNN models from scratch by utilizing small amount of labelled images or build a large in-house training dataset. The former could easily lead to overfitting problem while the latter would incur substantial financial and expert resources. Another concern associated with deep learning model was the enormous computational memory requirement, which was implicitly reflected through the selection of 2D slice-by-slice segmentation option in most knee cartilage segmentation models.
5 Evaluation of computational segmentation models
In 2010, SKI10 was announced in Grant Challenge Workshop organized by MICCAI (https://ski10.org) to promote a common evaluation framework among segmentation models by using a public dataset. A total of 170 research teams have registered and the best research team has reported an averaged total score of 75.73 (Ambellan et al. 2019). Besides, evaluation were localized into subregions in many cartilage segmentation studies (see Fig. 10). Given a broadly diversified types of segmentation models, it is hard to systematically evaluate their performance as a whole. Therefore, in following sub-sections, we analyzed performance of these models from the perspective of 1) deep learning against classical segmentation models, and 2) biomarkers in computational models.
(Left) Coronal view of DESS knee image showing division of femorotibial subregions i.e. central lateral femoral cartilage (cLFC), central tibial cartilage (cLTC), lateral tibia (LTB) and medial tibia (MTB). (Right) Cartilage plate maps detailing different subregions on femoral and tibial cartilage where “a” denotes anterior, “p” denotes posterior, “i” denotes interior, “e” denotes external and “c” denotes central (Eckstein and Peterfy 2016)
5.1 Performance of deep learning versus classical segmentation models
The standard practice to assess the performance of computational knee segmentation model is by comparing the segmentation results against groundtruth. Because groundtruth is often created by expert through manual segmentation, there is a scarcity of public groundtruth to assess different segmentation models in global domain. Among the statistical evaluation metrics (see Table 7), DSC was extensively applied metric in classical and deep learning segmentation models, especially cartilage segmentation to evaluate the degree of agreement between groundtruth and segmentation. Besides, two surface distance evaluation metrics were frequently used in bone segmentation i.e. average symmetric surface distance (ASD) and root-mean-square symmetric surface distance (RMSD). To compute the measurement, each surface boundary voxel of the segmentation was compared to the closest boundary voxel in groundtruth and Euclidean distance difference was derived and stored in a list.
A number of fully automatic segmentation models overlapped either 2D or 3D segmented cartilage (Ahn et al. 2016; Dodin et al. 2010; Folkesson et al. 2007; Liu et al. 2015; Öztürk and Albayrak 2016; Tamez-Peña et al. 2012; Zhang et al. 2013) or segmented bone (Ababneh et al. 2011; Bourgeat et al. 2007; Fabian et al. 2015; Fripp et al. 2007; Gandhamal et al. 2017; Wang et al. 2014) with groundtruth to validate the accuracy of their models. Overall, classical cartilage segmentation models’ DSC ranged from 70 to 88% while classical bone segmentation models’ DSC ranged from 90 to 97%. In semiautomatic cartilage segmentation models, both DSC (Gan et al. 2017, 2019; Liukkonen et al. 2017b; Shim et al. 2009a) and CV (Bae et al. 2009; Bowers et al. 2008; Brem et al. 2009; Duryea et al. 2007; Gougoutas et al. 2004; Lynch et al. 2000; Stammberger et al. 1999; Tang et al. 2006) were two equally important evaluation metrics. DSCs of semiautomatic cartilage segmentation models ranged from 80 to 94% and CV was measured from the perspective of different observers (inter-observer), within observer (intra-observer), different subjects, and different scans. The result details of CV could be referred to Table 4. Despite semiautomatic cartilage segmentation models have achieved better DSC results than fully automatic segmentation models, the number of knees in the former was small, attributed to the need of expert to supervise the segmentation. On the other hand, the highest DSC score attained by bone segmentation model was 97%. The high accuracy of segmented bone model enables researchers (Dam et al. 2015; Lee et al. 2014; Shan et al. 2014; Wang et al. 2014; Yin et al. 2010) to exploit the spatial relationship between bone and cartilage surface and facilitate subsequent cartilage segmentation.
Overall, deep learning-based cartilage and bone segmentation models attained DSC ranged from 80–90% and 97–98%, respectively. In both cases, deep learning models did not demonstrate apparent performance superiority compared to classical segmentation models. There are three key highlights about deep learning-based segmentation models. First, deep learning models demonstrated greater consistency compared to classical segmentation models. Besides, deep learning models were more robust to huge amount of image dataset. Tack and Zachow(2019) used 1378 subjects in their knee cartilage segmentation model while Ambellan et al. (2019) used 507 images in their knee bone segmentation model. Both were the largest sample size to date and it was unprecedented in classical segmentation models. Third, different research groups used their own groundtruth data during training even though their data originated from the OAI or MOST dataset. As a result, there was no regularization to assess and control the quality of annotated data. This also explained the varied accuracies among deep learning models, despite using similar architecture and amount of training data.
5.2 Biomarkers in computational segmentation models
Biomarker is defined as any anatomic, physiologic, or molecular parameter detachable with one or more imaging methods used to help establish the presence and/or severity of disease (Smith et al. 2003). In clinical knee OA research, morphological biomarkers are derived from cartilage and bone 3D models to replace traditional endpoint clinical trials in assessing and validating the morphology and functionality of cartilage tissue in vivo. Morphological biomarkers (see Table 8) were analyzed over a certain range of time point to identify the pattern of joint degradation. Notably, cartilage endures greater degradation at weight-bearing locations. Wluka et al. (2002) investigated cartilage volume loss at weight bearing lateral and medial tibial plateau regions in a two time points study. Based on 132 patients with symptomatic OA, tibial cartilage reported an annual volumetric loss of approximately 5% (Wluka et al. 2002). Pelletier et al. (2007) conducted a 24-months follow-up quantitative MRI subregional (different segments of femoral condyle and tibial plateau) study of 107 patients. Overall, the findings showed that medial region experienced greatest cartilage volume loss. Central region of the medial tibial plateau (cartilage loss: − 84.2 ± 72.4 mm/− 15.0 ± 12.0%) and of the medial femoral condyle (cartilage loss: − 87.9 ± 90.4 mm/− 12.0 ± 11.5%) (Pelletier et al. 2007). Meanwhile Eckstein et al. (2015) have analyzed cartilage thickness loss at tibiofemoral cartilage over 24 months (Eckstein et al. 2015).
5.2.1 Semiautomatic segmentation models
It is standard to test the responsiveness of a knee joint segmentation model by computing test–retest coefficient of variation (CV) via repeated measurements of biomarkers over certain time frame. A number of semiautomatic segmentation models (Akhtar et al. 2007; Bae et al. 2009; Brem et al. 2009; Carballido-Gamio et al. 2005; Cashman et al. 2002; Duryea et al. 2014; Duryea et al. 2007; Tang et al. 2006) were analyzed comprehensively in this manner. An early semiautomatic segmentation models by Waterton et al. (2000) has repeated the measurement of femoral cartilage volume over a time point range of three weeks. The assessment has reported a test–retest CV of 1.6% (Waterton et al. 2000). Brem et al. (2009) computed RMS CV of VC, ThC, AC, tAB to quantify test–retest reproducibility. Based on 12 knees, the paired analyses root mean square CV ranged from 0.9–1.2% for VC, 0.3–0.7% for AC, 0.6–2.7% for tAB and 0.8–1.5% for ThC. Duryea et al. (2014) tested the responsiveness of their segmentation processing tool by measuring the cartilage volume loss at localized fixed region, which reported standardized response mean (SRM) of − 0.52 at largest region (Duryea et al. 2014). However, the number of data used in these studies were usually small.
Moreover, semiautomatic segmentation results are influenced by experience of operators. These operators comprise of musculoskeletal experts, radiologists or clinicians with musculoskeletal subspecialty. Hence, conventional reproducibility analysis, which involves two to three experts, will be divided into inter- and intra-observer reproducibility. Intra-observer reproducibility measures the agreement of repeated results produced by each expert while inter-observer measures the agreement among result produced by experts. For example, reliability of graph cuts-based segmentation model (Bae et al. 2009) was validated on variation error of VC produced by two radiologists. The model reported high inter-observer reproducibility of 1.29 ± 1.05% and 1.67 ± 1.14% for radiologist 1 and 2 and intra-observer reproducibility of 1.31 ± 1.26% and 1.70 ± 1.72% for session 1 and 2, respectively. Duryea et al. (2007) measured the CV of their image segmentation software based on VC and ThC. The findings showed higher inter-observer variance (VC: 2.5–8.6%) (ThC: 1.9–5.2%) than intra-reader variance (VC: 1.6–2.5%) (ThC: 1.2–1.9%).
5.2.2 Fully automatic segmentation models
Only a few automatic knee segmentation models evaluated the longitudinal reproducibility of morphological biomarkers. Tamez-Pena et al. (2012) conducted comprehensive accuracy (in terms of mean difference) and test–retest precision evaluation of cartilage volume, thickness, and curvature biomarkers by using healthy and OA knees. Dam et al. (2015) evaluated the precision of cartilage volumes by using the OAI, CCBR and SKI10 dataset. Both models were tested against manual segmentation. In Tamez-Pena et al. (2012), the accuracy evaluation was localized into subregional areas: femur (F), femoral trochlea (FT), the central medial femur (cMF), the posterior medial femur (pMF), the central lateral femur (cLF), the posterior lateral femur (pLF), the medial tibia (MT), and the lateral tibia (LT). The mean accuracy for volume ranged from − 0.2% for the pLF to 4.1$ for the femur, the thickness accuracies ranged from − 2.2% for the cMF to 10.4% for the MT, the curvature accuracies ranged from − 5.2% for the MT to − 2.1% for the cMF. The large variance in thickness biomarker indicated that some degree of atlas bias were introduced into the segmentation process.
Since neural networks models have greater extraction power through hidden layers, some (Hafezi-Nejad et al. 2017; Shah et al. 2019) have trained the model as a longitudinal clinical tool to obtain insight about the progression of knee OA. Hafezi-Nejad et al. (2017) used a multilayer perceptron (MLP) model to predict the medial joint space loss (JSL) progression by using 24-months changes of cartilage volume loss in five knee plates and anthropometric parameters. The last layer would produce the prediction (importance value) of JSL progression based on association with cartilage degradation at different knee plates. Base on the findings, lateral femoral cartilage was the most predictive of medial JSL progression (average importance value: 0.191; range 0.177–0.204). By using a population of 3910 MR images, Shah et al. (2019) investigated the cartilage thickness change at four different knee points according to three demographic variables i.e. age, sex and body mass index. For instance, the use of neural network enables researchers to investigate the physiological variation of cartilage with respect to various anthropometric variables, and analyze their relationships through multivariate analysis.
6 Discussion and conclusion
Advancement of AI technology has spurred the rise of new machine learning techniques. Inspired by promising accuracy outcomes demonstrated by deep learning-based segmentation models, deep learning has been extended to a wide range of computer-aided diagnosis applications such as classification (Antony et al. 2017; Chang et al. 2018; Górriz et al. 2019; Norman et al. 2018b; Pedoia et al. 2019; Thomas et al. 2020; Tiulpin et al. 2019, 2018) and detection (Lim et al. 2019; Liu et al. 2018b) in radiographs and MRI-based knee OA studies. The ultimate goal is to detect and halt the progression of knee OA at early stage, where the cartilage degeneration remains reversible.
Classification is the process where an algorithm outputs the probability of a label for a given input image. Related knee OA-specific applications include grading of OA by radiography (Antony et al. 2017; Górriz et al. 2019; Norman et al. 2018b; Thomas et al. 2020; Tiulpin and Saarakkala 2019; Tiulpin et al. 2018), grading of OA by MRI (Pedoia et al. 2019), predicting the progression of OA by radiography (Tiulpin et al. 2019), and predicting knee pain by MRI (Chang et al. 2018). Based on the literature, quantification of OA severity via end-to-end deep neural network is vital to provide more precise computer-aided diagnosis to support clinicians in grading the severity of OA patients. Prior to deep learning, Ashinsky et al. (2015) has implemented Weighted Neighbour Distance using Compound Hierarchy of Algorithm Representing Morphology (WND-CHRM) algorithm (Shamir et al. 2008), an open source classical machine learning software dedicated to biological image analysis, to classify normal and pathological knee images.
Antony et al. (2017) utilized a FCN to localize the knee joint and trained a CNN model to classify the OA severity grade. The CNN model has apparently outperformed classical WND-CHRM, producing classification results of 60.3% and 29.3–34.8%, respectively. On a test set of 3,146 training images and 1,300 testing images from the OAI, and 2020 training images and 900 testing images, the jointly trained model has attained precision score of 0.68 for KL grade 0, 0.32 for KL grade 1, 0.53 for KL grade 2, 0.78 for KL grade 3 and 0.81 for KL grade 4. The essence of this work consolidated the baseline state-of-art for the application of deep learning models in this field, and opened up rooms for further enhancement to the model. Noteworthy, attention mechanism was implemented in subsequent classification-orientated deep learning models (Górriz et al. 2019; Norman et al. 2018b; Thomas et al. 2020; Tiulpin et al. 2018) to refine the weights of feature and improve the prediction. A latest attention-mechanism related model has achieved precision score of 0.73 for KL grade 0, 0.38 for KL grade 1, 0.71 for KL grade 2, 0. 82 for KL grade 3 and 0.87 for KL grade 4 (Thomas et al. 2020).
In addition to classification, an early detection model was developed by Lim et al. (2019) to predict the occurrence of OA in patients aged 50 years and above. The architecture consisted of a deep neural network with eight hidden layers and trained along with lifestyle- and health status-associated risk factors. Based on a sample size of 5479 subjects, the proposed model showed an Area Under Curve (AUC) of 76.8%. Nonetheless, the accuracy of this predictive model was limited to patients with determined OA disease, while patients under OA treatment were excluded from the model training. Hence, future models shall consider this limitation as well as other detail decision parameter such as physiological signals. On the other hand, a cartilage lesion detection model was developed by using two CNN models i.e. the former for cartilage segmentation and the latter to detect structural abnormalities on the segmented cartilage. To train the classification model, a total of 17,395 cartilage image patches were extracted from knee cartilage of 175 patients by a musculoskeletal radiologist; in which 2642 image patches were classified as cartilage lesions and the remaining 14,753 image patches were classified as normal cartilage. The model has reported a high AUC of 0.917 and 0.914 in two rounds of evaluations (Liu et al. 2018b).
Along with abovementioned deep learning-based classification and detection models, simulation of cartilage degeneration signifies another future direction of computer-aided diagnosis of knee OA. Herewith, we have presented important publications related to the development of knee cartilage degeneration simulation model (Liukkonen et al. 2017a; Mononen et al. 2019, 2016). Initial simulation algorithm (Mononen et al. 2016) was built on a computational finite element model, which took into account the stress distribution across the cartilage and change of collagen stiffness in cartilage, to estimate the alternations in cartilage tissue properties with time. Fibril reinforced poroviscoelastic (FRPVE) material was chosen to mimic the cartilage tissue. Patient-specific gait cycle experiment would provide the biomechanical stress loading information across different time points. The collagen fibril damage would happen at cartilage region where the tensile tissue stress exceeded a threshold limit of 5 MPa. The initial degeneration algorithm, however, was limited to tibiofemoral compartment and two subject groups i.e. normal weight and obese.
Since, a follow-up study was conducted to validate the performance of this cartilage degeneration stimulation algorithm by separating clinically healthy but obese subjects, diagnosed frim the baseline radiographs, into KL grade 2 and 3 based on the predicted level of cartilage degeneration (Liukkonen et al. 2017a). Twenty one subjects were involved in this study. In femoral cartilage model, an AUC of 0.94 and 0.84 was reported in predicting the cartilage KL grade 3 and 2, respectively. Meanwhile, in tibial cartilage model, AUC of 0.90 and 0.80 were reported in predicting the cartilage KL grade 3 and 2, respectively. In Mononen et al. (2019), a template-based approach was introduced to substitute the previous patient-specific approach. Anatomical dimensions were measured from 21 subjects to create templates from these subjects. Then, matching of template were conducted via minimum RMSE of anatomical dimension between the subject of interest and templates (multiple templates) or minimum RMSE of anatomical dimension between each subject and all templates (one template). An optimal template model was scaled to match anatomical dimension for the subject of interest and eventually, the scaled templated model was simulated along with the simplified gait loading. Interestingly, the approach with one template and the average meniscus support was statistically able to separate all KL grade groups to each other.
Existing research direction focuses on biomarkers to quantify the pattern of knee OA progression wherein nee segmentation serves as the cornerstone in knee OA research pipeline. Early segmentation models applied interaction from human expert for guidance but led to inter- and intra-observer ambiguity and heavy reliance on manual intervention. Then, substantial amount of attention has shifted to fully automatic segmentation models. Learning power of fully automatic segmentation models was limited to subjective feature selection, optimization and knowledge prior information. Consequently, these models failed to generalize to bigger dataset size. Nowadays, AI has transformed knee OA research direction toward prediction and early detection, given that deep learning has demonstrated great potential in terms of generalizability, robustness and versatility. Besides, it is noteworthy that advanced diagnostic applications are gradually becoming the future state-of-art. To embrace future challenges, more investigations are needed to validate the clinical applicability of deep learning models.
References
Ababneh SY, Prescott JW, Gurcan MN (2011) Automatic graph-cut based segmentation of bones from knee magnetic resonance images for osteoarthritis research. Med Image Anal 15:438–448. https://doi.org/10.1016/j.media.2011.01.007
Abdelsamea MM, Gnecco G, Gaber MM, Elyan E (2015) On the relationship between variational level set-based and SOM-based active contours. Comput Intell Neurosci 2015:109029. https://doi.org/10.1155/2015/109029
Adams R, Bischof L (1994) Seeded region growing. IEEE Trans Pattern Anal Mach Intell 16:641–647. https://doi.org/10.1109/34.295913
Ahmet S, Songül A (2020) Knee meniscus segmentation and tear detection from MRI: a review current medical. Imaging 16:2–15. https://doi.org/10.2174/1573405614666181017122109
Ahn C, Bui TD, Lee Y-W, Shin J, Park H (2016) Fully automated, level set-based segmentation for knee MRIs using an adaptive force function and template: data from the osteoarthritis initiative. BioMed Eng 15:99. https://doi.org/10.1186/s12938-016-0225-7
Akhtar S, Poh CL, Kitney RI (2007) An MRI derived articular cartilage visualization framework. Osteoarthr Cartil 15:1070–1085. https://doi.org/10.1016/j.joca.2007.03.009
Almajalid R, Shan J, Du Y, Zhang M (2019a) Identification of knee cartilage changing pattern. Appl Sci 9:1–14
Almajalid R, Shan J, Zhang M, Stonis G, Zhang M (2019b) Knee bone segmentation on three-dimensional MRI. In: IEEE 18th international conference on machine learning and applications (ICMLA), 16–19 Dec. 2019, pp 1725–1730. https://doi.org/10.1109/ICMLA.2019.00280
Ambellan F, Tack A, Ehlke M, Zachow S (2019) Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: data from the osteoarthritis initiative. Med Image Anal 52:109–118. https://doi.org/10.1016/j.media.2018.11.009
Antony J, McGuinness K, Moran K, O’Connor N Automatic detection of knee joints and quantification of knee osteoarthritis severity using convolutional neural networks. In: International conference on machine learning and data mining in pattern recognition, 2017. Lecture Notes in Computer Science. Springer, Cham. https://doi.org/10.1007/978-3-319-62416-7_27
Aprovitola A, Gallo L (2016) Knee bone segmentation from MRI: a classification and literature review. Biocybern Biomed Eng 36:437–449. https://doi.org/10.1016/j.bbe.2015.12.007
Ashinsky BG et al (2015) Machine learning classification of OARSI-scored human articular cartilage using magnetic resonance imaging. Osteoarthr Cartil 23:1704–1712. https://doi.org/10.1016/j.joca.2015.05.028
Ashinsky BG et al (2017) Predicting early symptomatic osteoarthritis in the human knee using machine learning classification of magnetic resonance images from the osteoarthritis initiative. J Orthop Res 35:2243–2250. https://doi.org/10.1002/jor.23519
Bae KT, Shim H, Tao C, Chang S, Wang JH, Boudreau R, Kwoh CK (2009) Intra- and inter-observer reproducibility of volume measurement of knee cartilage segmented from the OAI MR image set using a novel semi-automated segmentation method. Osteoarthr Cartil 17:1589–1597. https://doi.org/10.1016/j.joca.2009.06.003
Barr AJ, Campbell TM, Hopkinson D, Kingsbury SR, Bowes MA, Conaghan PG (2015) A systematic review of the relationship between subchondral bone features, pain and structural pathology in peripheral joint osteoarthritis. Arthritis Res Ther 17:228. https://doi.org/10.1186/s13075-015-0735-x
Bien N et al (2018) Deep-learning-assisted diagnosis for knee magnetic resonance imaging: development and retrospective validation of MRNet. PLOS Med 15:e1002699. https://doi.org/10.1371/journal.pmed.1002699
Bonaretti S, Gold GE, Beaupre GS (2020) pyKNEEr: an image analysis workflow for open and reproducible research on femoral knee cartilage. PLoS ONE 15:1–19. https://doi.org/10.1371/journal.pone.0226501
Bourgeat P, Fripp J, Stanwell P, Ramadan S, Ourselin S (2007) MR image segmentation of the knee bone using phase information. Med Image Anal 11:325–335. https://doi.org/10.1016/j.media.2007.03.003
Bowers ME, Trinh N, Tung GA, Crisco JJ, Kimia BB, Fleming BC (2008) Quantitative MR imaging using “LiveWire” to measure tibiofemoral articular cartilage thickness. Osteoarthr Cartil 16:1167–1173. https://doi.org/10.1016/j.joca.2008.03.005
Boykov Y, Funka-Lea G (2006) Graph cuts and efficient N-D image segmentation. Int J Comput Vision 70:109–131. https://doi.org/10.1007/s11263-006-7934-5
Boykov YY, Jolly M (2001) Interactive graph cuts for optimal boundary & region segmentation of objects in N-D images. In: Proceedings eighth IEEE international conference on computer vision. ICCV 2001, 7–14 July. vol 101, pp 105–112. https://doi.org/10.1109/ICCV.2001.937505
Brem MH et al (2009) Magnetic resonance image segmentation using semi-automated software for quantification of knee articular cartilage—initial evaluation of a technique for paired scans. Skelet Radiol 38:505–511. https://doi.org/10.1007/s00256-009-0658-1
Carballido-Gamio J, Majumdar S (2011) Atlas-based knee cartilage assessment. Magn Reson Med 66:575–581. https://doi.org/10.1002/mrm.22836
Carballido-Gamio J, Bauer JS, Keh-Yang L, Krause S, Majumdar S (2005) Combined image processing techniques for characterization of MRI cartilage of the knee. In: IEEE engineering in medicine and biology 27th annual conference, 17–18 Jan. 2006, pp 3043–3046. https://doi.org/10.1109/IEMBS.2005.1617116
Caselles V, Catté F, Coll T, Dibos F (1993) A geometric model for active contours in image processing. Numer Math 66:1–31. https://doi.org/10.1007/BF01385685
Caselles V, Kimmel R, Sapiro G (1997) Geodesic active contours. Int J Comput Vis 22:61–79. https://doi.org/10.1023/A:1007979827043
Cashman PMM, Kitney RI, Gariba MA, Carter ME (2002) Automated techniques for visualization and mapping of articular cartilage in MR images of the osteoarthritic knee: a base technique for the assessment of microdamage and submicro damage. IEEE Trans Nanobiosci 99:42–51. https://doi.org/10.1109/TNB.2002.806916
Chang GH, Felson DT, Qiu S, Capellini TD, Kolachalama VB (2018) Predicting bilateral knee pain from MR imaging using deep neural networks. bioRxiv:463497 https://doi.org/10.1101/463497
Charlier E et al (2016) Insights on molecular mechanisms of chondrocytes death in osteoarthritis. Int J Mol Sci 17:2146. https://doi.org/10.3390/ijms17122146
Cheng R et al (2020) Fully automated patellofemoral MRI segmentation using holistically nested networks: implications for evaluating patellofemoral osteoarthritis, pain, injury, pathology, and adolescent development. Magn Reson Med 83:139–153. https://doi.org/10.1002/mrm.27920
Cohen LD (1991) On active contour models and balloons. CVGIP: Image Underst 53:211–218. https://doi.org/10.1016/1049-9660(91)90028-N
Collins JE et al (2016) Semiquantitative imaging biomarkers of knee osteoarthritis progression: data from the foundation for the national institutes of health osteoarthritis biomarkers consortium. Arthritis Rheumatol 68:2422–2431. https://doi.org/10.1002/art.39731
Cootes TF, Taylor CJ (1992) Active shape models—‘Smart Snakes’. In: Hogg D, Boyle R (eds) BMVC92, London. Springer, London, pp 266–275
Cootes TF, Edwards GJ, Taylor CJ (2001) Active appearance models. IEEE Trans Pattern Anal Mach Intell 23:681–685. https://doi.org/10.1109/34.927467
Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20:273–297. https://doi.org/10.1007/BF00994018
Dalvi R, Abugharbieh R, Wilson D, Wilson DR (2007) Multi-contrast MR for enhanced bone imaging and segmentation. In: IEEE 29th engineering in medicine and biology society, Lyon, France, 22–26 Aug. 2007. IEEE, pp 5620–5623. https://doi.org/10.1109/IEMBS.2007.4353621
Dam E, Lillholm M, Marques J, Nielsen M (2015) Automatic segmentation of high- and low-field knee MRIs using knee image quantification with data from the osteoarthritis initiative. J Med Imaging 2:024001
Dargan S, Kumar M, Ayyagari MR, Kumar G (2019) A survey of deep learning and its applications: a new paradigm to machine learning. Arch Comput Methods Eng. https://doi.org/10.1007/s11831-019-09344-w
Davies-Tuck ML et al (2010) Development of bone marrow lesions is associated with adverse effects on knee cartilage while resolution is associated with improvement—a potential target for prevention of knee osteoarthritis: a longitudinal study. Arthritis Res Therapy 12:1–10. https://doi.org/10.1186/ar2911
Dodin P, Pelletier J, Martel-Pelletier J, Abram F (2010) Automatic human knee cartilage segmentation from 3-D magnetic resonance images. IEEE Trans Biomed Eng 57:2699–2711. https://doi.org/10.1109/TBME.2010.2058112
Dodin P, Martel-Pelletier J, Pelletier J-P, Abram F (2011) A fully automated human knee 3D MRI bone segmentation using the ray casting technique. Med Biol Eng Comput 49:1413–1424. https://doi.org/10.1007/s11517-011-0838-8
Duryea J et al (2007) Novel fast semi-automated software to segment cartilage for knee MR acquisitions. Osteoarthr Cartil 15:487–492. https://doi.org/10.1016/j.joca.2006.11.002
Duryea J et al (2014) Local area cartilage segmentation: a semiautomated novel method of measuring cartilage loss in knee osteoarthritis. Arthritis Care Res 66:1560–1565. https://doi.org/10.1002/acr.22332
Duryea J, Cheng C, Schaefer LF, Smith S, Madore B (2016) Integration of accelerated MRI and post-processing software: a promising method for studies of knee osteoarthritis. Osteoarthr Cartil 24:1905–1909. https://doi.org/10.1016/j.joca.2016.06.001
Ebrahimkhani S, Jaward MH, Cicuttini FM, Dharmaratne A, Wang Y, de Herrera AGS (2020) A review on segmentation of knee articular cartilage: from conventional methods towards deep learning. Artif Intell Med. https://doi.org/10.1016/j.artmed.2020.101851
Eckstein F, Peterfy C (2016) A 20 years of progress and future of quantitative magnetic resonance imaging (qMRI) of cartilage and articular tissues—personal perspective. Seminars Arthritis Rheumatism 45:639–647. https://doi.org/10.1016/j.semarthrit.2015.11.005
Eckstein F, Wirth W (2011) Quantitative cartilage imaging in knee osteoarthritis. Arthritis 2011:1–19. https://doi.org/10.1155/2011/475684
Eckstein F et al (2006) Proposal for a nomenclature for Magnetic Resonance Imaging based measures of articular cartilage in osteoarthritis. Osteoarthr Cartil 14:974–983. https://doi.org/10.1016/j.joca.2006.03.005
Eckstein F et al (2015) Brief report: cartilage thickness change as an imaging biomarker of knee osteoarthritis progression: data from the foundation for the national institutes of health osteoarthritis biomarkers consortium. Arthritis Rheumatol 67:3184–3189. https://doi.org/10.1002/art.39324
Englund M (2010) The role of biomechanics in the initiation and progression of OA of the knee Best Practice & Research. Clin Rheumatol 24:39–46. https://doi.org/10.1016/j.berh.2009.08.008
Fabian B, Tiziano R, Pletscher M (2015) Distal femur segmentation on MR images using random forests. In: Medical Image Analysis Laboratory. pp 1–6
Falcao AX, Udupa JK, Miyazawa FK (2000) An ultra-fast user-steered image segmentation paradigm: live wire on the fly. IEEE Trans Med Imaging 19:55–62. https://doi.org/10.1109/42.832960
Favero M, Ramonda R, Goldring MB, Goldring SR, Punzi L (2015) Early knee osteoarthritis. RMD Open 1:e000062. https://doi.org/10.1136/rmdopen-2015-000062
Folkesson J, Dam EB, Olsen OF, Pettersen PC, Christiansen C (2007) Segmenting articular cartilage automatically using a voxel classification approach. IEEE Trans Med Imaging 26:106–115. https://doi.org/10.1109/TMI.2006.886808
Fripp J, Crozier S, Warfield SK, Ourselin S (2007) Automatic segmentation of the bone and extraction of the bone–cartilage interface from magnetic resonance images of the knee. Phys Med Biol 52:1617–1631. https://doi.org/10.1088/0031-9155/52/6/005
Gan H-S, Sayuti K (2016) Comparison of improved semi-automated segmentation technique with manual segmentation: data from the osteoarthritis initiative. Am J Appl Sci 13:1068–1075. https://doi.org/10.3844/ajassp.2016.1068.1075
Gan H-S, Tan T-S, Wong L-X, Tham W-K, Sayuti KA, Abdul Karim AH, bin Abdul Kadir MR (2014a) Interactive knee cartilage extraction using efficient segmentation software: data from the osteoarthritis initiative. Bio-Med Mater Eng 24:3145–3157. https://doi.org/10.3233/BME-141137
Gan HS, Tan T, Karim AHA, Sayuti KA, Kadir MRA (2014b) Interactive medical image segmentation with seed precomputation system: data from the osteoarthritis initiative. In: IEEE conference on biomedical engineering and sciences (IECBES), 8–10 Dec 2014. pp 315-318. https://doi.org/10.1109/IECBES.2014.7047510
Gan HS, Tan T, Karim AHA, Sayuti KA, Kadir MRA (2014c) Multilabel graph based approach for knee cartilage segmentation: Data from the osteoarthritis initiative. In: IEEE conference on biomedical engineering and sciences (IECBES), 8–10 Dec 2014, pp 210–213. https://doi.org/10.1109/IECBES.2014.7047487
Gan H-S, Karim AHA, Sayuti KA, Tan T-S, Kadir MRA (2016) Analysis of parameters’ effects in semi-automated knee cartilage segmentation model: data from the osteoarthritis initiative. AIP Conf Proc 1775:030052. https://doi.org/10.1063/1.4965172
Gan H-S, Sayuti KA, Karim AHA (2017) Investigation of random walks knee cartilage segmentation model using inter-observer reproducibility: data from the osteoarthritis initiative. Bio-Med Mater Eng 28:75–85. https://doi.org/10.3233/BME-171658
Gan H, Rosidi RM, Hamidur H, Sayuti KA, Ramlee MH, Karim AHA, Salam BAA (2018) Binary seeds auto generation model for knee cartilage segmentation. In: International conference on intelligent and advanced system (ICIAS), 13–14 Aug. 2018, pp 1–5. https://doi.org/10.1109/ICIAS.2018.8540570
Gan H-S, Sayuti KA, Ramlee MH, Lee Y-S, Wan Mahmud WMH, Abdul Karim AH (2019) Unifying the seeds auto-generation (SAGE) with knee cartilage segmentation framework: data from the osteoarthritis initiative. Int J Comput Assist Radiol Surg 14:755–762. https://doi.org/10.1007/s11548-019-01936-y
Gandhamal A, Talbar S, Gajre S, Razak R, Hani AFM, Kumar D (2017) Fully automated subchondral bone segmentation from knee MR images: data from the osteoarthritis Initiative. Comput Biol Med 88:110–125. https://doi.org/10.1016/j.compbiomed.2017.07.008
Goceri E (2018) Formulas behind deep learning success. In: International conference on applied analysis and mathematical modeling, Istanbul, Turkey, 2018. p 156
Goceri E (2019) Challenges and recent solutions for image segmentation in the era of deep learning. In: 2019 ninth international conference on image processing theory, tools and applications (IPTA), 6–9 Nov 2019. pp 1–6. https://doi.org/10.1109/IPTA.2019.8936087
Goceri E, Goceri N (2017) Deep learning in medical image analysis: recent advances and future trends. In: 11th international conference on computer graphics. Visualization, computer vision and image processing, Lisbon, Portugal, 2017, pp 305–310
Goldring MB, Goldring SR (2010) Articular cartilage and subchondral bone in the pathogenesis of osteoarthritis. Ann N Y Acad Sci 1192:230–237. https://doi.org/10.1111/j.1749-6632.2009.05240.x
González G, Escalante-Ramírez B (2013) Knee cartilage segmentation using active shape models and contrast enhancement from magnetic resonance images vol 8922. IX International seminar on medical information processing and analysis. SPIE
González G, Escalante-Ramírez B (2014) Knee cartilage segmentation using active shape models and local binary patterns, vol 9138. SPIE Photonics Europe. SPIE
Górriz M, Antony J, McGuinness K, Giró-i-Nieto X, O’Connor NE (2019) Assessing knee OA severity with CNN attention-based end-to-end architectures. Paper presented at the Proceedings of The 2nd international conference on medical imaging with deep learning
Gougoutas AJ, Wheaton AJ, Borthakur A, Shapiro EM, Kneeland JB, Udupa JK, Reddy R (2004) Cartilage volume quantification via live wire segmentation. Acad Radiol 11:1389–1395. https://doi.org/10.1016/j.acra.2004.09.003
Grady L (2006) Random walks for image segmentation. IEEE Trans Pattern Anal Mach Intell 28:1768–1783. https://doi.org/10.1109/TPAMI.2006.233
Greenspan H, Bv G, Summers RM (2016) Guest Editorial Deep Learning in Medical Imaging: Overview and Future Promise of an Exciting New Technique. IEEE Trans Med Imaging. 35:1153–1159. https://doi.org/10.1109/TMI.2016.2553401
Guo Y, Jiang J, Hao S, Zhan S (2011) Distribution-based active contour model for medical image segmentation. In: International conference on image and graphics, 12–15 Aug 2011, pp 61–65. https://doi.org/10.1109/ICIG.2011.11
Hafezi-Nejad N et al (2017) Prediction of medial tibiofemoral compartment joint space loss progression using volumetric cartilage measurements: data from the FNIH OA biomarkers consortium. Eur Radiol 27:464–473. https://doi.org/10.1007/s00330-016-4393-4
Heimann T, Meinzer H-P (2009) Statistical shape models for 3D medical image segmentation: a review. Med Image Anal 13:543–563. https://doi.org/10.1016/j.media.2009.05.004
Heimann T, Morrison BJ, Styner MA, Niethammer M, Warfield SK (2010) Segmentation of knee images: a grand challenge. In: Proceedings MICCAI workshop on medical image analysis for the clinic, 2010. pp 207–214
Hesamian MH, Jia W, He X, Kennedy P (2019) Deep learning techniques for medical image segmentation: achievements and challenges. J Digit Imaging 32:582–596. https://doi.org/10.1007/s10278-019-00227-x
Hiligsmann M et al (2013) Health economics in the field of osteoarthritis: An Expert's consensus paper from the European Society for Clinical and Economic Aspects of Osteoporosis and Osteoarthritis (ESCEO) Seminars in Arthritis and Rheumatism 43:303–313. https://doi.org/10.1016/j.semarthrit.2013.07.003
Hunter DJ et al (2006) Increase in bone marrow lesions associated with cartilage loss: a longitudinal magnetic resonance imaging study of knee osteoarthritis. Arthritis Rheum 54:1529–1535. https://doi.org/10.1002/art.21789
Iranpour-Boroujeni T, Watanabe A, Bashtar R, Yoshioka H, Duryea J (2011) Quantification of cartilage loss in local regions of knee joints using semi-automated-segmentation software: analysis of longitudinal data from the Osteoarthritis Initiative (OAI). Osteoarthr Cartil 19:309–314. https://doi.org/10.1016/j.joca.2010.12.002
Jianbo S, Malik J (2000) Normalized cuts and image segmentation. IEEE Trans Pattern Anal Mach Intell 22:888–905. https://doi.org/10.1109/34.868688
Jolliffe IT (2002) Principal component analysis. Springer series in statistics, vol 2. Springer, New York
Kapoor M, Martel-Pelletier J, Lajeunesse D, Pelletier J-P, Fahmi H (2011) Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat Rev Rheumatol 7:33–42. https://doi.org/10.1038/nrrheum.2010.196
Kashyap S, Oguz I, Zhang H, Sonka M (2016) Automated segmentation of knee MRI using hierarchical classifiers and just enough interaction based learning: data from osteoarthritis initiative. In: Medical image computing and computer assisted intervention—MICCAI 2016, Athens, Greece, 2016. Springer, Berlin, pp 344–351. https://doi.org/10.1007/978-3-319-46723-8_40
Kashyap S, Zhang H, Rao K, Sonka M (2018) Learning-based cost functions for 3-D and 4-D multi-surface multi-object segmentation of knee MRI: data from the osteoarthritis initiative. IEEE Trans Med Imaging 37:1103–1113. https://doi.org/10.1109/TMI.2017.2781541
Kass M, Witkin A, Terzopoulos D (1988) Snakes: Active contour models. Int J Comput Vis 1:321–331. https://doi.org/10.1007/BF00133570
Khan A, Sohail A, Zahoora U, Qureshi AS (2020) A survey of the recent architectures of deep convolutional neural networks. Artif Intell Rev. https://doi.org/10.1007/s10462-020-09825-6
Kumar D, Gandhamal A, Talbar S, Hani AFM (2018) Knee articular cartilage segmentation from MR images: a review. ACM Comput Surv. https://doi.org/10.1145/3230631
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444. https://doi.org/10.1038/nature14539
Lee J-S, Chung Y-N (2005) Integrating edge detection and thresholding approaches to segmenting femora and patellae from magnetic resonance images. Biomed Eng Appl Basis Commun 17:1–11. https://doi.org/10.4015/S1016237205000020
Lee S, Park SH, Shim H, Yun ID, Lee SU (2011) Optimization of local shape and appearance probabilities for segmentation of knee cartilage in 3-D MR images. Comput Vis Image Understanding 115:1710–1720. https://doi.org/10.1016/j.cviu.2011.05.014
Lee J-G, Gumus S, Moon CH, Kwoh CK, Bae KT (2014) Fully automated segmentation of cartilage from the MR images of knee using a multi-atlas and local structural analysis method. Med Phys 41:092303. https://doi.org/10.1118/1.4893533
Lee H, Hong H, Kim J (2018) BCD-NET: a novel method for cartilage segmentation of knee MRI via deep segmentation networks with bone-cartilage-complex modeling. In: IEEE 15th international symposium on biomedical imaging (ISBI 2018), 4–7 April 2018, pp 1538–1541. https://doi.org/10.1109/ISBI.2018.8363866
Li G, Yin J, Gao J, Cheng TS, Pavlos NJ, Zhang C, Zheng MH (2013) Subchondral bone in osteoarthritis: insight into risk factors and microstructural changes. Arthritis Res Ther 15:223. https://doi.org/10.1186/ar4405
Lim J, Kim J, Cheon S (2019) A deep neural network-based method for early detection of osteoarthritis using statistical data. Int J Environ Res Public Health 16:1281. https://doi.org/10.3390/ijerph16071281
Litjens G et al (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88. https://doi.org/10.1016/j.media.2017.07.005
Liu Q, Wang Q, Zhang L, Gao Y, Shen D Multi-atlas context forests for knee MR Image segmentation. In: International workshop on machine learning in medical imaging, Munich, Germany, 2015. Springer International Publishing, pp 186–193
Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R (2018) Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn Reson Med 79:2379–2391. https://doi.org/10.1002/mrm.26841
Liu F et al (2018) Deep learning approach for evaluating knee MR images: achieving high diagnostic performance for cartilage lesion detection. Radiology 289:160–169. https://doi.org/10.1148/radiol.2018172986
Liukkonen MK, Mononen ME, Klets O, Arokoski JP, Saarakkala S, Korhonen RK (2017) Simulation of subject-specific progression of knee osteoarthritis and comparison to experimental follow-up data: data from the osteoarthritis initiative. Sci Rep 7:9177. https://doi.org/10.1038/s41598-017-09013-7
Liukkonen MK, Mononen ME, Tanska P, Saarakkala S, Nieminen MT, Korhonen RK (2017) Application of a semi-automatic cartilage segmentation method for biomechanical modeling of the knee joint. Comput Methods Biomech Biomed Eng 20:1453–1463. https://doi.org/10.1080/10255842.2017.1375477
Loeser RF, Goldring SR, Scanzello CR, Goldring MB (2012) Osteoarthritis: a disease of the joint as an organ. Arthritis Rheum 64:1697–1707. https://doi.org/10.1002/art.34453
Lorigo LM, Faugeras O, Grimson WEL, Keriven R, Kikinis R (1998) Segmentation of bone in clinical knee MRI using texture-based geodesic active contours. In: Medical image computing and computer assisted intervention—MICCAI 1998. Springer, Berlin, pp 1195–1204
Lundervold AS, Lundervold A (2019) An overview of deep learning in medical imaging focusing on MRI. Zeitschrift für Medizinische Physik 29:102–127. https://doi.org/10.1016/j.zemedi.2018.11.002
Lynch J, Zaim S, Zhao J, Stork A, Peterfy C, Genant H (2000) Cartilage segmentation of 3D MRI scans of the osteoarthritic knee combining user knowledge and active contours vol 3979. Medical Imaging 2000. SPIE
MacQueen JB (1967) Some methods for classification and analysis of multivariate observations. In: Proceedings of the fifth Berkeley symposium on mathematical statistics and probability. California: University of California Press, pp 281–297
Maier A, Syben C, Lasser T, Riess C (2019) A gentle introduction to deep learning in medical image processing. Zeitschrift für Medizinische Physik 29:86–101. https://doi.org/10.1016/j.zemedi.2018.12.003
Maldonado M, Nam J (2013) The role of changes in extracellular matrix of cartilage in the presence of inflammation on the pathology of osteoarthritis. Biomed Res Int 2013:284873–284873. https://doi.org/10.1155/2013/284873
Malladi R, Sethian JA, Vemuri BC (1995) Shape modeling with front propagation: a level set approach. IEEE Trans Pattern Anal Mach Intell 17:158–175. https://doi.org/10.1109/34.368173
Man GS, Mologhianu G (2014) Osteoarthritis pathogenesis—a complex process that involves the entire joint J Med. Life 7:37–41
Mononen ME, Tanska P, Isaksson H, Korhonen RK (2016) A novel method to simulate the progression of collagen degeneration of cartilage in the knee: data from the osteoarthritis initiative. Sci Rep 6:21415. https://doi.org/10.1038/srep21415
Mononen ME, Liukkonen MK, Korhonen RK (2019) Utilizing atlas-based modeling to predict knee joint cartilage degeneration: data from the osteoarthritis initiative. Ann Biomed Eng 47:813–825. https://doi.org/10.1007/s10439-018-02184-y
Mortensen EN, Barrett WA (1998) Interactive segmentation with intelligent scissors. Gr Models Image Process 60:349–384. https://doi.org/10.1006/gmip.1998.0480
Neogi T (2012) Clinical significance of bone changes in osteoarthritis. Ther Adv Musculoskeletal Dis 4:259–267. https://doi.org/10.1177/1759720X12437354
Neogi T et al (2009) Cartilage loss occurs in the same subregions as subchondral bone attrition: a within-knee subregion-matched approach from the multicenter osteoarthritis study. Arthritis Care Res 61:1539–1544. https://doi.org/10.1002/art.24824
Neogi T et al (2013) Magnetic resonance imaging-based three-dimensional bone shape of the knee predicts onset of knee osteoarthritis: data from the osteoarthritis initiative. Arthritis Rheum 65:2048–2058. https://doi.org/10.1002/art.37987
Norman B, Pedoia V, Majumdar S (2018) Use of 2D U-Net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology 288:177–185. https://doi.org/10.1148/radiol.2018172322
Norman B, Pedoia V, Noworolski A, Link T, Majumdar S (2018) Applying densely connected convolutional neural networks for staging osteoarthritis severity from plain radiographs. J Digit Imaging 32:471–477. https://doi.org/10.1007/s10278-018-0098-3
Öztürk CN, Albayrak S (2016) Automatic segmentation of cartilage in high-field magnetic resonance images of the knee joint with an improved voxel-classification-driven region-growing algorithm using vicinity-correlated subsampling. Comput Biol Med 72:90–107. https://doi.org/10.1016/j.compbiomed.2016.03.011
Pakin SK, Tamez-Pena J, Totterman S, Parker K (2002) Segmentation, surface extraction, and thickness computation of articular cartilage, vol 4684. Medical Imaging 2002. SPIE
Palazzo C, Nguyen C, Lefevre-Colau M-M, Rannou F, Poiraudeau S (2016) Risk factors and burden of osteoarthritis. Ann Phys Rehabil Med 59:134–138. https://doi.org/10.1016/j.rehab.2016.01.006
Panfilov E, Tiulpin A, Klein S, Nieminen MT, Saarakkala S (2019) Improving robustness of deep learning based knee MRI segmentation: mixup and adversarial domain adaptation. In: IEEE International conference on computer vision workshop (ICCVW), Seoul, Korea, pp 450–459
Pang J, Li P, Qiu M, Chen W, Qiao L (2015) Automatic articular cartilage segmentation based on pattern recognition from knee MRI images. J Digit Imaging 28:695–703. https://doi.org/10.1007/s10278-015-9780-x
Paranjape CS et al (2019) A new stress test for knee joint cartilage. Sci Rep 9:2283. https://doi.org/10.1038/s41598-018-38104-2
Park SH et al (2009) Fully automatic 3-D segmentation of knee bone compartments by iterative local branch-and-mincut on MR images from osteoarthritis initiative (OAI). In: IEEE 16th international conference on image processing (ICIP), 7–10 Nov. 2009. pp 3381–3384. https://doi.org/10.1109/ICIP.2009.5413874
Pedoia V, Majumdar S, Link TM (2016) Segmentation of joint and musculoskeletal tissue in the study of arthritis. Magn Reson Mater Phys Biol Med 29:207–221. https://doi.org/10.1007/s10334-016-0532-9
Pedoia V, Lee J, Norman B, Link T, Majumdar S (2019) Diagnosing osteoarthritis from T2 Maps using deep learning: an analysis of the entire osteoarthritis initiative baseline cohort. Osteoarthr Cartil 27:1002–1010. https://doi.org/10.1016/j.joca.2019.02.800
Pelletier J-P et al (2007) Risk factors associated with the loss of cartilage volume on weight-bearing areas in knee osteoarthritis patients assessed by quantitative magnetic resonance imaging: a longitudinal study. Arthritis Res Ther 9:R74. https://doi.org/10.1186/ar2272
Peterfy CG, Schneider E, Nevitt M (2008) The osteoarthritis initiative: report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthr Cartil 16:1433–1441. https://doi.org/10.1016/j.joca.2008.06.016
Peuna A et al (2018) Variable angle gray level co-occurrence matrix analysis of T2 relaxation time maps reveals degenerative changes of cartilage in knee osteoarthritis: Oulu knee osteoarthritis study. J Magn Reson Imaging 47:1316–1327. https://doi.org/10.1002/jmri.25881
Prasoon A, Petersen K, Igel C, Lauze F, Dam E, Nielsen M (2013) Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. In: Medical image computing and computer assisted intervention—MICCAI 2013. Springer, Berlin, pp 246–253
Quinlan JR (1986) Induction of decision trees. Mach Learn 1:81–106. https://doi.org/10.1007/BF00116251
Raghu M, Schmidt E (2020) A survey of deep learning for scientific discovery. arXiv Preprint 0:1–48
Raj A, Vishwanathan S, Ajani B, Krishnan K, Agarwal H (2018) Automatic knee cartilage segmentation using fully volumetric convolutional neural networks for evaluation of osteoarthritis. In: IEEE 15th international symposium on biomedical imaging (ISBI 2018), 4–7 April 2018, pp 851–854. https://doi.org/10.1109/ISBI.2018.8363705
Rini C, Perumal B, Rajasekaran MP (2020) Automatic knee joint segmentation using Douglas-Rachford splitting method. Multimed Tools Appl 79:6599–6621. https://doi.org/10.1007/s11042-019-08303-8
Rish I (2001) An empirical study of the naive Bayes classifier. In: IJCAI 2001 workshop on empirical methods in artificial intelligence, vol 22, pp 41–46
Riza S, Marlinawati D, Fahmi M (2019) COMSeg technique for MRI knee cartilage segmentation. Int Rev Appl Sci Eng 10:1–9. https://doi.org/10.1556/1848.2019.0018
Roemer FW et al (2010) A comparison of dedicated 1.0 T extremity MRI vs large-bore 1.5 T MRI for semiquantitative whole organ assessment of osteoarthritis: the MOST study. Osteoarthr Cartil. 18:168–174. https://doi.org/10.1016/j.joca.2009.08.017
Rohlfing T, Brandt R, Menzel R, Russakoff DB, Maurer CR (2005) Quo vadis, atlas-based segmentation? In: Suri JS, Wilson DL, Laxminarayan S (eds) Handbook of biomedical image analysis: Volume III: registration models. Springer US, Boston, MA, pp 435–486. https://doi.org/10.1007/0-306-48608-3_11
Rokach L (2010) Ensemble-based classifiers. Artif Intell Rev 33:1–39. https://doi.org/10.1007/s10462-009-9124-7
Roughley PJ, Mort JS (2014) The role of aggrecan in normal and osteoarthritic cartilage Journal of Experimental Orthopaedics 1:8. https://doi.org/10.1186/s40634-014-0008-7
Schaefer LF et al (2017) Quantitative measurement of medial femoral knee cartilage volume - analysis of the OA Biomarkers Consortium FNIH Study cohort Osteoarthritis and cartilage 25:1107–1113. https://doi.org/10.1016/j.joca.2017.01.010
Schmid J, Magnenat-Thalmann N (2008) MRI bone segmentation using deformable models and shape priors. In: Medical image computing and computer assisted intervention—MICCAI 2008, 2008. Springer, Berlin, pp 119–126
Seim H, Kainmueller D, Lamecker H, Bindernagel M, Malinowski J, Zachow S (2010) Model-based auto-segmentation of knee bones and cartilage in MRI data Proc Med Image Anal
Sengupta S et al (2020) A review of deep learning with special emphasis on architectures, applications and recent trends Knowledge-Based Systems 194:105596 https://doi.org/10.1016/j.knosys.2020.105596
Serre T (2019) Deep Learning: The Good, the Bad, and the Ugly Annual Review of Vision Science 5:399–426 https://doi.org/10.1146/annurev-vision-091718-014951
Shah RF, Martinez AM, Pedoia V, Majumdar S, Vail TP, Bini SA (2019) Variation in the Thickness of Knee Cartilage. The Use of a Novel Machine Learning Algorithm for Cartilage Segmentation of Magnetic Resonance Images The Journal of Arthroplasty 34:2210–2215. https://doi.org/10.1016/j.arth.2019.07.022
Shamir L, Orlov N, Eckley DM, Macura T, Johnston J, Goldberg IG (2008) Wndchrm – an open source utility for biological image analysis Source Code for Biology and Medicine 3:13. https://doi.org/10.1186/1751-0473-3-13
Shan L, Charles C, Niethammer M (2012a) Automatic Atlas-based Three-label Cartilage Segmentation from MR Knee Images Proc Workshop Math Methods Biomed Image Analysis:241–246. https://doi.org/10.1109/mmbia.2012.6164757
Shan L, Charles C, Niethammer M Automatic multi-atlas-based cartilage segmentation from knee MR images. In: IEEE 9th International Symposium on Biomedical Imaging (ISBI 2012), 2–5 May 2012 2012b. pp 1028–1031. https://doi.org/10.1109/ISBI.2012.6235733
Shan L, Zach C, Charles C, Niethammer M (2014) Automatic atlas-based three-label cartilage segmentation from MR knee images. Med Image Anal 18:1233–1246. https://doi.org/10.1016/j.media.2014.05.008
Sharif B, Garner R, Hennessy D, Sanmartin C, Flanagan WM, Marshall DA (2017) Productivity costs of work loss associated with osteoarthritis in Canada from 2010 to 2031. Osteoarthr Cartil 25:249–258. https://doi.org/10.1016/j.joca.2016.09.011
Sharma AR, Jagga S, Lee S-S, Nam J-S (2013) Interplay between cartilage and subchondral bone contributing to pathogenesis of osteoarthritis. Int J Mol Sci 14:19805–19830. https://doi.org/10.3390/ijms141019805
Shen D, Wu G, Suk H-I (2017) Deep learning in medical image analysis. Annu Rev Biomed Eng 19:221–248. https://doi.org/10.1146/annurev-bioeng-071516-044442
Shim H, Chang S, Tao C, Wang J-H, Kwoh CK, Bae KT (2009) Knee cartilage: efficient and reproducible segmentation on high-spatial-resolution MR images with the semiautomated graph-cut algorithm method. Radiology 251:548–556. https://doi.org/10.1148/radiol.2512081332
Shim H, Kwoh CK, Yun ID, Lee SU, Bae K (2009b) Simultaneous 3D segmentation of three bone compartments on high resolution knee MR images from osteoarthritis initiative (OAI) using graph cuts, vol 7259. SPIE Medical Imaging. SPIE
Shrestha A, Mahmood A (2019) Review of deep learning algorithms and architectures. IEEE Access 7:53040–53065. https://doi.org/10.1109/ACCESS.2019.2912200
Singh S, Wang L, Gupta S, Goli H, Padmanabhan P, Gulyas B (2020) 3D deep learning on medical images: a review. arXiv Preprint:1–13
Smith JJ, Sorensen AG, Thrall JH (2003) Biomarkers in imaging: realizing radiology’s future. Radiology 227:633–638. https://doi.org/10.1148/radiol.2273020518
Sokolove J, Lepus CM (2013) Role of inflammation in the pathogenesis of osteoarthritis: latest findings and interpretations. Ther Adv Musculoskeletal Dis 5:77–94. https://doi.org/10.1177/1759720X12467868
Solloway S, Hutchinson CE, Waterton JC, Taylor CJ (1997) The use of active shape models for making thickness measurements of articular cartilage from MR images. Magn Reson Med 37:943–952. https://doi.org/10.1002/mrm.1910370620
Stammberger T, Eckstein F, Michaelis M, Englmeier K-H, Reiser M (1999) Interobserver reproducibility of quantitative cartilage measurements: comparison of B-spline snakes and manual segmentation. Magn Reson Imaging 17:1033–1042. https://doi.org/10.1016/S0730-725X(99)00040-5
Stewart HL, Kawcak CE (2018) The importance of subchondral bone in the pathophysiology of osteoarthritis. Front Vet Sci 5:178–178. https://doi.org/10.3389/fvets.2018.00178
Tack A, Zachow S (2019) Accurate automated volumetry of cartilage of the knee using convolutional neural networks: data from the osteoarthritis initiative. In: IEEE 16th international symposium on biomedical imaging (ISBI 2019), Venice, Italy. IEEE, pp 40–43. https://doi.org/10.1109/ISBI.2019.8759201
Tamez-Peña JG, Farber J, González PC, Schreyer E, Schneider E, Totterman S (2012) Unsupervised segmentation and quantification of anatomical knee features: data from the osteoarthritis initiative. IEEE Trans Biomed Eng 59:1177–1186. https://doi.org/10.1109/TBME.2012.2186612
Tan C, Yan Z, Zhang S, Li K, Metaxas DN (2019) collaborative multi-agent learning for MR knee articular cartilage segmentation. In: Shen D et al (eds) Medical image computing and computer assisted intervention—MICCAI 2019, Shenzhen, China, 2019. Springer Berlin Heidelberg, pp 282–290
Tang J, Millington S, Acton ST, Crandall J, Hurwitz S (2006) Surface extraction and thickness measurement of the articular cartilage from MR images using directional gradient vector flow snakes. IEEE Trans Biomed Eng 53:896–907. https://doi.org/10.1109/TBME.2006.872816
Thaha R, Jogi SP, Rajan S, Mahajan V, Venugopal VK, Mehndiratta A, Singh A (2020) Modified radial-search algorithm for segmentation of tibiofemoral cartilage in MR images of patients with subchondral lesion. Int J Comput Assist Radiol Surg 15:403–413. https://doi.org/10.1007/s11548-020-02116-z
Thengade A, Rajurkar A (2019) A comprehensive survey of articular cartilage segmentation methods on knee MRI. Int J Adv Sci Technol 27:148–159
Thomas KA et al (2020) Automated classification of radiographic knee osteoarthritis severity using deep neural networks radiology. Artif Intell 2:e190065. https://doi.org/10.1148/ryai.2020190065
Tiulpin A, Saarakkala S (2019) Automatic grading of individual knee osteoarthritis features in plain radiographs using deep convolutional neural networks. arXiv Preprint:1–14
Tiulpin A, Thevenot J, Rahtu E, Lehenkari P, Saarakkala S (2018) Automatic knee osteoarthritis diagnosis from plain radiographs: a deep learning-based approach. Sci Rep 8:1727. https://doi.org/10.1038/s41598-018-20132-7
Tiulpin A et al (2019) Multimodal machine learning-based knee osteoarthritis progression prediction from plain radiographs and clinical data. Sci Rep 9:20038. https://doi.org/10.1038/s41598-019-56527-3
Vilimek D, Kubicek J, Penhaker M, Oczka D, Augustynek M, Cerny M (2019) Current automatic methods for knee cartilage segmentation: a review. In: 8th European workshop on visual information processing (EUVIP), 28–31 Oct. 2019, pp 117–122. https://doi.org/10.1109/EUVIP47703.2019.8946132
Vina ER, Kwoh CK (2018) Epidemiology of osteoarthritis: literature update. Curr Opin Rheumatol 30:160–167. https://doi.org/10.1097/BOR.0000000000000479
Wang Q, Wu D, Lu L, Liu M, Boyer KL, Zhou SK (2014) Semantic context forests for learning-based knee cartilage segmentation in 3D MR images. In: International conference on medical image computing and computer-assisted intervention: MICCAI Cham. Medical Computer Vision. Large data in medical imaging. Springer International Publishing, pp 105–115
Warner SC, Valdes AM (2016) The genetics of osteoarthritis: a review. J Funct Morphol Kinesiol 1:140–153
Waterton JC et al (2000) Diurnal variation in the femoral articular cartilage of the knee in young adult humans. Magn Reson Med 43:126–132. https://doi.org/10.1002/(sici)1522-2594(200001)43:1%3c126::aid-mrm15%3e3.0.co;2-#
Williams TG et al (2010a) Automatic segmentation of bones and inter-image anatomical correspondence by volumetric statistical modelling of knee MRI. In: IEEE International symposium on biomedical imaging: from nano to macro, 14–17 April 2010, pp 432–435. https://doi.org/10.1109/ISBI.2010.5490316
Williams TG et al (2010b) Measurement and visualisation of focal cartilage thickness change by MRI in a study of knee osteoarthritis using a novel image analysis tool. Br J Radiol 83:940–948. https://doi.org/10.1259/bjr/68875123
Wluka AE, Stuckey S, Snaddon J, Cicuttini FM (2002) The determinants of change in tibial cartilage volume in osteoarthritic knees. Arthritis Rheum 46:2065–2072. https://doi.org/10.1002/art.10460
Wluka AE et al (2009) Bone marrow lesions predict increase in knee cartilage defects and loss of cartilage volume in middle-aged women without knee pain over 2 years. Ann Rheum Dis 68:850–855. https://doi.org/10.1136/ard.2008.092221
Wu Z, Leahy R (1993) An optimal graph theoretic approach to data clustering: theory and its application to image segmentation. IEEE Trans Pattern Anal Mach Intell 15:1101–1113. https://doi.org/10.1109/34.244673
Xu Z, Niethammer M (2019) DeepAtlas: joint semi-supervised learning of image registration and segmentation. In: Shen D et al (eds) Medical image computing and computer assisted intervention—MICCAI 2019, Cham, 2019. Springer, Berlin, pp 420–429
Yin Y, Zhang X, Williams R, Wu X, Anderson DD, Sonka M (2010) LOGISMOS-layered optimal graph image segmentation of multiple objects and surfaces: cartilage segmentation in the knee joint. IEEE Trans Med Imaging 29:2023–2037. https://doi.org/10.1109/TMI.2010.2058861
Zhang K, Lu W, Marziliano P (2013) Automatic knee cartilage segmentation from multi-contrast MR images using support vector machine classification with spatial dependencies. Magn Reson Imaging 31:1731–1743. https://doi.org/10.1016/j.mri.2013.06.005
Zhang W, Ouyang H, Dass CR, Xu J (2016) Current research on pharmacologic and regenerative therapies for osteoarthritis. Bone Res 4:15040–15040. https://doi.org/10.1038/boneres.2015.40
Zhang B, Zhang Y, Cheng H-D, Xian M, Gai S, Cheng O, Huang K (2018) Computer-aided knee joint magnetic resonance image segmentation—a survey. CoRR abs/1802.04894:1–10
Zhou Z, Zhao G, Kijowski R, Liu F (2018) Deep convolutional neural network for segmentation of knee joint anatomy. Magn Reson Med 80:2759–2770. https://doi.org/10.1002/mrm.27229
Zhou T, Ruan S, Canu S (2019) A review: deep learning for medical image segmentation using multi-modality fusion. Array 3–4:100004. https://doi.org/10.1016/j.array.2019.100004
Acknowledgements
The authors acknowledge the valuable help provided by Prashant Shukla Kumar in collecting the literature materials.
Funding
The study was funded by Fundamental Research Grant Scheme (FRGS) (project title: Graph Transformed Deep ‘Interactive’ Learning Framework in Medical Image Segmentation, grant no: FRGS/1/2018/ICT02/UNIKL/02/4) provided by Ministry of Education, Malaysia (MoE).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gan, HS., Ramlee, M.H., Wahab, A.A. et al. From classical to deep learning: review on cartilage and bone segmentation techniques in knee osteoarthritis research. Artif Intell Rev 54, 2445–2494 (2021). https://doi.org/10.1007/s10462-020-09924-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10462-020-09924-4