Abstract
Knee osteoarthritis (OA) is a degenerative joint disease that is prevalent in advancing age. The pathology of OA disease is still unclear, and there are no effective interventions that can completely alter the OA disease process. Magnetic resonance (MR) image evaluation is sensitive for depicting early changes of knee OA, and therefore important for early clinical intervention for relieving the symptom. Automated cartilage segmentation based on MR images is a vital step in experimental longitudinal studies to follow-up the patients and prospectively define a new quantitative marker from OA progression. In this paper, we develop a deep learning–based coarse-to-fine approach for automated knee bone, cartilage, and meniscus segmentation with high computational efficiency. The proposed method is evaluated using two-fold cross-validation on 507 MR volumes (81,120 slices) with OA from the Osteoarthritis Initiative (OAI)1 dataset. The mean dice similarity coefficients (DSCs) of femoral bone (FB), tibial bone (TB), femoral cartilage (FC), and tibial cartilage (TC) separately are 99.1%, 98.2%, 90.9%, and 85.8%. The time of segmenting each patient is 12 s, which is fast enough to be used in clinical practice. Our proposed approach may provide an automated toolkit to help computer-aided quantitative analyses of OA images.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Knee osteoarthritis (OA) is a common degenerative joint disease that affects a large number of people and increases in incidence with advancing age [1]. With the aging of the population, more and more people suffer from the problems caused by knee OA [2]. Among people over 60 years old, the rate for men and women with symptomatic knee OA is 10% and 13%, respectively [3]. Furthermore, OA is the main cause of morbidity and disability, resulting in high socioeconomic costs. As a most common form of arthritis, OA was estimated to cost the US $336 billion, or 3% of the gross domestic product in 2004 [4, 5]. In the USA, more than 30 million adults have radiological evidence of OA [6]. By 2030, an estimated 20% of Americans (about 70 million people) may be at increased risk for this disease [4, 5]. The main symptoms of knee OA are pain, stiffness, and swelling. These symptoms adversely affect activities of daily living and overall function. The pathology of OA disease is still unclear, and there are no interventions that can effectively modify the OA disease process [7]. We can only mitigate the symptoms by anti-inflammatory drugs, joint replacement, etc. To better understand this disease, the OA imitative (OAI)1 consortium collected and generated substantial data sets including magnetic resonance (MR) and computed tomography (CT) imaging data, genotyping data, and clinical data for evaluating potential biomarkers and characterizing OA incidence and progression. In clinical studies, the diagnosis and severity assessment of OA are mainly based on radiologic findings. For example, the measurement of cartilage in MR and CT images is a primary way of demonstrating the structural progression of OA and is also used to evaluate the effectiveness of therapeutic approaches.
MR imaging is a non-invasive technology that can generate 2D and 3D images of intra-articular soft-tissue structures, including cartilage, which is commonly used to find the structural changes within the knee joint and, specifically, in articular cartilage [8]. However, obtaining accurate and reproducible quantitative measurements from MR scans is burdensome due to the structure and morphology of the knee as well as the nature of MR imaging [9]. There are several challenges in manually segmentation and extraction of MR images. First, it is time-consuming and labor-consuming work. It may take up to 6 h for a clinical reader to manually segment each series of 3-dimensional (3D) knee MR [10]. Secondly, the results could be subjective due to the clinicians’ knowledge and experience. Furthermore, operators who use cartilage segmentation software often need extensive training [11], which costs more time and money.
Over the past years, researchers have developed different approaches for reducing the workload of measuring knee cartilage on MR images. These approaches include segmenting alternate MR slices or confining measurements to partial regions of cartilage [12,13,14,15,16]. Besides, some computer-aided algorithms (e.g., active contours, B-splines) have also been developed to assist with cartilage segmentation for MR images [17,18,19]. Unfortunately, these methods lack sufficient accuracy and reliability in detecting small cartilage changes [12, 20]. Du et al. [10] proposed a novel method to directly predict knee osteoarthritis progression on MRI. In their paper, the Cartilage Damage Index (CDI) information [21, 22] is computed from 36 informative locations on the tibiofemoral cartilage compartment from 3D MR images, and PCA analysis is utilized to process the feature set. This approach is not fully automatic because it requires the user to give the slice numbers of the most medial and lateral images. Ambellan et al. [23] incorporated 3D Statistical Shape Models (SSMs) as well as 2D and 3D CNNs to segment even highly pathological knee structures. They used SSM method as adjustment and postprocessing to polish the segmentation results based on CNNs. However, SSM method is complex and inefficient. The authors reported that using their implementation, it would take 43 weeks to segment the full OAI database on a single computational node (around 8.7 min per patient) [23]. Therefore, the previous methods in both Du et al. [10] and Ambellan et al. [23] are time-consuming and impractical to be used in routine clinical practice. Thus, there remains an urgent need for researchers to develop a segmentation method that has both high accuracy and computational efficiency.
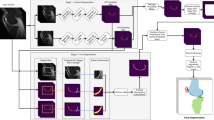
To solve this problem, we develop an automated time-efficient segmentation algorithm based on UNet ++ architecture [24], which is a new segmentation architecture consisting of nested and dense skip connections. UNet ++ in many cases can achieve more excellent performance than U-Net [25], the baseline approach used in various medical image segmentation tasks [26, 27]. In this paper, we develop a coarse-to-fine framework [37, 38] that uses a predicted segmentation mask to shrink the input region. This is motivated by the fact that a smaller input region often leads to more accurate segmentation results. Specifically, we first use an UNet ++ network to localize the region of interest regions (ROIs) and crop the corresponding bounding boxes. Then we send the shrunken image into another UNet ++ network for the fine segmentation. The ROIs include bone and cartilage on OA MR images, which separately are femoral bone (FB), tibial bone (TB), and the respective femoral and tibial cartilage (FC, TC) as well as meniscus (see Fig. 1).
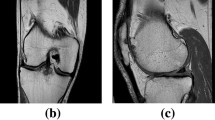
(a) Schematic view of knee joint. (b) and (c) Demonstrative images from the sagittal and coronal directions of a 3D magnetic resonance (MR) knee data, respectively. They show a knee joint, including femoral bone (FB), tibial bone (TB), femoral cartilage (FC), tibial cartilage (TC), and meniscus (MC)
The main contribution to our approach is that we develop a coarse-to-fine framework by combining localization information of bone and cartilage to improve the segmentation performance. Compared with the current state-of-the-art approach for OA image segmentation [23], our proposed approach can achieve higher segmentation accuracy on three tissues and has much less time cost (around 43 times less).
Materials and Methods
Data Collection
We use OAI ZIB Dataset from Ambellan et al. [23]. This dataset consists of 507 3D MR data (81,120 slices) in dual-echo steady-state (DESS) sequence from the OAI database for which manual segmentations were carried out thoroughly by experienced users at Zuse Institute Berlin2 starting from automatic segmentations employing [28]. The data cover the full spectrum of OA grades with a strong tendency towards severe cases. The details of the dataset can be seen in Table 1. Our approach is a 2D method based on horizontal images (size 384 × 384) as indicated in Table 1.
Methods
The overview of the proposed framework is shown in Fig. 2. First, we use an UNet ++ network to detect the region of interest regions (ROIs) and crop the corresponding bounding boxes. Then we send the shrunk input region into another UNet ++ network for the fine segmentation. We segment FB, FC, TB, and TC separately and obtain the meniscus tissue based on the segmentation results of bones and cartilages. We also design a postprocessing method to remove the false positives.
Bone and Cartilage Segmentation with UNet++
Recently, increasing state-of-the-art models based on deep learning are proposed for medical image segmentation [29] and most of these models are variants of U-Net [25] or fully convolutional networks (FCN) [30]. Both U-Net and FCN are encoder-decoder architectures containing skip connections, which can merge low-level and high-level features. However, these encoder-decoder architectures for image segmentation may encounter two limitations. First, there is no way to determine the optimal depth of an encoder-decoder network. It always varies from one task to another, depending on the application complexity and the amount of annotated data available for training. Second, the design of skip connections used in an encoder-decoder network is experiential rather than based on solid theory, and how to design the most effective skip connections architectures lacks theoretical support [24]. To overcome these two limitations, Zhou et al. proposed UNet ++ [24], a new segmentation architecture based on nested and dense skip connections.
As shown in Fig. 3, UNet ++ is composed of U-Net with different depths, and its decoders are densely connected with the same resolution through redesigned hop connections. The architecture changes introduced in UNet ++ realize the following advantages. First of all, UNet ++ is not easy to choose network depth because it embeds different depth U-Nets in its architecture. All of these U-Net parts share an encoder, and their decoders are interleaved with each other. Through in-depth supervision training of UNet ++ , all U-Nets are trained at the same time and benefit from shared image representation. This design not only improves the overall segmentation performance but also realizes the model pruning in the reasoning process. Secondly, UNet ++ is not constrained by unnecessary restrictive jump connections, in which only the same proportion of feature maps from the encoder and decoder can be fused. The redesigned hop join introduced in UNet ++ presents different proportions of feature maps at the decoder node, allowing the aggregation layer to decide how to fuse the various feature maps carried by skipping connection with the feature mapping of the decoder. The redesigned jump connection is implemented in UNet ++ , which is realized by connecting the decoders of U-Net in the same resolution. Compared with the traditional U-Net architecture, this significant improvement of U-Net ++ is due to the advantages of redesigned skip connection and extended decoder, which together enable image features to gradually aggregate horizontally and vertically over the network. More details about UNet ++ can be seen in [24].
The backbone of UNet ++ can be any feature-extraction convolution neural network, such as VGG [31] series and ResNet [32] series. We choose VGG16 [31] as the backbone here for convenience. We segment one target at a time, which separately is FB, TB, FC, and TC at each time and the loss function is binary cross-entropy.
Postprocessing for Removing False Positives
By visualizing the segmentation result on each slice of every patient, we find that some false positives exist on specific regions close to those slices which contain minimum object content. For example, for 3D patient data, if the first slice containing the target (including bone or cartilage) is the 18th slice, the model tends to predict false positives from the 15th slice to the 17th slice (as shown in Fig. 4). The main reason may be that since the target often occupies a small region on these slices where it first appears, the network could be easily confused by the background region. To reduce false positives, the minimum size of the target is calculated and set to be the threshold. Those predicted slices whose object size is less than the threshold will be automatically zeroed (negative). For instance, if the minimum size of TC is a, all the predictions whose size is less than a will be zeroed. By using this simple postprocessing method, the final segmentation performance is improved (see Table 2).
Meniscus Segmentation
The medial and lateral meniscus are two thicker wedge-shaped pads of fibrocartilage attached to the top of the tibia (shin bone), called the tibial plateau. Each meniscus is curved in a C-shape, with the front part of the cartilage called the anterior horn and the back part called the posterior horn (see Fig. 1). A torn meniscus is one of the most common knee injuries. Any activity that causes you to forcefully twist or rotate your knee, especially when putting your full weight on it, can lead to a torn meniscus. In older adults, degenerative changes of the knee can contribute to a torn meniscus with little or no trauma. MRI is the test of choice to confirm the diagnosis of a torn meniscus, and segmenting meniscus is the key step for clinical diagnosis. As shown in Fig. 5, the meniscus lies between the tissue FC and TC, and it will disappear if FC and TC merge. In other words, in a knee with severe OA, the meniscus will be abraded at first and the thickness of the meniscus can be used to evaluate the severity of OA. However, the data we attain from OAI ZIB do not include the annotation of meniscus tissues and there is no other labeled dataset available. Therefore, we have to segment the meniscus in a compromised way. By observing Fig. 5, we can see that meniscus could be annotated by doing some addition and subtraction between images and ground truths (also predictions) using the positional relationship of bone and cartilage. Specifically, we first localize the meniscus (see Fig. 6f) by using cartilage ground truth (Fig. 6c and d) because the meniscus regions always lie between the FC and TC. Then meniscus Fig. 6g is obtained by multiplying Fig. 6f and e (the difference between original image (Fig. 6a) and corresponding ground truth (Fig. 6b). Finally, the ground truth of the meniscus is obtained (Fig. 6h). The process can be seen in Eq. 1.
where x denotes the original image (Fig. 6a), y’ denotes the prediction of bones and cartilages (Fig. 6b), yc’ denotes the prediction of cartilages (Fig. 6c), and fbx represents taking bounding box operation (Fig. 6d). S(x,y’) represents using x and y’ to segment meniscus (Fig. 6g).
Experimental Settings
In the training stage, the ground-truth annotation is used as the input mask. We train the models separately for each stage. The coarse-scaled models are trained on specific slices to prevent the model from being heavily impacted by the background. Those chosen slices should be those in which the number of pixels of the object is greater than a certain value. For FB and TB segmentation tasks, the objects should occupy at least 300 pixels, while for FC and TC segmentation tasks, the number is 100. For the fine-scaled models, we crop each slice according to the minimal 2D box covering the knee, add a frame around it, and fill it up with the original image data. The top, bottom, left, and right margins of the frame are fixed values of 30. This strategy, known as data augmentation, helps to regularize the network and prevent over-fitting. Before fed into the fine stage UNet ++ , the images are resized to [192,192]. To avoid overfitting, we adopt a dynamic learning rate (learning rate = 0.01, decay = 0.0005, momentum = 0.9) and an early stopping strategy in the training stage. The optimization is Adam [33]. All the networks are trained and tested by a 32 GB-RAM NVIDIA Tesla V100 GPU.
Results
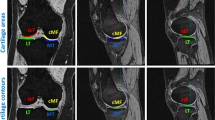
To compare our proposed approach with the current state-of-the-art method in Eckstein and Wirth [12], we follow the cross-validation strategy in Eckstein and Wirth [12] and split the dataset into 2 fixed folds, each of which contains approximately the same number of samples. We apply cross-validation, i.e., training the model on 1 out of 2 subsets and testing it on the remaining one. We measure the segmentation accuracy by computing the dice similarity coefficient (DSC) for each sample. This is a similarity metric between the prediction voxel set Z and the ground-truth set Y, with the mathematical form of DSC (Z, Y) = 2 ×|Z ∩ Y|/|Z| +|Y|. We report the average DSC score of over 507 testing cases. The results can be seen in Table 2.
As shown in Table 2, our approach outperforms some other state-of-the-art approaches for three tissues segmentation (FB, FC, TC) and is quite rapid. The effectiveness of our proposed postprocessing approach is also confirmed in Table 2. Compared with Ambellan’s [12] approach which combines 2D and 3D U-Net with SSM for adjustment or postprocessing, our proposed approach has better performance, which demonstrates the strong capability of deep neural network (DNN) to extract image features and that classical image preprocessing or postprocessing is less important with the development of DNN. A DNN approach could achieve higher accuracy with much less time than classical image processing approaches. In addition, we can also from Table 2 see that the accuracy of knee bones (FB, TB) is much higher (over 10%) than cartilages in all approaches. The reason is that bones have a larger size and higher contrast, which is easier to segment. The qualitative results can be seen in Fig. 7.
Discussion
Despite the promising results shown in this study, some limitations need to be acknowledged. On the one hand, because the size of knee bone (FB and TB) is much bigger than cartilage’s (FC and TC), to segment these four tissues at the same time will encounter the problem of category imbalance. Therefore, our method segments each tissue at a time, which is not ideal. We need to improve it by using some specific loss function like focal loss [34]. On the other hand, due to the lack of available public meniscus data currently, the progress of OA image processing algorithms is to some extent hindered. Therefore, it is of great meaning to construct a meniscus dataset. Although we could segment meniscus without ground truth in a compromise way, the results were not satisfactory. We will manually annotate around 2000 slices meniscus data from OAI dataset and make it available to public, which will be an important contribution for facilitating various algorithms development for meniscus image segmentation. At the same time, we will try to incorporate our proposed model into an end-to-end learning framework to minimize a global energy function for improving the performance.
In our future work, we will collect more clinical OA patient data such as clinical data from Johnston County Osteoarthritis Project (JoCoOA) [35] funded by CDC to validate our proposed approach for OA bone and cartilage segmentation. We will attempt to further improve our approach in several ways below. (1) We will combine an attention mechanism proposed in [36] with UNet ++ model, which we name attention UNet ++ . (2) We will use a computational method to directly measure the thickness of meniscus. We could regress the boundary of FC and TC and draw the tangent lines of boundary to calculate the distance d of these two tangent lines. As mentioned in section meniscus segmentation, the value of d reflects the severity of OA and smaller d value represents severer OA disease. We can model the d and time t, which can reveal the relationship between the severity of OA and time. This approach is more time-saving and accurate than labeling meniscus data to train a segmentation model. (3) Because the OAI dataset also provides electronic medical records (EMRs) of each patient, we will develop a classifier based on the segmentation results to stage the severity of knee OA (measured by Kellgren-Lawrence (K-L) score) of each patient and validate it with the EMRs provided by OAI. Besides, we will use extend our approach to a time series–based method to mimic the developments of OA and predict the patients’ current stage by their latest MR scans.
Conclusion
In this paper, we propose a novel coarse-to-fine approach for knee bone and cartilage segmentation by combining two UNet ++ models. The approach is evaluated on the OA ZIB dataset consisting of 507 3D volumes. Compared with the current state-of-the-art segmentation approach [12] on data from the OAI, our proposed approach can achieve higher segmentation accuracy in three tissues (FB, FC, TC) and has much less time cost (around 43 times less). Our approach is fast enough to be used in clinical practice. Furthermore, based on the segmentation results of bones and cartilages, we segment the meniscus from the OAI dataset without ground truth for the first time. Our approach is intuitive but effective: first use a deep learning model to shrink the input region, which could remove much interference from the background, and then use another model to finish the fine segmentation. We anticipate that our approach can provide a promising computerized toolkit to help enhance computer-aided quantitative analyses of OA MR images.
References
R. C. Lawrence et al., Estimates of the prevalence of arthritis and other rheumatic conditions in the United States: Part II, Arthritis & Rheumatism, vol. 58, no. 1, pp. 26-35, 2008.
K. D. Brandt, Diagnosis and nonsurgical management of osteoarthritis. Professional Communications, 2010.
Y. Zhang and J. M. Jordan, Epidemiology of osteoarthritis, Clinics in geriatric medicine, vol. 26, no. 3, pp. 355-369, 2010.
C. R. Chu, A. A. Williams, C. H. Coyle, and M. E. Bowers, Early diagnosis to enable early treatment of pre-osteoarthritis, Arthritis research & therapy, vol. 14, no. 3, p. 212, 2012.
E. Yelin, S. Weinstein, and T. King, The burden of musculoskeletal diseases in the United States, in Seminars in arthritis and rheumatism, 2016, vol. 46, no. 3, p. 259.
A. Mendy, J. Park, and E. R. Vieira, Osteoarthritis and risk of mortality in the USA: a population-based cohort study, International Journal of Epidemiology, vol. 47, no. 6, pp. 1821-1829, 2018.
D. Bhatia, T. Bejarano, and M. Novo, Current interventions in the management of knee osteoarthritis, Journal of pharmacy & bioallied sciences, vol. 5, no. 1, p. 30, 2013.
M. T. Nieminen, V. Casula, M. T. Nevalainen, and S. Saarakkala, Osteoarthritis year in review 2018: imaging, Osteoarthritis and cartilage, vol. 27, no. 3, pp. 401-411, 2019.
H. Shim, S. Chang, C. Tao, J.-H. Wang, C. K. Kwoh, and K. T. Bae, Knee cartilage: efficient and reproducible segmentation on high-spatial-resolution MR images with the semiautomated graph-cut algorithm method, Radiology, vol. 251, no. 2, pp. 548-556, 2009.
Y. Du, R. Almajalid, J. Shan, and M. Zhang, A novel method to predict knee osteoarthritis progression on MRI using machine learning methods, IEEE transactions on nanobioscience, vol. 17, no. 3, pp. 228-236, 2018.
J. Jaremko, R. Cheng, R. Lambert, A. Habib, and J. Ronsky, Reliability of an efficient MRI-based method for estimation of knee cartilage volume using surface registration, Osteoarthritis and cartilage, vol. 14, no. 9, pp. 914-922, 2006.
F. Eckstein and W. Wirth, Quantitative cartilage imaging in knee osteoarthritis, Arthritis, vol. 2011, 2011.
J. Fripp, S. Crozier, S. K. Warfield, and S. Ourselin, Automatic segmentation and quantitative analysis of the articular cartilages from magnetic resonance images of the knee, IEEE transactions on medical imaging, vol. 29, no. 1, pp. 55-64, 2009.
Z. Javaid, M. G. Boocock, P. J. McNair, and C. P. Unsworth, Contour interpolated radial basis functions with spline boundary correction for fast 3D reconstruction of the human articular cartilage from MR images, Medical Physics, vol. 43, no. 3, pp. 1187-1199, 2016.
J. G. Lee, S. Gumus, C. H. Moon, C. K. Kwoh, and K. T. Bae, Fully automated segmentation of cartilage from the MR images of knee using a multi‐atlas and local structural analysis method, Medical physics, vol. 41, no. 9, p. 092303, 2014.
Y. Yin, X. Zhang, R. Williams, X. Wu, D. D. Anderson, and M. Sonka, LOGISMOS—layered optimal graph image segmentation of multiple objects and surfaces: cartilage segmentation in the knee joint, IEEE transactions on medical imaging, vol. 29, no. 12, pp. 2023-2037, 2010.
P. M. Cashman, R. I. Kitney, M. A. Gariba, and M. E. Carter, Automated techniques for visualization and mapping of articular cartilage in MR images of the osteoarthritic knee: a base technique for the assessment of microdamage and submicro damage, IEEE transactions on nanobioscience, vol. 99, no. 1, pp. 42-51, 2002.
H. Z. Tameem and U. S. Sinha, Automated image processing and analysis of cartilage MRI: enabling technology for data mining applied to osteoarthritis, in AIP conference proceedings, 2007, vol. 953, no. 1, pp. 262–276: American Institute of Physics.
G. Vincent, C. Wolstenholme, I. Scott, and M. Bowes, Fully automatic segmentation of the knee joint using active appearance models, Medical Image Analysis for the Clinic: A Grand Challenge, vol. 1, p. 224, 2010.
V. Pedoia, S. Majumdar, and T. M. Link, Segmentation of joint and musculoskeletal tissue in the study of arthritis, Magnetic Resonance Materials in Physics, Biology and Medicine, vol. 29, no. 2, pp. 207-221, 2016.
M. Zhang et al., Development of a rapid knee cartilage damage quantification method using magnetic resonance images, BMC musculoskeletal disorders, vol. 15, no. 1, p. 264, 2014.
M. Zhang, J. B. Driban, L. L. Price, G. H. Lo, E. Miller, and T. E. McAlindon, Development of a rapid cartilage damage quantification method for the lateral tibiofemoral compartment using magnetic resonance images: data from the osteoarthritis initiative, BioMed research international, vol. 2015, 2015.
F. Ambellan, A. Tack, M. Ehlke, and S. Zachow, Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: Data from the Osteoarthritis Initiative, Medical image analysis, vol. 52, pp. 109-118, 2019.
Z. Zhou, M. M. R. Siddiquee, N. Tajbakhsh, and J. Liang, Unet++: Redesigning skip connections to exploit multiscale features in image segmentation, IEEE transactions on medical imaging, vol. 39, no. 6, pp. 1856-1867, 2019.
O. Ronneberger, P. Fischer, and T. Brox, U-net: Convolutional networks for biomedical image segmentation, in International Conference on Medical image computing and computer-assisted intervention, 2015, pp. 234–241: Springer.
B. Park, H. Park, S. M. Lee, J. B. Seo, and N. Kim, Lung segmentation on HRCT and volumetric CT for diffuse interstitial lung disease using deep convolutional neural networks, Journal of Digital Imaging, vol. 32, no. 6, pp. 1019-1026, 2019.
G. Singadkar, A. Mahajan, M. Thakur, and S. Talbar, Deep deconvolutional residual network based automatic lung nodule segmentation, Journal of Digital Imaging, pp. 1–7, 2020.
H. Seim, D. Kainmueller, H. Lamecker, M. Bindernagel, J. Malinowski, and S. Zachow, Model-based auto-segmentation of knee bones and cartilage in MRI data, Proc. Medical Image Analysis for the Clinic: A Grand Challenge. Bejing, China, pp. 215–223, 2010.
M. H. Hesamian, W. Jia, X. He, and P. Kennedy, Deep learning techniques for medical image segmentation: achievements and challenges, Journal of digital imaging, vol. 32, no. 4, pp. 582-596, 2019.
J. Long, E. Shelhamer, and T. Darrell, Fully convolutional networks for semantic segmentation, in Proceedings of the IEEE conference on computer vision and pattern recognition, 2015, pp. 3431–3440.
K. Simonyan and A. Zisserman, Very deep convolutional networks for large-scale image recognition, arXiv preprint arXiv:1409.1556, 2014.
K. He, X. Zhang, S. Ren, and J. Sun, Deep residual learning for image recognition, in Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 770–778.
D. P. Kingma and J. Ba, Adam: A method for stochastic optimization, arXiv preprint arXiv:1412.6980, 2014.
T.-Y. Lin, P. Goyal, R. Girshick, K. He, and P. Dollár, Focal loss for dense object detection, in Proceedings of the IEEE international conference on computer vision, 2017, pp. 2980–2988.
J. M. Jordan et al., Prevalence of knee symptoms and radiographic and symptomatic knee osteoarthritis in African Americans and Caucasians: the Johnston County Osteoarthritis Project, The Journal of rheumatology, vol. 34, no. 1, pp. 172-180, 2007.
O. Oktay et al., Attention u-net: learning where to look for the pancreas, arXiv preprint arXiv:1804.03999, 2018.
D. Dreizin, Y. Zhou, Y. Zhang, N. Tirada, and A. L. Yuille, Performance of a deep learning algorithm for automated segmentation and quantification of traumatic pelvic hematomas on CT, Journal of digital imaging, vol. 33, no. 1, pp. 243-251, 2020.
Q. Yu, L. Xie, Y. Wang, Y. Zhou, E. K. Fishman, and A. L. Yuille, Recurrent saliency transformation network: Incorporating multi-stage visual cues for small organ segmentation, in Proceedings of the IEEE conference on computer vision and pattern recognition, 2018, pp. 8280–8289.
Acknowledgements
We thank Zuse Institute Berlin for kindly providing annotations.
Funding
This work was supported in part by the National Institutes of Health (NIH) under Grant 1R01DE027027-02 and Grant 1U01 AR069395-03 (XZ).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Deng, Y., You, L., Wang, Y. et al. A Coarse-to-Fine Framework for Automated Knee Bone and Cartilage Segmentation Data from the Osteoarthritis Initiative. J Digit Imaging 34, 833–840 (2021). https://doi.org/10.1007/s10278-021-00464-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-021-00464-z